Abstract
Leucine-rich α2-glycoprotein (LRG) is a plasma protein in which leucine-rich repeats (LRRs) were first discovered. Although the physiological function of LRG is not known, increases in the serum level of LRG have been reported in various diseases. In this study, we found that LRG was induced by recombinant human IL-6 in human hepatoma HepG2 cells. The induction of LRG by IL-6 was up-regulated synergistically with either IL-1β or TNFα in a pattern similar to those for type 1 acute-phase proteins. We also found that lipopolysaccharide (LPS) administered intraperitoneally to mice enhanced dose-dependently the expression of LRG mRNA in the liver as well as those for mouse major acute-phase proteins. These results strongly suggest that LRG was a secretory type 1 acute-phase protein whose expression was up-regulated by the mediator of acute-phase response.
Keywords: Leucine-rich α2-glycoprotein, Leucine-rich repeats, Acute-phase, Interleukin 6, Inflammation, Lipopolysaccharide, Serum amyloid
During an acute inflammation, cytokines, which are released by various cells including macrophages, fibroblasts, and endothelial cells, stimulate the synthesis and secretion of a set of plasma proteins, acute-phase proteins (APPs), by the liver [1]. Major APPs, which increased at least 1000-fold higher than normal levels, include serum amyloid A protein (SAA) and either C-reactive protein (CRP) in human or its homolog in mice, serum amyloid P component (SAP). APPs are considered to be non-specific innate immune components involved in the restoration of homeostasis and the restraint of microbial growth before the activation of acquired immunity. The concentrations of circulating APPs, such as CRP and SAA, are related to the severity of the disorder and the extent of tissue damage and thus provide the diagnostic and prognostic information.
Leucine-rich α2-glycoprotein (LRG) has been isolated from the human serum by Haupt and Baudner in 1977 [2], and its amino acid sequence has been determined in 1985 [3]. LRG consists of 312 amino acid residues, 66 of which are leucines. This protein contains 8 repeating consensus sequences, each of which consists of 24 amino acid residues and exhibits a periodic pattern in the occurrence of leucine, proline, and asparagines. This consensus sequence, termed leucine-rich repeat (LRR), has since been identified as an LRR-containing domain, now appeared in over 55,000 sequences in PFAM database [4]. Most LRRs are 20–30 amino acids long and the repeat number ranges from 2 to 52. The primary function of LRRs appears to be to provide a versatile structural framework of the formation of protein–protein interactions [5]. Although many LRR proteins have been found to function in the plant immune response and the mammalian innate immune response, the physiological function of LRG is still unknown.
We found that a β-type phospholipase A2 (PLA2) inhibitory protein (PLIβ), which had been isolated from the blood plasma of a venomous snake Agkistrodon blomhoffii siniticus (renamed to Gloydius brevicaudus according to the present taxonomy), had significant sequence homology with human LRG [6]. This finding has led to the hypothesis that human LRG might be a human homolog of the PLIβ and function as a PLA2 inhibitory protein. To test this possibility, mouse LRG was purified from the sera by sequential chromatography on Sephadex G-200, Q-Sepharose, Phenyl Sepharose, and Mono Q columns, but this isolated mouse LRG did not show any PLA2 inhibitory activities (Shirai et al., unpublished results).
In 2002, it has been reported that LRG could be a marker of granulocytic differentiation and its expression was up-regulated during neutrophil differentiation [7]. Northern blot analysis revealed a high level of LRG expression in liver with a much lower level in heart and minimally detectable levels of the expression in spleen and lung [7]. Recently, with the development of proteomic analysis with 2-D electrophoresis and mass spectrometry, it has also been revealed that the expression of LRG increased in various diseases. In inflammation of the small intestine of cystic fibrosis mouse, the amount of LRG increased about 21-fold greater than that for normal mouse, and this up-regulation was associated with an increased number of neutrophils in the intestine [8]. In the sera of patients with graft-versus-host disease (GVHD), lung cancer, and pancreas cancer, the amount of LRG increased 3.5-fold, 6-fold, and 3-fold, respectively [9], [10], [11], [12]. In the cerebrospinal fluid of patients with idiopathic normal pressure hydrocephalus, the amounts of LRG also increased about 3-fold [13].
In this study, we found that the expression of LRG in HepG2 human hepatoma cells was induced by IL-6, IL-1β, and TNFα. We also found that the LRG expression was enhanced by the induction of acute inflammation in mice through the injection of lipopolysaccharide (LPS) in a dose-dependent manner. Therefore, like CRP and SAA, LRG might serve as a diagnostic or prognostic biomarker in some inflammatory conditions.
Materials and methods
Materials. Recombinant human IL-6, IL-1β, and TNFα were purchased from Sigma (St. Louis, MO, USA), Roche (Basel, Switzerland), and R&D SYSTEMS (Minneapolis, MN, USA), respectively. A human hepatoblastoma cell line, HepG2, was obtained from Human Science Research Resources Bank (Osaka, Japan). Male ICR mice (10 week old) were obtained from Tokyo Laboratory Animals Science (Tokyo, Japan). LPS (Escherichia coli endotoxin 0111:B4, L2630) was purchased from Sigma.
Cell culture and cytokine stimulation. HepG2 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal-bovine serum and 50 U/ml penicillin–streptomycin. Cells were seeded at 5 × 105 cells/well in 48-well plates and stimulated with various combinations of cytokines for 72 h afterward under subconfluent conditions.
Real-time quantitative RT-PCR. Total RNA was isolated from HepG2 cells or the pieces of liver by means of QuickGene RNA cultured cell kit S (Fujifilm, Tokyo, Japan) or RNeasy mini Kit (QIAGEN, Venlo, Netherlands). cDNA was synthesized with ExScript RT reagent kit (Takara, Shiga, Japan). Quantitative real-time PCR was performed on a Thermal Cycler Dice (Takara) using SYBR Premix Ex Taq (Takara) according to the manufacturer’s protocol. Primer sets used for mouse GAPDH, LRG, SAA, and SAP, and human GAPDH, LRG, SAA, and type II PLA2 are listed in Table S1. Gene expression levels were normalized according to the level of GAPDH expression.
Production of recombinant human LRG and generation of rabbit polyclonal antibodies against human LRG. Human LRG cDNA was isolated by PCR cloning from the cDNA prepared from HepG2 cells. An NdeI restriction site and a BamH1 site were introduced to the LRG cDNA by PCR using the primers shown in Table S2. The PCR product was digested with NdeI and BamH1 and inserted between the NdeI and BamHI sites of pET16b (Invitrogen, Carlsbad, CA, USA). The resulting expression plasmids containing the LRG cDNA were transformed into E. coli BL21 (DE3) pLys (Invitrogen). After the induction by 1 mM IPTG for 4 h at 37 °C, E. coli cells were harvested by centrifugation, suspended in 50 mM Tris–HCl buffer (pH 7.5), and the suspension was sonicated. The cell extract was centrifuged, and the resulting inclusion bodies were collected and washed with the same buffer containing 4% Triton X-100 followed by pure water. Since the protein in the pellet gave a single band corresponding to the recombinant LRG protein on SDS–PAGE, this pellet was used as the antigen to obtain anti-LRG antibody.
Rabbit polyclonal antibodies against human LRG were raised against the recombinant LRG protein by subcutaneously immunization of rabbits with the protein (100 μg), which had been emulsified in Freund’s incomplete adjuvant (Nacarai Tesque, Kyoto, Japan). The polyclonal IgG was purified from the immunized rabbit serum by affinity chromatography using a protein G (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) column.
Immunodetection. Proteins secreted into the culture media were concentrated by using Microcon Ultracel YM-3 (Millipore, Bedford, MA, USA), separated by SDS–PAGE, and transferred electrophoretically onto a PVDF membrane (Bio-Rad, Hercules, CA, USA). The membranes were blocked in Blocking One solution (Nacarai Tesque), and then probed with the rabbit anti-LRG polyclonal antibody, prepared as described above. After washing, the proteins on the blot were detected by using the anti-rabbit IgG donkey antibody conjugated to horseradish peroxidase with ECL plus detection reagents (GE Healthcare Bio-Sciences).
In vivo studies. To induce acute inflammation, ICR mice were injected intraperitoneally with 0.05–2.0 mg/kg LPS (E. coli endotoxin 0111:B4, L2630, Sigma) dissolved in 50 μl PBS. Liver samples were obtained at 6 h after LPS injection and preserved in RNA later solution (QIAGEN) at −80 °C.
Statistical analysis. Statistical significance was determined with GraphPad Prism software (GraphPad Software, San Diego, CA, USA).
Results and discussion
Time course study of LRG expression after stimulation with proinflammatory cytokines
To evaluate whether LRG is one of the acute-phase proteins (APPs), we first investigated the time course of LRG expression in HepG2 cells after the stimulations with proinflammatory cytokines, IL-6, IL-1β, and, TNFα. As shown in Fig. 1 B and C, the amounts of LRG mRNA increased slightly and gradually at 12–24 h after the stimulation with the IL-1β and TNFα, respectively. On the contrary, the amounts of LRG mRNA abruptly increased 5-fold at 2 h after the stimulation with IL-6, reached its maximum at 6 h, and markedly decreased less than the basal level at 24 h (Fig. 1D). IL-1β and TNFα had synergistic actions on the expression of LRG mRNA induced by IL-6 (Fig. 1E and F, respectively). The amount of LRG mRNA induced by using a combination of IL-6 and IL-1β, and that for a combination of IL-6 and TNFα were 2 and 1.5-fold greater, respectively, than that induced by IL-6 alone. Furthermore, a marked decrease in the expression of LRG mRNA, found at 24 h after the stimulation with IL-6, was attenuated by coexisting IL-1β or TNFα. IL-6 was known to be the major regulator of APP synthesis in human hepatocytes [1], [14]. These results show that the LRG expression in HepG2 cells is up-regulated by IL-6, and IL-6 acts synergistically with coexisting IL-1β or TNFα.
Fig. 1.

Time course study on the LRG mRNA expressions in HepG2 cells stimulated with proinflammatory cytokines. HepG2 cells were stimulated with (A) none, (B) 10 ng/ml IL-1β alone, (C) 10 ng/ml TNFα alone, (D) 10 ng/ml IL-6 alone, (E) 10 ng/ml IL-6 and 10 ng/ml IL-1β, or (F) 10 ng/ml IL-6 and 10 ng/ml TNFα. The expressions of LRG and GAPDH mRNA were measured by real-time quantitative RT-RCR at 0, 2, 4, 6, 12, 24, and 48 h after the cytokine stimulation. The expression levels of LRG mRNA were corrected with that of GAPDH mRNA. Values are means ± SD of two independent determinations.
The secretion of LRG protein into the culture media at 48 h after the stimulations was confirmed by Western blots of the culture media (Fig. 2 A). LRG could be detected immunologically in the media at 48 h after the stimulation with IL-6, and the corresponding band was intensified after the stimulation by the coexistence of IL-1β or TNFα. Fig. 2B compares the expressions of LRG, SAA, and type II PLA2 mRNAs in HepG2 cells at 4 h after the stimulations with IL-6 alone, IL-6 and IL-1β, and IL-6 and TNFα. IL-6 induced the expression of LRG mRNA in a manner similar to those for known APPs, SAA, and PLA2 mRNAs.
Fig. 2.
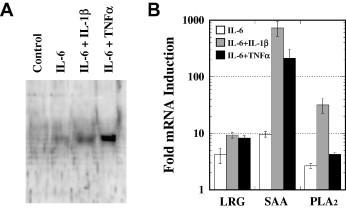
Synergistic effects of IL-1β and TNFα on LRG induction by IL-6 in HepG2 cells. (A) Western blot analysis of the culture media of HepG2 cells at 48 h after stimulation with 10 ng/ml IL-6 alone, 10 ng/ml IL-6 and 10 ng/ml IL-1β, and 10 ng/ml IL-6 and 10 ng/ml TNFα. (B) Comparison of the expressions of LRG, SAA, and PLA2 mRNAs in HepG2 cells at 4 h after stimulations with 10 ng/ml IL-6 alone, 10 ng/ml IL-6 and 10 ng/ml IL-1β, and 10 ng/ml IL-6 and 10 ng/ml TNFα. The amounts of the respective mRNAs were measured by real-time quantitative RT-RCR. The relative expression levels of each mRNA were corrected with that of GAPDH mRNA and then compared with that for an unstimulated control sample. Values are means ± SD of two independent determinations.
APPs can be classified into two types according to their inducers [15]. Type 1 APPs, such as SAA, CRP, PLA2, and α1-acid glycoprotein require the IL-6 stimulation in conjunction with IL-1 to exhibit their maximal expressions, whereas type 2 APPs such as fibrinogen and α2-macroglobulin are fully induced by IL-6 alone [16]. Although the synergistic effects of IL-1β on the expression of LRG mRNA was less prominent than those for the expressions of SAA and PLA2 mRNAs, the present result strongly suggests that LRG belongs to type 1 APPs.
Up-regulation of LRG expression in an experimental mouse endotoxemia model
LPS, a major constituent of the outer membrane of Gram-negative bacteria, is known to induce productions of proinflammatory cytokines by macrophages and this leads to the productions of APPs by the liver. LRG expression was examined in mouse liver at 6 h after intraperitoneal injections of various concentrations of LPS. As shown in Fig. 3 , LPS was found to induce a dose-dependent enhancement of the expression of LRG mRNA as well as those for the mRNAs of major APPs in mouse, SAA and SAP. The expression of LRG mRNA was most strongly correlated with the dose of LPS (r = 0.51, P = 0.006) in comparison with those of the mRNAs of SAA (r = 0.47, P = 0.012) and SAP (r = 0.35, P = 0.071). The expression of LRG mRNA was highly correlated with that of SAA mRNA (r = 0.60, P = 0.0007). Although we could not quantify the LRG concentrations in the sera of LPS-treated mice in the present study, it is reasonable to conclude that the dose-dependent increase in the level of hepatic LRG mRNA results in the dose-dependent increase in the serum level of LRG protein.
Fig. 3.

The expressions of (A) LRG, (B) SAA, and (C) SAP mRNAs in the liver of mouse at 6 h after intraperitoneal injection of various concentrations of LPS. The amounts of the respective mRNAs were measured by real-time quantitative RT-PCR. The relative expression levels of each mRNA were corrected with that of GAPDH mRNA. Horizontal bars represent the mean ± SEM.
LRG levels have been reported to increase in the sera of patients infected with bacteria including Haemophilus influenzae type b, Salmonella, and Streptococcus pyogenes [17] and of the patients with severe acute respiratory syndrome (SARS) [18]. Recently, the concentration of LRG in the sera was determined by ELISA employing cytochrome c (Cyt c) as the capturing ligand and found to increase significantly in the sera of patients infected with bacteria (toxic shock syndrome, TSS) [19]. But it was shown that LRG level in the sera of the patients with TSS did not correlate with the CRP level [19]. This contrasts with our present result that the expressions of LRG and SAA mRNAs in the mouse liver significantly correlated to each other, since the CRP and SAA levels are known to show a close relationship in a wide range of clinical conditions.
Possible physiological function of LRG
The physiological function of LRG is not known. Therefore, we assumed that LRG might function as a PLA2 inhibitory protein and thus bind a PLA2 molecule as a natural ligand on the basis of our previous study on the PLIβ which was isolated from the sera of venomous snake [6]. However, the purified mouse LRG did not show any PLA2 inhibitory activities (Shirai et al., unpublished results). Since PLIβ from the venomous snake sera binds specifically basic PLA2 included in its own venom and inhibit its enzymatic activity, it was suggested that PLIβ directly participate in the clearance of venom PLA2 for self-protection against its own venom. Recently, Cummings et al. found that LRG bound strongly to Cyt c [20] and, therefore, Cyt c might be one of the natural ligands of LRG. Since high concentrations of extracellular Cyt c have been reported to induce arthritis [21], LRG might directly participate in the clearance of Cyt c which may be released from the apoptotic neutrophils after bacterial infections.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2009.03.104.
Appendix A. Supplementary data
References
- 1.Heinrich P.C., Castell J.V., Andus T. Interleukin-6 and the acute phase response. Biochem. J. 1990;265:621–636. doi: 10.1042/bj2650621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haupt H., Baudner S. Isolierung und charakterisierung eines bisher unbekannten leucinreichen 3.1S-α2-glykoproteins aus human serum. Hoppe-Seyler Z. Physiol. Chem. 1977;358:639–646. [PubMed] [Google Scholar]
- 3.Takahashi N., Takahashi Y., Putnam F.W. Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc. Natl. Acad. Sci. USA. 1985;82:1906–1910. doi: 10.1073/pnas.82.7.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsushima N., Tanaka T., Enkhbayar P., Mikami T., Taga M., Yamada K., Kuroki Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics. 2007;8:124. doi: 10.1186/1471-2164-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobe B., Kajava A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 6.Okumura K., Ohkura N., Inoue S., Ikeda K., Hayashi K. A novel phospholipase A2 inhibitor with leucine-rich repeats from the blood plasma of Agkistrodon blomhoffii siniticus. Sequence homologies with human leucine-rich alpha2-glycoprotein. J. Biol. Chem. 1998;273:19469–19475. doi: 10.1074/jbc.273.31.19469. [DOI] [PubMed] [Google Scholar]
- 7.O’Donnell L.C., Druhan L.J., Avalos B.R. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J. Leukoc. Biol. 2002;72:478–485. [PubMed] [Google Scholar]
- 8.Norkina O., Kaur S., Ziemer D., De Lisle R.C. Inflammation of the cystic fibrosis mouse small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G1032–G1041. doi: 10.1152/ajpgi.00473.2003. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Clouthier S.G., Galchev V., Misek D.E., Duffner U., Min C.K., Zhao R., Tra J., Omenn G.S., Ferrara J.L., Hanash S.M. Intact-protein-based high-resolution three-dimensional quantitative analysis system for proteome profiling of biological fluids. Mol. Cell. Proteomics. 2005;4:618–625. doi: 10.1074/mcp.M400126-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Kakisaka T., Kondo T., Okano T., Fujii K., Honda K., Endo M., Tsuchida A., Aoki T., Itoi T., Moriyasu F., Yamada T., Kato H., Nishimura T., Todo S., Hirohashi S. Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D-DIGE): up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer. J. Chromatogr. B. 2007;852:257–267. doi: 10.1016/j.jchromb.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okano T., Kondo T., Kakisaka T., Fujii K., Yamada M., Kato H., Nishimura T., Gemma A., Kudoh S., Hirohashi S. Plasma proteomics of lung cancer by a linkage of multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis. Proteomics. 2006;6:3938–3948. doi: 10.1002/pmic.200500883. [DOI] [PubMed] [Google Scholar]
- 12.Deng R., Lu Z., Chen Y., Zhou L., Lu X. Plasma proteomic analysis of pancreatic cancer by 2-dimensional gel electrophoresis. Pancreas. 2007;34:310–317. doi: 10.1097/MPA.0b013e31802f2483. [DOI] [PubMed] [Google Scholar]
- 13.Li X., Miyajima M., Jiang C., Arai H. Expression of TGF-betas and TGF-beta type II receptor in cerebrospinal fluid of patients with idiopathic normal pressure hydrocephalus. Neurosci. Lett. 2007;413:141–144. doi: 10.1016/j.neulet.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 14.Castell J.V., Gomez-Lechon M.J., David M., Hirano T., Kishimoto T., Heinrich P.C. Recombinant human interleukin-6 (IL-6/BSF-2/HSF) regulates the synthesis of acute phase proteins in human hepatocytes. FEBS Lett. 1988;232:347–350. doi: 10.1016/0014-5793(88)80766-x. [DOI] [PubMed] [Google Scholar]
- 15.Baumann H., Gauldie J. The acute phase response. Immunol. Today. 1994;15:74–80. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 16.Baumann H., Gauldie J. Regulation of hepatic acute phase plasma protein genes by hepatocyte stimulating factors and other mediators of inflammation. Mol. Biol. Med. 1990;7:147–159. [PubMed] [Google Scholar]
- 17.Bini L., Magi B., Marzocchi B., Cellesi C., Berti B., Raggiaschi R., Rossolini A., Pallini V. Two-dimensional electrophoretic patterns of acute-phase human serum proteins in the course of bacterial and viral diseases. Electrophoresis. 1996;17:612–616. doi: 10.1002/elps.1150170333. [DOI] [PubMed] [Google Scholar]
- 18.Chen J.H., Chang Y.W., Yao C.W., Chiueh T.S., Huang S.C., Chien K.Y., Chen A., Chang F.Y., Wong C.H., Chen Y.J. Plasma proteome of severe acute respiratory syndrome analyzed by two-dimensional gel electrophoresis and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:17039–17044. doi: 10.1073/pnas.0407992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weivoda S., Andersen J.D., Skogen A., Schlievert P.M., Fontana D., Schacker T., Tuite P., Dubinsky J.M., Jemmerson R. ELISA for human serum leucine-rich alpha-2-glycoprotein-1 employing cytochrome c as the capturing ligand. J. Immunol. Methods. 2008;336:22–29. doi: 10.1016/j.jim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummings C., Walder J., Treeful A., Jemmerson R. Serum leucine-rich alpha-2-glycoprotein-1 binds cytochrome c and inhibits antibody detection of this apoptotic marker in enzyme-linked immunosorbent assay. Apoptosis. 2006;11:1121–1129. doi: 10.1007/s10495-006-8159-3. [DOI] [PubMed] [Google Scholar]
- 21.Pullerits R., Bokarewa M., Jonsson I.M., Verdrengh M., Tarkowski A. Extracellular cytochrome c, a mitochondrial apoptosis-related protein, induces arthritis. Rheumatology. 2005;44:32–39. doi: 10.1093/rheumatology/keh406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


