Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) 7a is an accessory protein with no known homologues. In this study, we report the interaction of a SARS-CoV 7a and small glutamine-rich tetratricopeptide repeat-containing protein (SGT). SARS-CoV 7a and human SGT interaction was identified using a two-hybrid system screen and confirmed with interaction screens in cell culture and cellular co-localization studies. The SGT domain of interaction was mapped by deletion mutant analysis and results indicated that tetratricopeptide repeat 2 (aa 125-158) was essential for interaction. We also showed that 7a interacted with SARS-CoV structural proteins M (membrane) and E (envelope), which have been shown to be essential for virus-like particle formation. Taken together, our results coupled with data from studies of the interaction between SGT and HIV-1 vpu indicated that SGT could be involved in the life-cycle, possibly assembly of SARS-CoV.
Keywords: SARS-CoV, ORF 7a, SGT, TPR, Protein–protein interaction, Accessory protein
Coronaviruses are members of the Coronaviridae family in the order Nidovirales. This family of viruses contains genomes of ∼30 kb that include non-structural (pp1ab) and structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]), making them the largest known RNA viruses. Interspersed among the structural proteins are group-specific proteins that differ in location and composition between the three coronavirus groups. The group-specific genes are not well characterized and the vast majority has, as yet, no known function. Initial research into the functions of these genes has shown that they are non-essential and dispensable for virus growth in cell culture [1]. However, more recent studies have shown that the accessory genes are required for in vivo infection in the natural host [2], [3], [4].
Severe acute respiratory syndrome virus (SARS-CoV) encodes for eight potential open reading frames (ORFs), i.e., ORF 3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b, with no known homologues [5], [6], [7]. Of these, SARS 3a, 3b, 6, 7a, and 9b have been detected in SARS-CoV infected cells, as well as in clinical samples [8], indicating possible in vivo functions. SARS-CoV 7a (previously designated U122 in [9] and X4 in [10]) is the first ORF of subgenomic RNA 7 [6] and has been shown to be expressed in SARS-CoV infected Vero E6 cells [9], [11]. It has been shown to localize to the Golgi apparatus and the endoplasmic reticulum (ER) and is recycled between the ER and Golgi complex via the intermediate compartments [9], [10], [11], where coronaviruses are known to bud [12]. Additionally, 7a has been shown to interact specifically with another unique SARS-CoV protein encoded by ORF3a (also called U274; [13]), which has been reported to be a novel SARS-CoV minor structural protein [14], [15]. Interestingly, overexpression of 7a results in caspase-dependent apoptosis in Vero E6, as well as in cells from the lung, kidney, and liver. Therefore, based on the clinical observation of apoptosis in tissues from different organs, it appears as though 7a contributes to apoptosis during SARS-CoV infection and possibly to the pathogenesis of SARS-CoV [16]. Recently, the structure of the N-terminal domain of 7a was resolved and reported to be similar in fold and topology to members of the Ig superfamily [11]. Taken together, results indicate that 7a could play an important role in the viral infection cycle, but the exact biological function(s) of 7a is yet to be determined.
Identification of host proteins interacting with SARS-CoV 7a could be vital in elucidating its possible functions. It could also provide crucial insight into the biology and pathogenicity of SARS-CoV. This is the first report of a cellular protein that interacts with SARS-CoV 7a. We used a yeast two-hybrid system to screen a B-lymphocyte cDNA expression library for cellular proteins capable of interacting with SARS-CoV 7a. The principal cDNA identified from the screen encoded a ∼37.1 kDa peptide identified as human small glutamine-rich tetratricopeptide repeat (hSGT) containing protein. Our results indicated that 7a interacted with both human, as well as African Green monkey SGT (mSGT) from Vero E6 cells. The conceptual amino acid sequence of mSGT was determined and compared to the hSGT sequence; the amino acid (aa) sequence identity was found to be >99%. Protein–protein interaction was confirmed by co-immunoprecipitation and immunofluorescent staining in Vero E6 cells, while the SGT domains involved in the interaction were mapped by deletion mutant analysis. Tetratricopeptide repeat (TPR) 2 of hSGT (aa 125-158) was shown to be crucial for this interaction. The biological significance of the interaction between SARS-CoV 7a and SGT needs to be elucidated.
Materials and methods
Two-hybrid system library screen. The yeast reporter strain AH109 [GAL4 2H-3] (Clontech) was used for the two-hybrid selection. Plasmid pAS2-7aΔ96-122 (Table 1 ) was used as bait and a pACT-cDNA library (human lymphocyte MATCHMAKER, Clontech) was used as the source of prey genes. Yeast cells were grown on YPD or on synthetic minimal medium (0.67% yeast nitrogen base, the appropriate auxotrophic supplements, 2% agar [for plates]) supplemented with 2% dextrose. Yeast was transformed with appropriate plasmids by the lithium acetate method and transformants were selected on synthetic minimal medium. The bait plasmid and the pB42AD cDNA library were introduced into the yeast strain AH109 [GAL4 2H-3]. Two-hybrid screen and interaction assays were performed essentially as described in the protocol (Clontech) in the presence of 2% galactose and 80 mg of 5-bromo-4-chloro-3-indolyl-d-galactopyranoside per liter. Prey plasmids were selected from yeast colonies giving a positive signal according to the manufacturer’s protocol. False positives were eliminated by re-transforming the host AH109 [GAL4 2H-3] strain with pACT-cDNA library plus bait plasmid. Additionally, the pACT-cDNA was transfected in yeast strain PJ69-2A to check for autoactivation. The positive clones that contained cDNAs encoding 7a-interacting proteins were sequenced and analyzed using BLAST.
Table 1.
Plasmids used in this study
| Plasmid | Description (protein expressed) | Source/reference |
|---|---|---|
| pAS2-1 | Gal4 DNA-binding domain in a 2 μm TRP1 yeast shuttle vector | Clontech Inc. |
| pAS2-1-7aΔ96-122 | Deletion of C-terminus (ORF7a aa 1-95) | This work |
| pACT2 | Gal4 activation domain in a 2 μm LEU2 yeast shuttle vector | Clontech Inc. |
| pXJ40-flag | Mammalian expression vector for tagging proteins with flag epitope at the N-terminus | [17] |
| pXJ40-3′HA | Mammalian expression vector for tagging proteins with HA at the C- terminus | GLAXO laboratory, IMCBa |
| pXJ40-flagSGT | Full-length SGT, flag-tagged at N-terminus (SGT aa 1-313) | This work |
| pXJ40-flagSGTΔC | Deletion of C-terminus (SGT aa 1-192) | This work |
| pXJ40-flagSGTΔC3 | Deletion of C-terminus and TPR-3 (SGT aa 1-158) | This work |
| pXJ40-flagSGTΔC3-2 | Deletion of C-terminus and TPR-2 and 3 (SGT aa 1-124) | This work |
| pXJ40-flagSGTΔN1-2 | Deletion of N-terminus and TPR-1 and 2 (SGT aa 159-313) | This work |
| pXJ40-7a-HA | Full-length ORF7a, HA-tagged at C-terminus (aa 1-122) | [13] |
| pXJ40-3′7a-myc | Full-length ORF7a, myc-tagged at C-terminus (aa 1-122) | This work |
IMCB, Institute of Molecular and Cell Biology, Singapore.
Mammalian cell lines and DNA constructs. African Green monkey kidney epithelial (Vero E6) cells (American Type Culture Collection, Manassas, USA) were maintained as described previously [9]. A cDNA clone expressing full-length 7a-HA were prepared as described previously [13]. A 7a-myc construct was prepared as in [13], but instead the amplicon was cloned into pXJ40-3′myc; all 7a proteins were epitope-tagged at the C-terminus. Full length, as well as truncations of hSGT cDNA, tagged with an N-terminus flag epitope for expression in mammalian cells is summarized in Table 1. All cDNA sequences were confirmed by sequencing.
Antibodies used. Glutathione S-transferase-hSGT (GST-hSGT) fusion protein was constructed to raise SGT-specific antibodies in mice and rabbits. The full-length hSGT was digested from pXJ40-flag-hSGT with BamHI and XhoI and cloned into the compatible sites of pGEX-4T-1. Fusion proteins were expressed in BL-21 Escherichia coli by induction with isopropyl-1-thio-l-d-galactopyranoside (IPTG) at 37 °C for 3 h. Subsequently, GST-hSGT was purified using Glutathione Sepharose beads (Pharmacia) and eluted with excess glutathione. Purified GST-hSGT was then used to raise polyclonal antibodies in mice and rabbits, as described in [18]. Polyclonal (Santa Cruz) and monoclonal anti-HA (Roche) and anti-flag (Sigma) antibodies were used according to the manufacturer’s instructions. All procedures on animals were done in accordance with the regulations of the Animal Research Ethics Committee, Singapore.
Transient expression and Western blotting. Vero E6 cells were transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocols. Unless stated otherwise, 1 μg of plasmid cDNA was used for transfection into Vero E6 cells in transient expression studies; full-length flag-hSGT was used at 0.25 μg. Western blotting was done as described in [13].
Immunoprecipitation. Cell lysates were extracted from transfected Vero E6 cells as described above. Typically, 150 μg of whole cell lysates was incubated with either rabbit anti-flag or rabbit anti-SGT antibody conjugated to Protein A-agarose beads (Roche) for 16 h at 4 °C with end-over-end mixing. Following incubation, the beads were collected and complexes were washed three times with IP buffer. The bound proteins were eluted by boiling in SDS sample buffer and Western blotted as discussed above.
Sequencing of the African Green monkey kidney epithelial SGT. Total cellular RNA was extracted from Vero E6 cells using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA was prepared from 1.0 μg total RNA using the SuperScript II RNase Reverse Transcriptase kit (Invitrogen). Subsequently, a 1:10 dilution of the first-strand cDNA was used in a PCR according to standard protocols. The primary nucleotide sequence of African Green monkey kidney epithelial SGT (mSGT) was determined by automated sequencing and compared to the hSGT sequence (NCBI Accession No. NP_003012) using CLUSTAL X [19]. The conceptual amino acid sequence of mSGT was compared to hSGT and comparisons were visualized using GENEDOC software [20].
Immunoflourescence. Transiently transfected Vero E6 cells were grown on coverslips. At about 16 h posttransfection, the medium was removed and the coverslips fixed in methanol at −20 °C. After 5 min, the coverslips were lifted out and completely air-dried. Fixed cells were incubated with the primary antibody combination of mouse anti-HA and rabbit anti-flag at room temperature for 1 h. Mouse anti-HA and rabbit anti-flag antibody were used at a dilution of 1:200. Following washing, cells were incubated with the secondary antibody combination of FITC-conjugated goat anti-mouse and Rh-conjugated anti-rabbit antibodies at room temperature for 1 h (Santa Cruz Biochemicals, USA). Following extensive washing, the coverslips were mounted on glass slides and viewed.
Results and discussion
Identification of cellular proteins interacting with SARS-CoV 7a
Biological processes are dependent on the direct physical interaction between different proteins. Therefore, the identification of host proteins that interact with viral accessory proteins could help elucidate possible functions of these unique viral proteins. In this study we used a yeast two-hybrid screen to identify host proteins that interacted with 7a. Truncated 7a (aa 1-96) protein—excluding the transmembrane-containing C-terminus to improve solubility of the expressed protein—was used as bait and 10 positive interacting candidates were identified. Following co-transformation and an autoactivation check to confirm the positive phenotypes, five clones were retained. The positive cDNA clones were sequenced and analyzed using BLAST; four of the positive clones contained complete ORFs that showed nucleotide sequence identity to human (h) SGT. hSGT identified in our screens encodes for a potential ORF of 313 amino acids (aa) with a predicted protein molecular mass of ∼37.1 kDa and contained three 34 aa tetratricopeptide repeat (TPR) motifs.
TPR motifs were first identified as protein interaction modules in cell division cycle proteins in yeast [21], [22]. They have now been shown to be ubiquitous and present in a number of functionally distinct polypeptides from a variety of different species. Different protein–protein interactions are mediated by these motifs and the majority of TPR motif-containing proteins have been shown to be involved in processes as diverse as cell cycle control, transcription and splicing events, protein transport and protein folding, to name a few [23]. The TPR-containing protein, hSGT, was first identified and described as a cellular binding partner for the non-structural (NS) protein of autonomous parvovirus H-1. Interestingly, both H1-virus infection and transient expression of the NS protein result in modification (most likely phosphorylation) of hSGT [24]. A subsequent study showed that hSGT interacts with HIV-I Vpu and Gag proteins, with Callahan and co-workers postulating that hSGT plays a role in HIV-1 virus assembly or release [25]. Other binding partners of SGT include the growth hormone receptor [26], myostatin [27], heat shock cognate protein [28], and heat shock protein [29], and it has been speculated that SGT could also have a cochaperone function. Recently, it has been shown that SGT is present throughout the cell cycle and that depletion there-of leads to an increase in the mitotic index [30]. On the contrary, Wu and colleagues report that SGT is a pro-apoptotic factor and that knockdown of SGT expression in a hepatocarcinoma cell line protects against apoptotic stimuli [31]. It is clear that SGT has diverse functions within the different cell types and possibly plays a role in the life cycle of at least two human viruses.
SGT interacts with SARS-CoV 7a in Vero E6 cells
The mouse anti-SGT antibody specifically detected endogenous SGT from Vero E6 cells, as well as transfected flag-hSGT (Fig. 1 A) and the polyclonal rabbit SGT antibody specifically immunoprecipitated endogenous Vero E6 SGT (Fig. 1B). Co-immunoprecipitation studies were used to confirm the interaction between SGT and 7a-HA in Vero E6 cells (Fig. 1B). Cells were co-transfected with pXJ40-flag-hSGT and pXJ40-7aHA, as described elsewhere. For 7a-HA interaction with endogenous Vero E6 SGT, only pXJ40-7aHA was transfected. Total protein extracts were immunoprecipitated with either rabbit anti-flag (lane 4) or rabbit anti-hSGT (lane 5) antibody conjugated to Protein A-agarose beads and Western blotted; an unrelated antibody (rabbit anti-myc, lane 6) was used as negative antibody control. Western blots and IP blots were detected with mouse anti-SGT and mouse anti-HA antibodies (Fig. 1B). SARS-CoV 7a-HA co-immunoprecipitated with both flag-hSGT (lane 4), as well as with endogenous SGT from Vero E6 cells (lane 5). 7a-HA was not detected when the unrelated antibody (lane 6) was used for immunoprecipitation. Our results showed that 7a-HA interacted specifically with SGT from both human and monkey cells.
Fig. 1.

mαSGT antibody detects human and Vero E6 SGT. (A) Mouse anti-SGT specifically detects endogenous SGT from Vero E6 cells (mSGT), as well as transfected flag-hSGT. (B) Co-immunoprecipitation of flag-hSGT and mSGT with 7a-HA. Lysates from Vero E6 cells transfected with flag-hSGT and/or 7a-HA were extracted and immunoprecipitated (IP) with rabbit anti-flag (lane 4), rabbit anti-SGT (lane 5) or unrelated rabbit antibody (lane 6) conjugated to Protein-A beads, respectively. IP samples (lanes 4–6), together with direct lysates (lanes 1–3) were analyzed by 15% SDS–PAGE and Western blotting with the appropriate mouse anti-SGT or mouse anti-HA antibody.
To explain the interaction of 7a with both hSGT as well as African Green monkey SGT (mSGT), we determined the nucleotide sequence of mSGT using cDNA from Vero E6 cells. The deduced primary amino acid sequence was compared to hSGT using CLUSTAL X [19] and visualized with GENEDOC [20]. mSGT showed >96% nucleotide and >99% aa sequence identity with hSGT (Fig. 2 ). This high sequence identity could explain why 7a-HA interacted with SGT from two distinct organisms.
Fig. 2.

Comparison of the amino acid sequence of monkey SGT with a human homologue. Conceptual amino acid sequence of mSGT was compared to the amino acid sequence of hSGT (NCBI Accession No. NP_003012) using CLUSTAL X. Results were visualized with GENEDOC software. Identical residues are represented with black shading. H. sapiens, Homo sapiens; C. aethiops, Cercopithecus aethiops.
SARS-CoV 7a co-localizes with flag-hSGT
The sub-cellular localization of 7a-HA and flag-hSGT in Vero E6 cells was studied. Vero E6 cells were transfected with pXJ40-flag-hSGT and pXJ40-7aHA (Fig. 3 ). At 16 h post-transfection, cells were fixed with methanol and stained with both mouse anti-HA and rabbit anti-flag antibodies. As a negative control untransfected Vero E6 cells were treated in the same way. Whereas 7a-HA showed a perinuclear localization (Fig. 3, middle panel), flag-hSGT was distributed to both the nucleus and cytoplasm of transfected cells (left panel) and untransfected cells did not stain (not shown) with the antibodies. The cellular distribution of SGT is consistent with a report by Cziepluch and co-workers [24] who found that untagged rat SGT is detectable in cytoplasm and nucleus of FREJ4 cells. Thus, 7a-HA was found to partially co-localize with flag-hSGT in Vero E6 cells (right panel). This partial co-localization of flag-hSGT and 7a-HA indicated that SARS-CoV 7a could interact with SGT in Vero E6 cells.
Fig. 3.
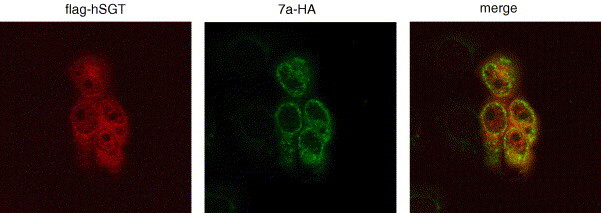
Co-localization of flag-hSGT and 7a-HA in Vero E6 cells. Vero E6 cells were grown on coverslips and transfected with pXJ3′-7a-HA (middle panel) and pXJ40flag-hSGT (left panel). At 16 h posttransfection, the cells were fixed and co-stained with both rabbit anti-flag and mouse anti-HA. Following washes the cells were treated with FITC-conjugated anti-mouse secondary antibody and rhodamine-conjugated (Rh) anti-rabbit antibody. Primary antibodies were used at dilutions of 1:200 and secondary antibodies at 1:100. Coverslips were mounted on glass slides and viewed by immunofluorescence. Panels on the left represent cells stained with FITC, those in the centre panels with Rh and composite photos are shown in the right panels.
hSGT TPR2 is essential for the interaction with 7a
Human small glutamine-rich tetratricopeptide contains three 34 aa TPR motifs that have been shown to mediate protein–protein interactions. In fact, recently Liou and Wang [32] investigated the functional importance of the different regions of SGT. They confirmed previous reports that the N-terminus of hSGT (aa 1-90) is responsible for self-dimerization of the protein and that the TPR domain (aa 91-192) is important for interaction with different proteins. Furthermore, in in vitro studies the glutamine-rich C-terminus portion (aa 193-313) could interact with short peptide segments consisting of consecutive non-polar amino acids. Thus, to determine whether the TPR motifs played a role in the interaction between hSGT and SARS-CoV 7a, four hSGT deletion mutants were created (Fig. 4 A). Binding of the hSGT mutants with 7a-HA was studied using immunoprecipitation assays in Vero E6 cells. Results showed that full-length flag-hSGT (aa 1-313), flag-hSGTΔC (aa 1-192), and flag-hSGTΔC3 (aa 1-158) interacted with 7a-HA in Vero E6 cells. On the other hand, flag-hSGTΔN1-2 (aa 159-313) and flag-hSGTΔC3-2 (aa 1-125) did not interact with 7a-HA (Fig. 4B). Also, the negative control flag-GST did not interact with 7a-Ha. Since only full-length and SGT mutants that contain TPR2 interacted with 7a, our results showed that TPR2 (aa 125-158) was essential for the interaction.
Fig. 4.

TPR2 is essential for the interaction with SARS-CoV 7a-HA. (A) Full-length flag-hSGT (aa 1-313) and deletion mutants flag-hSGTΔN1-2 (aa 159-313), flag-hSGTΔC (aa 1-192), flag-hSGTΔC-3 (aa 1-158), and flag-hSGTΔC3-2 (aa 1-125) are shown. All hSGT proteins were flag-tagged at the N-terminus. The locations of the TPR motifs 1, 2, and 3 are represented by shaded boxes. The name of each clone is shown on the left and results of the co-IP are shown on the right. (B) Flag-tagged full-length or mutant hSGT were co-transfected into Vero-E6 cells with 7a-HA, respectively. Total proteins were extracted and co-immunoprecipitated with mouse anti-flag beads. IP complexes (upper panel) and total cell lysates (middle and lower panel) were Western blotted and probed with rabbit anti-flag or rabbit anti-HA antibodies.
7a-HA interacted with SARS-CoV M and E
7a-HA has previously been shown to interact with the SARS-CoV minor structural protein 3a [13]. Co-immunoprecipitation studies in Vero E6 cells were used to determine whether 7a-myc also interacted with the SARS-CoV structural proteins (nucleocapsid) N, M, and E (Fig. 5 ). In this study, the HA-tagged proteins HA-N, M-HA, and E-HA were used as previously described in [13]. Whereas, the N protein is responsible for packaging the RNA into the nucleocapsid, the M and E proteins are responsible for virus assembly. In fact, the co-expression of M and E in a baculovirus expression system was sufficient for the assembly of virus-like particles [33]. Results showed that 7a-myc (aa 1-122) indeed interacted with HA-M (lane 4) and E-HA (lane 6), but not with HA-N (lane 2). There was no interaction between the negative control myc-GST and HA-N, M-HA or E-HA (lanes 1, 3, and 5).
Fig. 5.

7a-myc interacts with SARS-CoV structural proteins. Total cell lysates from cells co-transfected with myc-GST or 7a-myc with either HA-N, E-HA or M-HA were immunoprecipitated with myc-polyclonal antibody conjugated to Protein A-beads. IP complexes (upper panel) and total cell lysates (middle and lower panel) were Western blotted and probed with mouse anti-myc or mouse anti-HA antibodies.
The interaction of SGT with the HIV-1 accessory protein vpu and major structural protein gag has previously been reported [25]. The accessory protein vpu has been shown to be essential in regulating viral particle release and viral load [34]. Interestingly, overexpression of SGT and subsequent association with vpu protein reduces the titre of virus released from HIV-1 infected cells [25]. The authors also reported that the in vivo interaction between gag and SGT was abrogated when vpu was overexpressed in cells, indicating an intricate relationship between SGT, the accessory protein and the major structural protein. In this study, we reported the interaction of SARS-CoV 7a with hSGT. To our knowledge, this is the first report of the interaction between a SARS-CoV accessory protein and a cellular protein. The interaction was confirmed by co-immunoprecipitation and immunofluorescent studies in VeroE6 cells. We concluded that TPR2 of hSGT (aa 125-158) was essential for the interaction between SGT and SARS-CoV 7a. We also showed that 7a interacted with SARS-CoV M and E. Based on recent reports, it is clear that the SARS-CoV accessory proteins are probably involved in various viral processes in vivo, including pathogenicity and infectivity. In showing that 7a interacted with SGT, a protein with many diverse and essential functions, our findings provided further evidence in support of this hypothesis. Our data raised the possibility that the interaction between SGT and 7a, and the latter’s interaction with M and E which have been shown to be sufficient for VLP formation, could play a role in virus assembly or release from the cell. Understanding the biological significance of the interaction between SGT and 7a could possibly lead to the discovery of novel therapeutics in treating SARS-CoV infection.
Acknowledgments
We thank the BRC, Singapore for technical support. We also thank Professor Nito Panganiban, Molecular Genetics and Microbiology, University of New Mexico for the generous gift of hSGT antibody that was used in the initial work to verify the specificity of our hSGT antibody. This work was supported by grants from the Agency for Science, Technology and Research (A*STAR), Singapore. We thank Yook-Wah Choi, Puay Yoke Tham, Daphne Chan, Choong-Tat Keng, and Kuo-Ming Lip for technical assistance.
Footnotes
The sequence reported in this paper has been assigned the GenBank Accession No. AY847013.
References
- 1.Ontiveros E., Kuo L., Masters P.S., Perlman S. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology. 2001;289:230–238. doi: 10.1006/viro.2001.1167. [DOI] [PubMed] [Google Scholar]
- 2.de Haan C.A., Volders H., Koetzner C.A., Masters P.S., Rottier P.J. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 2002;76:12491–12502. doi: 10.1128/JVI.76.24.12491-12502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ortego J., Sola I., Almazan F., Ceriani J.E., Riquelme C., Balasch M., Plana J., Enjuanes L. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology. 2003;308:13–22. doi: 10.1016/S0042-6822(02)00096-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 7.Tan Y.J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan W.S., Wu C., Chow S.C., Cheung T., To K.F., Leung W.K., Chan P.K., Lee K.C., Ng H.K., Au D.M., Lo A.W. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Mod. Pathol. 2005 doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fielding B.C., Tan Y.J., Shuo S., Tan T.H., Ooi E.E., Lim S.G., Hong W., Goh P.Y. Characterization of a unique group-specific protein (U122) from the severe acute respiratory syndrome (SARS) coronavirus. J. Virol. 2004;78:7311–7318. doi: 10.1128/JVI.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y.Y., Shuang B., Tan Y.X., Meng M.J., Han P., Mo X.N., Song Q.S., Qiu X.Y., Luo X., Gan Q.N., Zhang X., Zheng Y., Liu S.A., Wang X.N., Zhong N.S., Ma D.L. The protein X4 of severe acute respiratory syndrome-associated coronavirus is expressed on both virus-infected cells and lung tissue of severe acute respiratory syndrome patients and inhibits growth of Balb/c 3T3 cell line. Chin. Med. J. 2005;118:267–274. [PubMed] [Google Scholar]
- 11.Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-Coronavirus Orf7a accessory protein. Structure (Camb.) 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X., Lim S.G., Hong W., Tan Y.J. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G., Hong W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manser E., Huang H.Y., Loo T.H., Chen X.Q., Dong J.M., Leung T., Lim L. Expression of constitutively active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H., Chou C.F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.J. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.K.B. Nicholas, H.B. Nicholas Jr., GeneDoc: a tool for editing and annotating multiple sequence alignments, Distributed by the author (1997).
- 21.Hirano T., Kinoshita N., Morikawa K., Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 22.Sikorski R.S., Boguski M.S., Goebl M., Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 23.Blatch G.L., Lassle M. Review: the tetratricopeptide repeat: a structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Cziepluch C., Kordes E., Poirey R., Grewenig A., Rommelaere J., Jauniaux J.C. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1. J. Virol. 1998;72:4149–4156. doi: 10.1128/jvi.72.5.4149-4156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callahan M.A., Handley M.A., Lee Y.H., Talbot K.J., Harper J.W., Panganiban A.T. Functional interaction of human immunodeficiency virus type 1 Vpu and Gag with a novel member of the tetratricopeptide repeat protein family. J. Virol. 1998;72:5189–5197. doi: 10.1128/jvi.72.6.5189-5197.1998. Erratum in: J. Virol. 72 (1998) 8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schantl J.A., Roza M., De Jong A.P., Strous G.J. Small glutamine-rich tetratricopeptide repeat-containing protein (SGT) interacts with the ubiquitin-dependent endocytosis (UbE) motif of the growth hormone receptor. Biochem. J. 2003;373:855–863. doi: 10.1042/BJ20021591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Zhang Q., Zhu D. hSGT interacts with the N-terminal region of myostatin. Biochem. Biophys. Res. Commun. 2003;311:877–883. doi: 10.1016/j.bbrc.2003.10.080. [DOI] [PubMed] [Google Scholar]
- 28.Wu S.J., Liu F.H., Hu S.M., Wang C. Different combinations of the heat-shock cognate protein 70 (hsc70) C-terminal functional groups are utilized to interact with distinct tetratricopeptide repeat-containing proteins. Biochem. J. 2001;359:419–426. doi: 10.1042/0264-6021:3590419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angeletti P.C., Walker D., Panganiban A.T. Small glutamine-rich protein/viral protein U-binding protein is a novel cochaperone that affects heat shock protein 70 activity. Cell Stress Chaperones. 2002;7:258–268. doi: 10.1379/1466-1268(2002)007<0258:sgrpvp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winnefeld M., Rommelaere J., Cziepluch C. The human small glutamine-rich TPR-containing protein is required for progress through cell division. Exp. Cell. Res. 2004;293:43–57. doi: 10.1016/j.yexcr.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Shen H., Wang Y., Li Z., Yin H., Zong H., Jiang J., Gu J. Overexpression of small glutamine-rich TPR-containing protein promotes apoptosis in 7721 cells. FEBS Lett. 2005;579:1279–1284. doi: 10.1016/j.febslet.2004.12.092. [DOI] [PubMed] [Google Scholar]
- 32.Liou S.-T., Wang C. Small glutamine-rich tetratricopeptide repeat-containing protein is composed of three structural units with distinct functions. Acta Biochem. Biophys. 2005;435:253–263. doi: 10.1016/j.abb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 33.Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 2004;318:833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bour S., Strebel K. The HIV-1 Vpu protein: a multifunctional enhancer of viral particle release. Microbes Infect. 2003;5:1029–1039. doi: 10.1016/s1286-4579(03)00191-6. [DOI] [PubMed] [Google Scholar]


