Abstract
A novel aptameric enzyme subunit (AES) which can detect target DNAs has been developed. AES is an enzyme-inhibiting aptamer bearing a target-molecule binding site which can allosterically control enzymatic activity. The thrombin-inhibiting aptamer bearing a G-quartet structure was chosen as the enzyme-inhibiting aptamer. The stem-and-loop structure, which contains a strand complementary to the target DNA, was inserted into the G-quartet structure of the thrombin-inhibiting aptamer to disrupt the G-quartet structure through the hybridization of the target DNA with the complementary strand in the AES. The disruption of the G-quartet structure led to a decrease of the inhibitory activity of the whole aptameric complex. Using this designed aptamer, we were able to detect target DNAs by measuring the thrombin activity in a homogeneous solution without bound/free separation, and the lower detection limit was 20 nM.
Keywords: Homogeneous DNA sensing, Enzyme-inhibiting DNA aptamers, Aptameric enzyme subunit, SARS detection, Thrombin-inhibiting aptamer
The detection of specific DNA sequences is very useful for the clinical diagnosis of various diseases, the detection of toxic microorganisms in food or in the environment, and fundamental research in molecular biology. Most DNA sensing systems are based on the detection of the hybridization of the target DNA with the immobilized probe DNA (DNA chip); however, it takes time to detect the target DNA using this method, since the separation of the hybridized DNA molecules from the non-hybridized ones is usually required. Especially for the diagnosis of pathogenic bacteria or viruses, a rapid DNA sensing system is necessary.
To detect target DNAs quickly and easily, a homogeneous detection system which does not require the bound/free separation is ideal. A homogeneous detection system which uses a molecular recognition element that generates a detection signal when binding to the target molecule is required. Recently, such functional molecular recognition elements have been reported for DNA sensing; for example, molecular beacons [1], double-stranded probes [2], excimer fluorescence [3], [4], fluorescent polymers [5], DNAzymes [6], [7], DNA-fused nanoparticles [8], and inhibitor–DNA–enzyme modules [9], [10] have been developed. These novel sensor systems mainly rely on fluorescence resonance energy transfer (FRET) to generate a detection signal. FRET is useful for biosensors. However, enzyme detection of the hybridization between the probe DNA and the target DNA can improve sensitivity, since the enzyme can amplify the signal. At present, there are DNA sensor systems which use enzymes bound to the probe DNA and their inhibitors that can detect the target DNA by measuring the enzymatic activity in a homogeneous solution [9], [10]. However, the chemical modification of the enzyme using the DNA probe is sometimes difficult; if we can develop a DNA probe which induces a change in the enzymatic activity, which can then be used as a detection signal upon hybridization with the target DNA, it would make the DNA sensing system simpler and easier to integrate into a wide range of applications. With the aim of constructing such a DNA probe, we focused on aptamers.
Aptamers are nucleic acid ligands obtained through the systematic evolution of ligands by exponential enrichment (SELEX). Many aptamers which recognize various molecules have been reported [11], [12], [13], [14] and some of them show strong binding at K d values in the nanomolar range, especially to protein. Some aptamers with high enzyme-binding activity also show high inhibitory activity and can therefore be used as pharmaceuticals [15]. DNA aptamers are easily and inexpensively synthesized; therefore, it is easy to add a probe DNA to a DNA aptamer. Additionally, DNA aptamers have a distinctive advantage, in that they are single-stranded DNAs, and therefore the way their structure changes is easy to design. Signal-generating molecules [16], [17], [18], [19], [20], [21], [22], [23], which acquire their properties as a result of such changes, work like molecular beacons. Molecular beacons generate signals, usually fluorescence emission, as a result of the structural changes caused by the hybridization with the target DNA.
We developed an aptameric enzyme subunit (AES), which is a DNA aptamer composed of an enzyme-inhibiting aptamer and a target-molecule-binding aptamer [24]. We designed an AES whose structure is such that the enzyme-inhibiting aptamer site changes when the target molecule binds to the target-molecule-binding aptamer site. The structural change of the enzyme-inhibiting aptamer site induces a change in the inhibitory activity of the AES, which enables us to detect a target molecule by measuring the enzymatic activity of the whole aptameric complex in a homogeneous solution without bound/free separation.
Here, we present a novel AES that can detect its target DNA using a DNA aptamer that inhibits the coagulation activity of thrombin [25]. The detection mechanism is schematized in Fig. 1 . This thrombin-inhibiting aptamer folds into a G-quartet structure, which is very important for the inhibition of thrombin activity. We inserted the stem-and-loop structure bearing a sequence complementary to the target DNA at the loop region into the G-quartet structure so that it would be destabilized by the hybridization of the target DNA with its complementary sequence in the aptamer. We constructed some AESs and investigated the changes in thrombin activity which resulted from the hybridization of these AESs with their target DNAs.
Fig. 1.
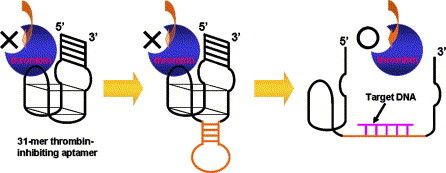
Schematic of the DNA sensing system, in which a thrombin-inhibiting aptamer is fused with a stem-and-loop structure. The thrombin-inhibiting aptamer into which a stem-and-loop structure is inserted forms a G-quartet structure, since the stem site is formed by the annealing of both of its sides. When the target DNA hybridizes with the loop site of the stem-and-loop structure, the stem site is dehybridized, which leads to the disruption of the G-quartet structure of the thrombin-inhibiting aptamer site, thus reducing its thrombin-inhibiting activity.
Materials and methods
Materials. Human thrombin was purchased from Wako Chemicals (Osaka, Japan) and human fibrinogen was purchased from Wako Chemicals and Sigma (Missouri, USA). Dimethyl sulfate and 2-mercaptoethanol were purchased from TOKYO KASEI KOGYO (Tokyo, Japan), ethidium bromide was purchased form Sigma, and tRNA from baker’s yeast was purchased from Roche Diagnostics (Basel, Switzerland). All oligonucleotides were obtained from Invitrogen (California, USA). The other reagents were of analytical grade.
Design of AESs. The DNA sequences used in this study are shown in Table 1 . We used a 31-mer thrombin-inhibiting aptamer as the enzyme-inhibiting aptamer. The stem-and-loop structure bearing the probe DNA sequence was inserted into the 3′-end T-T loop of the G-quartet structure of the 31-mer thrombin-inhibiting aptamer.
Table 1.
The sequences of the oligonucleotides used in this study
| 31-mer thrombin-inhibiting aptamer | 5′ CACTGGTAGGTTGGTGTGGTTGGGGCCAGTG 3′ |
| AES 1 | 5′ CACTGGTAGGTTGGTGTGGTGCGAGAAGTTAAGACCTATGCTCGCTGGGGCCAGTG 3′ |
| AES 2 | 5′ CACTGGTAGGTTGGTGTGGTTGCGAGAAGTTAAGACCTATGCTCGCTTGGGGCCAGTG 3′ |
| 15-mer target DNA | 5′ CATAGGTCTTAACTT 3′ |
| 15-mer control DNA | 5′ ATTGCTATCGTACAT 3′ |
| AES SARS 1 | 5′ CACTGGTAGGTTGGTGTGGTTGGACGACGAATTCATGATCACGTCCTTGGGGCCAGTG 3′ |
| 15-mer SARS DNA | 5′ TGATCATGAATTCGT 3′ |
| Stem and loop SARS 1 | 5′TGGACGACGAATTCATGATCACGTCCT 3′ |
| AES SARS 2 | 5′ CACTGGTAGGTTGGTGTGGTTCGGACGCACGAATTCATGATCAACATCCCTACGTCCGTTGGGGCCAGTG 3′ |
| AES SARS 3 | 5′ CACTGGTAGGTTGGTGTGGTTGCGGACGCACGAATTCATGATCAACATCCCTACGTCCGCTTGGGGCCAGTG 3′ |
| AES SARS 4 | 5′ CACTGGTAGGTTGGTGTGGTTCGCGGACGCACGAATTCATGATCAACATCCCTACGTCCGCGTTGGGGCCAGTG 3′ |
| 25-mer SARS DNA | 5′ TAGGGATGTTGATCATGAATTCGTG 3′ |
The stem-and-loop sequence inserted into the 31-mer thrombin-inhibiting aptamer is underlined. The 15-mer target DNA, the 15-mer SARS, and the 25-mer SARS are complementary strands of the loop sequence of AES 1, 2, and AES SARS 1 and AES SARS 2, 3, and 4, respectively. The stem-and-loop SARS 1 sequence is the oligonucleotide inserted into AES SARS 1.
For AES 1 and 2, we used the DNA sequence of the molecular beacons as the stem-and-loop structure, which has 5-bp-stem and 15-mer-loop oligonucleotides [1]. For the series of SARS AESs, the loop sequences were designed to contain a sequence complementary to a portion of the genome sequence of the severe acute respiratory syndrome (SARS) coronavirus (nucleotide positions 15581–15595 for AES SARS 1 and 15572–15596 for AES SARS 2, 3, and 4). These genome sequences were amplified using a pair of primers whose sequences have been published by the World Health Organization (WHO), and the stem sites were redesigned using the mfold program [26]. We also inserted two additional T bases between the thrombin-inhibiting aptamer and the stem-and-loop structure in all AESs except AES 1, since the diameter of the DNA double helix is different from the distance between two Gs of the G-quartet structure (approximately 17 and 20 Å, respectively). The putative secondary structure of the AES SARS 1 is shown in Fig. 2 .
Fig. 2.
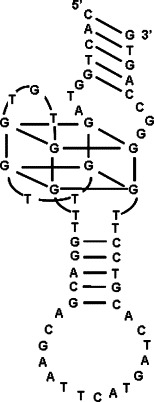
Presumed structure of AES SARS 1.
For the native PAGE experiment with AES SARS 1, AES SARS 1 was modified with fluorescein isothiocyanate (FITC) at its 5′-end. For the measurement of the CD spectra of AES SARS 1, a stem-and-loop structure to be inserted into the 31-mer thrombin-inhibiting aptamer on AES SARS 1 was synthesized. For the DMS footprinting of AES SARS 1, AES SARS 1, and the 31-mer thrombin-inhibiting aptamer were modified with FITC at their 5′-end.
Measurement of the inhibitory effect of the AESs on thrombin. Fifty microliters of folding buffer (50 mM Tris–HCl, 5 mM KCl, pH 8.0) containing 4 μM AES and 4 μM target DNA or control DNA (final concentration: 1μM) was heat-denatured at 95 °C for 3 min and cooled to 25 °C over a period of 30 min. Following the heat treatment, the AES solution was mixed with 50 μl of the measurement buffer (50 mM Tris–HCl, 5 mM KCl, and 100 mM NaCl, pH 8.0) containing thrombin (final concentration: 54 nM), and the mixture was incubated for 5 min at 37 °C. After 5 min, 100 μl of the AES solution containing thrombin was added to 100 μl of the measurement buffer containing fibrinogen (final concentration: 1 mg/ml). The clotting time was measured using an automated fibrometer (Amelung KC-4 A Micro Coagulation Analyzer) at 37 °C. In the experiment on the influence of the target DNA concentrations on AES SARS 1, we used 50 nM AES SARS 1, 5.4 nM thrombin, and 1 mg/ml fibrinogen, which was purchased from Sigma.
Native PAGE of the oligonucleotides. FITC-labeled 15-mer SARS DNA (8.3 μM) and AES SARS 1 (8.3 μM) were mixed with the folding buffer, and the mixture was heat-treated as described above. Following the heat treatment, 6 μl of the mixture was electrophoresed on 11% polyacrylamide gel in 1× TBE buffer (89 mM Tris, 89 mM H3BO3, and 2 mM EDTA) at room temperature. Fluorescence imaging of FITC was carried out on a Typhoon 8600 (Amersham Biosciences, New Jersey, USA), and the oligonucleotides were also stained with ethidium bromide.
CD spectrum measurement of AES SARS 1. The CD spectra of the thrombin-inhibiting aptamer, AES SARS 1, AES SARS 1 hybridized with the 15-mer SARS DNA, the stem-and-loop structure, and the stem-and-loop structure hybridized with the 15-mer SARS DNA were measured. All oligonucleotides, at a final concentration of 25 μM, were heat-treated in the folding buffer as described above, and then 100 μl of the AES solution was added to 300 μl of the measurement buffer. The CD spectra were measured with a J-720 spectropolarimeter (Jasco, Tokyo, Japan) at 200–320 nm using a 0.1-cm path-length cuvette at 25 °C.
DMS footprinting. FITC-labeled AES SARS 1 (10 μM) was heat-treated as described above in the presence or absence of 10 μM 15-mer SARS DNA or 15-mer control DNA. Dimethyl sulfate (DMS) was added to give a final concentration of 0.1% (v/v) in a total volume of 100 μl, and the solutions were incubated at room temperature for 10 min. The methylation reaction was halted by adding 5 μl of the stop solution (1.5 M sodium acetate, pH 7.0, 1 M 2-mercaptoethanol, and 100 μg/ml tRNA). The reactants were precipitated using ethanol, dissolved in 100 μl of 10% piperidine, and incubated at 90 °C for 30 min. After the piperidine cleavage, the sample solution was lyophilized and separated on 7-M urea–20% polyacrylamide gel, and FITC fluorescence imaging was carried out on a Typhoon 8600 (Amersham Biosciences, New Jersey, USA).
Results and discussion
Change in the inhibitory activity of the AESs after hybridization with the target DNA
We used a 31-mer thrombin-inhibiting aptamer as the enzyme-inhibiting aptamer; this aptamer was screened using an evolution-mimicking algorithm and a library that contains a 15-mer prototype thrombin-inhibiting aptamer with 8-mer oligonucleotides added to both the 5′- and the 3′-ends [25]. Therefore, the 31-mer thrombin-inhibiting aptamer contains the consensus sequence of the 15-mer thrombin-inhibiting aptamer that has already been determined to fold into a G-quartet structure [27], [28], [29], [30], which is important for the inhibition of thrombin activity. Thus, the 31-mer thrombin-inhibiting aptamer used in this study was expected to form a G-quartet structure.
There are three loops in the 15-mer thrombin-inhibiting aptamer connected to the G-quartet structure: a T-G-T loop, a 3′-end T-T loop, and a 5′-end T-T loop. The T-G-T loop contacts thrombin directly [31], therefore, the stem-and-loop structure was inserted into the 31-mer thrombin-inhibiting aptamer at the 3′-end T-T loop or 5′-end T-T loop. We designed the stem-and-loop structure inserted into the 31-mer thrombin-inhibiting aptamer in such a way that the conformational change in stem-and-loop structure would lead to a conformational change in the thrombin-inhibiting aptamer, which would in turn cause a decrease in the inhibitory effect of the whole complex on thrombin.
First, a stem-and-loop structure reported to be a molecular beacon [1] was used, since this DNA sequence has the ability to form a stem-and-loop structure composed of a 5-bp stem and a 15-mer loop. This structure is known to be destroyed during hybridization of its target DNA with the loop part. The diameter of a DNA double helix (approximately 20 Å) is longer than the distance between two Gs of the G-quartet structure (approximately 17 Å). Therefore, we expected that the difference would cause the distortion and destabilization of the G-quartet structure if the stem-and-loop structure were to be directly connected to the G-quartet structure. Therefore, we constructed two kinds of thrombin-inhibiting aptamers: one was a thrombin-inhibiting aptamer which has the stem-and-loop structure inserted at the 3′-end T-T loop directly (AES 1), and the other was a thrombin-inhibiting aptamer, to which a stem-and-loop structure with two additional T bases, one at the 3′-end and one at the 5′-end, was attached at the 3′-end of the T-T loop of the G-quartet structure (AES 2).
The thrombin-inhibiting activities of these AESs were determined by measuring the coagulation activity of thrombin in their presence. The clotting time was approximately 10 s in the presence of only thrombin (Fig. 3 A, bar 1); the addition of the 31-mer thrombin-inhibiting aptamer resulted in the extension of the clotting time to approximately 150 s, since it inhibited thrombin activity (bar 2). The clotting time was approximately 19 s in the presence of AES 1 (data not shown); however, it was approximately 45 s in the presence of AES 2, which has two additional T bases insertion (bar 3). This result indicates that the two additional T bases insertion prevented the G-quartet structure from becoming distorted. Although the inhibitory activity of AES 2 did not change in the presence of the 15-mer control DNA, which does not hybridize with it (bar 4), the clotting time for the AES 2 changed from 45 to 26 s as a result of the addition of its 15-mer target DNA (bar 5). This suggests that the hybridization of the target DNA with the loop site of AES 2 led to the destabilization of the thrombin-inhibiting aptamer site, causing a decrease in the inhibitory effect of the whole complex on thrombin.
Fig. 3.
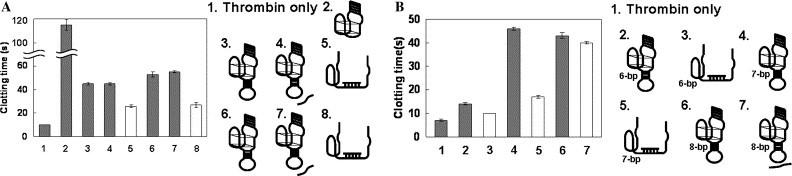
Inhibition of clotting by the addition of DNA. These experiments were performed using 1 μM AES, 1 μM target DNA, 1 μM control DNA, 54 nM thrombin, and 1 mg/ml fibrinogen at 37 °C. (A) The inhibitory activity of AES 2 and AES SARS 1. Bar 1, thrombin only; bar 2, 31-mer thrombin-inhibiting aptamer; bar 3, AES 2; bar 4, AES 2 with a 15-mer control DNA; bar 5, AES 2 with a 15-mer target DNA; bar 6, AES SARS 1; bar 7, AES SARS 1 with a 15-mer control DNA; bar 8, AES SARS 1 with a 15-mer SARS DNA. (B) The inhibitory activity of AES SARS 2, 3, and 4. Bar 1, thrombin only; bar 2, AES SARS 2; bar 3, AES SARS 2 with a 25-mer SARS DNA; bar 4, AES SARS 3; bar 5, AES SARS 3 with a 25-mer SARS DNA; bar 6, AES SARS 4; bar 7, AES SARS 4 with a 25-mer SARS DNA. The black bars indicate the absence of the target DNA and the white bars indicate the presence of the target DNA. The clotting times are given as averages of three experiments and error bars are provided.
Next, we investigated whether this system is applicable to target DNAs with other sequences and other lengths. First, we designed AES SARS 1, which has a 5-bp-stem and a 15-mer-loop inserted into 3′-side T-T loop site, and the loop sequence was designed to contain a sequence complementary to a portion of the SARS coronavirus genome (nucleotide positions 15581–15595) [32], [33], as well as two additional T bases insertion. The stem sequence was chosen so that its melting temperature (T m) was slightly higher than of the hybrid between the loop and the target DNA (stem: 52.0 °C; loop, 55.9 °C) using the mfold program [26]. We analyzed a character of the AES SARS 1. AES SARS 1 as well as AES 2 inhibited the activity of thrombin, and the clotting time decreased from 53 to 27 s when the 15-mer SARS DNA was added (Fig. 3A, bars 6–8). These results suggest that it is possible to detect other target DNA sequences using this DNA sensing system.
We also constructed a thrombin-inhibiting aptamer which has a stem-and-loop structure with two additional T bases inserted at its 5′-end T-T loop. This AES had the same characteristics as AES 2; that is, the inhibitory effect on thrombin decreased when its target DNA was added (data not shown). This indicates that the stem-and-loop structure can be inserted into either the 5′-end or the 3′-end of the T-T loop of the 31-mer thrombin-inhibiting aptamer.
Finally, we designed AES SARS 2, 3, and 4, all of which have a 25-mer loop sequence, containing a sequence complementary to a portion of SARS coronavirus genome (nucleotide positions 15572–15596), inserted into the 31-mer thrombin-inhibiting aptamer. Each of these AESs has two additional T bases insertion, and a 6-, 7- or 8-bp stem was used, respectively. AES SARS 2 had little inhibitory effect, and the inhibitory activity of AES SARS 4 did not decrease even when its target DNA was added. AES SARS 3 had a much higher inhibitory effect, and the clotting time decreased from 46 to 17 s when its target DNA was added (Fig. 3B). For these AESs, the local T m of the stem-and-loop structure were predicted by the mfold program as follows: 67.9 °C at the loop site, and 55.6, 62.9, and 69.8 °C at the 6-, 7-, and 8-bp stem sites, respectively. Namely, when the T m of the stem site was lower than that of the loop site, the AES did not fold into a G-quartet structure, and when the T m of stem site was higher than that of the loop site, the AES structure seemed not to be disrupted by the hybridization with the target DNA. These results are similar to those obtained for molecular beacons, which suggests that the dehybridization of the stem site induced by the hybridization between the target DNA and the loop site is related to the disruption of the G-quartet structure.
We constructed three types of AESs (AES 2, AES SARS 1, and AES SARS 3) for different target DNAs using a thrombin-inhibiting aptamer. We speculated that these AESs would inhibit the enzymatic activity of thrombin, and their inhibitory effects would decrease when their target DNAs were added. The G-quartet structure of the thrombin-inhibiting aptamer plays an important role in the inhibitory action of the aptameric complex on thrombin. In other words, these AESs would be destroyed when they hybridized with their target DNAs because they fold into G-quartet structures. To confirm our hypothesis, we investigated whether AES SARS 1 hybridizes with its target DNA and whether its G-quartet structure is destroyed when its target DNA is added.
Hybridization of AES SARS 1 with its target DNA
The hybridization of AES SARS 1 with its target DNA was confirmed by native PAGE on 11% polyacrylamide gel using FITC-labeled target DNA. The oligonucleotides were detected by both ethidium bromide staining and FITC fluorescence. As shown in Fig. 4 , in the presence of AES SARS 1, while the band of the 15-mer control DNA did not shift (lane 3), the band of the 15-mer SARS DNA shifted to the AES SARS 1 band (lane 4). This result indicates that most AES SARS 1 hybridized with the 15-mer SARS DNA in this experimental condition.
Fig. 4.
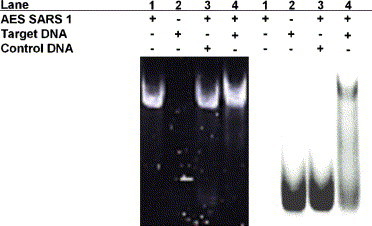
Native PAGE of DNA complexes. The oligonucleotides (50 pmol) were electrophoresed on 11% polyacrylamide gel in 1× TBE buffer at room temperature. DNA was detected by ethidium bromide (left) and fluorescence (right); both figures show same gel. Lane 1, AES SARS 1; lane 2, FITC-labeled 15-mer SARS DNA; lane 3, AES SARS 1 with FITC-labeled 15-mer control DNA; lane 4, AES SARS 1 with FITC-labeled 15-mer SARS DNA.
Structural change of AES SARS 1 upon hybridization with its target DNA
We expected the 31-mer thrombin-inhibiting aptamer used in this study to form a G-quartet structure, which would be important for the inhibitory activity of the aptamer complex. Therefore, we speculated that the AESs will have a G-quartet structure and that it would be destroyed upon hybridization with the target DNAs, judging from the fact that the inhibitory activity of the AESs decreased upon hybridization with their respective target DNAs. In order to investigate whether this structural change occurs or not, we measured the CD spectra of AES SARS 1 in the presence or absence of its target DNA and analyzed them using DMS footprinting assay.
CD spectra
CD spectra of the 31-mer thrombin-inhibiting aptamer, AES SARS 1 and the stem-and-loop structure which was inserted into the 31-mer thrombin-inhibiting aptamer of AES SARS 1 were measured. The CD spectrum of the 31-mer thrombin-inhibiting aptamer had three positive peaks, one near 295 nm, one near 250 nm, and one near 210 nm, and two negative peaks, one near 265 nm and another near 235 nm (Fig. 5 ). This CD spectrum is almost identical to that of the 15-mer thrombin-inhibiting aptamer [34], [35] which folds into an anti-parallel G-quartet structure, as previously determined by NMR and X-ray structural analysis. This suggests that the 31-mer thrombin-inhibiting aptamer folds into an anti-parallel G-quartet structure.
Fig. 5.
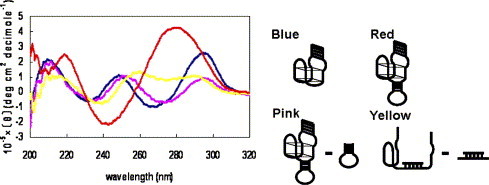
CD spectra of AES SARS 1. Oligonucleotides at a concentration of 25 μM were used. The 31-mer thrombin-inhibiting aptamer (blue), AES SARS 1 (red), the CD spectrum of the stem-and-loop SARS 1 subtracted from that of AES SARS 1 (pink), the CD spectrum of the stem-and-loop SARS 1 hybridized with a 15-mer SARS DNA subtracted from that of AES SARS 1 hybridized with the 15-mer SARS DNA (yellow). The CD spectra were measured using a 0.1-cm path-length cuvette at 25 °C.
The CD spectrum of AES SARS 1 had peaks different from that of the 31-mer thrombin-inhibiting aptamer; however, the CD spectrum produced by subtracting the spectrum of the stem-and-loop DNA sequence from that of AES SARS 1 had peaks similar to the CD spectrum of the 31-mer thrombin-inhibiting aptamer. When comparing the CD spectrum produced by subtracting the spectrum of the stem-and-loop DNA sequence from that of AES SARS 1 with that of the 31-mer thrombin-inhibiting aptamer, we can see that the peaks near 265 and 250 nm shifted to 273 and 253 nm, respectively, and that the positive peak at 295 nm was weak. The anti-parallel G-quartet structure had a positive peak near 295 and 250 nm, and a negative peak at 265 nm, and these peaks disappeared due to the destabilization of the G-quartet structure [36]. This suggests that AES SARS 1 folds into a G-quartet structure, but that the G-quartet structure of AES SARS 1 is unstable compared with the structure of the 31-mer thrombin-inhibiting aptamer. This hypothesis is supported by the results for AES SARS 1, which showed lower thrombin-inhibiting activity than the 31-mer thrombin-inhibiting aptamer, since the stability of the G-quartet structure of the 31-mer thrombin-inhibiting aptamer relates to the thrombin-inhibiting activity.
The CD spectrum produced by subtracting the spectrum of the stem-and-loop structure hybridized with its target DNA from that of AES SARS 1 hybridized with its target DNA was very different from the CD spectrum produced by subtracting the spectrum of the stem-and-loop DNA sequence from that of AES SARS 1. It does not have the positive peaks near 295 and 250 nm and the negative peak near 265 nm characteristic of the G-quartet structure. This suggests that the G-quartet structure of AES SARS 1 is destroyed during hybridization with the target DNA.
DMS footprinting
We also investigated whether the G-quartet structure of AES SARS 1 is destroyed during hybridization with the target DNA, using a DMS footprinting assay. Although the N7 of guanine is methylated by dimethyl sulfate (DMS), an oligonucleotide folded into a G-quartet structure is protected from N7 methylation since the N7 of one guanine forms a hydrogen bond with the N2 of another guanine to form the G-quartet structure. The DMS footprinting assay was performed on FITC-labeled AES SARS 1, FITC-labeled AES SARS 1 hybridized with its target DNA, and FITC-labeled 31-mer thrombin-inhibiting aptamer containing potassium ions or not. In the 31-mer thrombin-inhibiting aptamer, the potassium ions are necessary for G-quartet structure folding. Therefore, as shown Fig. 6 , the guanine residues composing the G-quartet structure (G9, G10, G13, G14, G18, G19, G22, and G23) were exposed to N7 methylation in the absence of potassium ions (lanes 1 and 2). In AES SARS 1, although G49 and G50 (residues corresponding to G22 and G23 in the 31-mer thrombin-inhibiting aptamer, respectively) were not detected, G9, G10, G13, G14, G18, and G19 (residues corresponding to G9, G10, G13, G14, G18, and G19 in the 31-mer thrombin-inhibiting aptamer, respectively) were protected from N7 methylation (lanes 3 and 5). The N7 methylation of the guanine residues of AES SARS 1 did not change the residues in the presence of the 15-mer control DNA; however, they were not protected from N7 methylation in the presence of the target DNA (lane 6). These results indicate that AES SARS 1 has a G-quartet structure and that it is destroyed during hybridization with the target DNA.
Fig. 6.
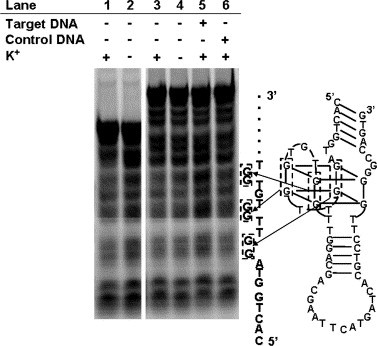
DMS footprinting assays of AES SARS 1. Lane 1, 31-mer thrombin-inhibiting aptamer with K+; lane 2, 31-mer thrombin-inhibiting aptamer without K+; lane 3, AES SARS 1 with K+; lane 4, AES SARS 1 without K+; lane 5, AES SARS 1 hybridized with its target DNA in the presence of K+; lane 6, AES SARS 1 containing the 15-mer control DNA in the presence of K+. The methylated guanines in the G-quartet structure are shown in the secondary structure attached to AES SARS 1. The experimental conditions are described in the Materials and methods.
Calibration graph for the AES SARS 1 target DNA
We made a calibration graph using 5.4 nM thrombin, 1 mg/ml fibrinogen, and 50 nM AES SARS 1. Under these conditions, the clotting time was approximately 100 s in the presence of thrombin only (data not shown). The addition of 50 nM AES SARS 1 led to the extension of the clotting time to approximately 150 s. A decrease in the clotting time from 150 to 110 s was observed with the increase in the concentration of the target DNA to 50 nM, and when target DNA at concentrations higher than 50 nM was added, the clotting time became saturated (Fig. 7 ). The thrombin-inhibiting activity was not influenced by the addition of the control DNA. This calibration graph suggests that the target DNA of AES SARS 1 was selectively detected by measuring the thrombin enzymatic activity in a homogeneous solution and that the lower limit of the target DNA detection was 20 nM.
Fig. 7.
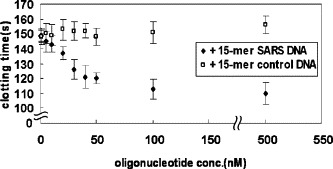
Calibration graph of the 15-mer SARS DNA using AES SARS 1. These experiments were performed using 50 nM AES SARS 1, 5.4 nM thrombin, and 1 mg/ml fibrinogen. The clotting times are given as averages of three experiments and error bars are provided.
When we used 1 μM AES SARS 1, the dynamic range was from 0.2 to 1 μM (data not shown). The clotting time was saturated when AES SARS 1 and its target DNA reached the same concentration. This result indicates that AESs bind to their target DNAs at a ratio of one to one and that the dynamic range can be changed by changing the concentration of the AESs.
Although we carried out an investigation of the amount of thrombin/AES complex after the addition of the target DNA, smear bands were observed and there should be many types of AES/target DNA complexes (data not shown). It was difficult to estimate the amount of thrombin/AES complex, so we are not sure whether they are separated or not. The only thing we can say is that the G-quartet structure was rendered unstable by the target-DNA hybridization, and that the thrombin inhibition by AES showed a dependence on the target DNA concentration.
We used a 31-mer thrombin-inhibiting aptamer as the enzyme-inhibiting aptamer and used an automated fibrometer to measure the thrombin activity. Many hospitals have this machine, since it is used for the quantitative determination of blood fibrinogen. Therefore, this DNA sensing system can be immediately used for the diagnosis of viruses and pathogens.
As a DNA sensing system, the AES constructed from a thrombin-inhibiting aptamer was not as sensitive as the inhibitor–DNA–enzyme module. We believe that if an AES can be constructed from a suitable enzyme-inhibiting aptamer, the sensitivity can be improved. There are two requirements which a suitable enzyme-inhibiting aptamer must fulfill: it must have a high affinity to its target enzyme, and it must have enzyme-inhibiting activity that can amplify the sensing signal.
In this DNA sensing system, the ratio of AES molecules whose structure is changed due to the hybridization with the target DNA is very important for the detection of the target DNA, since this system is based on the measurement of the ratio of structurally changed AES molecules. Therefore, the optimization of the AES concentration can alter the dynamic range of this DNA sensor. Namely, to achieve a lower detection limit, it is necessary to use a lower concentration of AES. In the measurement of the detection limit using AES SARS 1, we used 50 nM AES SARS 1, since the 31-mer thrombin-inhibiting aptamer binds to thrombin with a K d of 34 nM [25]. However, if an enzyme-inhibiting aptamer which has a higher affinity for its target enzyme is used, it can show inhibitory activity at lower AES concentrations, which would lead to the improvement of the AES sensitivity.
In conclusion, when using a thrombin-inhibiting aptamer which contains a sequence complementary to the target DNA, the structure of the designed aptamer is destroyed during hybridization with the target DNA, and it is possible to detect the target DNA by measuring the thrombin activity in a homogeneous solution without B/F separation. In this study, we constructed a new AES whose function changes upon hybridization with the target DNA. This DNA detection system is very simple compared to other reported DNA sensing systems, because it is simply an unmodified oligonucleotide; the only preparatory procedure is the insertion of a stem-and-loop structure into the aptamer, and so the system can be applied to any aptamer. We had already developed different types of AESs by connecting a thrombin-inhibiting aptamer to target-molecule-binding aptamers, one of which was used for adenosine detection [24]. With those AES systems, target molecules can be detected by measuring the thrombin activity. Therefore, we believe that it is possible to detect many target DNAs, RNAs, low-mass molecules, and proteins in a common assay using AESs.
Acknowledgment
This work was supported by a Grant-in-Aid for the 21st Century COE program “Future Nano-Materials” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
References
- 1.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Li Q., Luan G., Guo Q., Liang J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002;30:E5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masuko M., Ohtani H., Ebata K., Shimadzu A. Optimization of excimer-forming two-probe nucleic acid hybridization method with pyrene as a fluorophore. Nucleic Acids Res. 1998;26:5409–5416. doi: 10.1093/nar/26.23.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isacsson J., Cao H., Ohlsson L., Nordgren S., Svanvik N., Westman G., Kubista M., Sjoback R., Sehlstedt U. Rapid and specific detection of PCR products using light-up probes. Mol. Cell. Probes. 2000;14:321–328. doi: 10.1006/mcpr.2000.0321. [DOI] [PubMed] [Google Scholar]
- 5.Ho H.A., Boissinot M., Bergeron M.G., Corbeil G., Dore K., Boudreau D., Leclerc M. Colorimetric and fluorometric detection of nucleic acids using cationic polythiophene derivatives. Angew. Chem., Int. Ed. 2002;41:1618–1621. doi: 10.1002/1521-3773(20020503)41:9<1548::aid-anie1548>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Wang D.Y., Sen D. A novel mode of regulation of an RNA-cleaving DNAzyme by effectors that bind to both enzyme and substrate. J. Mol. Biol. 2001;310:723–734. doi: 10.1006/jmbi.2001.4811. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Y., Pavlov V., Niazov T., Dishon A., Kotler M., Willner I. Beacons for the detection of DNA and telomerase activity. J. Am. Chem. Soc. 2004;126:7430–7431. doi: 10.1021/ja031875r. [DOI] [PubMed] [Google Scholar]
- 8.Elghanian R., Storhoff J.J., Mucic R.C., Letsinger R.L., Mirkin C.A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 9.Saghatelian A., Guckian K.M., Thayer D.A., Ghadiri M.R. DNA detection and signal amplification via an engineered allosteric enzyme. J. Am. Chem. Soc. 2003;125:344–345. doi: 10.1021/ja027885u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavlov V., Shlyahovsky B., Willner I. Fluorescence detection of DNA by the catalytic activation of an aptamer/thrombin complex. J. Am. Chem. Soc. 2005;127:6522–6523. doi: 10.1021/ja050678k. [DOI] [PubMed] [Google Scholar]
- 11.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 13.Feigon J., Dieckmann T., Smith F.W. Aptamer structures from A to zeta. Chem. Biol. 1996;3:611–617. doi: 10.1016/s1074-5521(96)90127-1. [DOI] [PubMed] [Google Scholar]
- 14.Hesselberth J., Robertson M.P., Jhaveri S., Ellington A.D. In vitro selection of nucleic acids for diagnostic applications. J. Biotechnol. 2000;74:15–25. doi: 10.1016/s1389-0352(99)00005-7. [DOI] [PubMed] [Google Scholar]
- 15.Rusconi C.P., Scardino E., Layzer J., Pitoc G.A., Ortel T.L., Monroe D., Sullenger B.A. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto R., Baba T., Kumar P.K. Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells. 2000;5:389–396. doi: 10.1046/j.1365-2443.2000.00331.x. [DOI] [PubMed] [Google Scholar]
- 17.Hamaguchi N., Ellington A.D., Stanton M. Aptamer beacons for the direct detection of proteins. Anal. Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 18.Stojanovic M.N., de Prada P., Landry D.W. Fluorescent sensors based on aptamer self-assembly. J. Am. Chem. Soc. 2000;122:11547–11548. doi: 10.1021/ja0022223. [DOI] [PubMed] [Google Scholar]
- 19.Stojanovic M.N., de Prada P., Landry D.W. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 20.Nutiu R., Li Y. Structure-switching signaling aptamers. J. Am. Chem. Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 21.Jhaveri S., Kirby R., Conrad R., Maglott E., Bowser M., Kennedy R., Glick G., Ellington A.D. Designed signaling aptamers that transduce molecular recognition to changes in fluorescence intensity. J. Am. Chem. Soc. 2000;122:2469–2473. [Google Scholar]
- 22.Jhaveri S., Rajendran M., Ellington A.D. In vitro selection of signaling aptamers. Nat. Biotechnol. 2000;18:1293–1297. doi: 10.1038/82414. [DOI] [PubMed] [Google Scholar]
- 23.Stojanovic M.N., Kolpashchikov D.M. Modular aptameric sensors. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida W., Sode K., Ikebukuro K. Aptameric enzyme subunit for biosensing based on enzymatic activity measurement. Anal. Chem. 2006;78:3296–3303. doi: 10.1021/ac060254o. [DOI] [PubMed] [Google Scholar]
- 25.Ikebukuro K., Okumura Y., Sumikura K., Karube I. A novel method of screening thrombin-inhibiting DNA aptamers using an evolution-mimicking algorithm. Nucleic Acids Res. 2005;33:e108. doi: 10.1093/nar/gni108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 28.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultze P., Macaya R.F., Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 30.Kuryavyi V., Majumdar A., Shallop A., Chernichenko N., Skripkin E., Jones R., Patel D.J. A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)–G(syn)–G(anti)–G(anti) tetrads flanked by a G-(T-T) triad and a T-T-T triple. J. Mol. Biol. 2001;310:181–194. doi: 10.1006/jmbi.2001.4759. [DOI] [PubMed] [Google Scholar]
- 31.Padmanabhan K., Padmanabhan K.P., Ferrara J.D., E Sadler J., Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:7651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 32.Rota P.A. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 33.Marra M.A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 34.Smirnov I., Shafer R.H. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 35.Dapic V., Abdomerovic V., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyoshi D., Nakao A., Toda T., Sugimoto N. Effect of divalent cations on antiparallel G-quartet structure of d(G4T4G4) FEBS Lett. 2001;496:128–133. doi: 10.1016/s0014-5793(01)02416-4. [DOI] [PubMed] [Google Scholar]


