Abstract
Two candidate small interfering RNAs (siRNAs) corresponding to severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike gene were designed and in vitro transcribed to explore the possibility of silencing SARS-CoV S gene. The plasmid pEGFP-optS, which contains the codon-optimized SARS-CoV S gene and expresses spike-EGFP fusion protein (S-EGFP) as silencing target and expressing reporter, was transfected with siRNAs into HEK 293T cells. At various time points of posttransfection, the levels of S-EGFP expression and amounts of spike mRNA transcript were detected by fluorescence microscopy, flow cytometry, Western blot, and real-time quantitative PCR, respectively. The results showed that the cells transfected with pEGFP-optS expressed S-EGFP fusion protein at a higher level compared with those transfected with pEGFP-S, which contains wildtype SARS-CoV spike gene sequence. The green fluorescence, mean fluorescence intensity, and SARS-CoV S RNA transcripts were found significantly reduced, and the expression of SARS-CoV S glycoprotein was strongly inhibited in those cells co-transfected with either EGFP- or S-specific siRNAs. Our findings demonstrated that the S-specific siRNAs used in this study were able to specifically and effectively inhibit SARS-CoV S glycoprotein expression in cultured cells through blocking the accumulation of S mRNA, which may provide an approach for studies on the functions of SARS-CoV S gene and development of novel prophylactic or therapeutic agents for SARS-CoV.
Keywords: SARS-CoV, S glycoprotein, RNA interference, siRNA, Gene silencing
The coronaviruses are a diverse group of enveloped positive-strand RNA viruses that can cause respiratory, enteric, and neurologic diseases in their host species. The recently identified severe acute respiratory syndrome-associated coronavirus (SARS-CoV) has been proved as a new group of coronaviruses [1], [2], [3], which consists of four major structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N). The S protein, with 1255 amino acids in size, is a surface glycoprotein and contains signal peptide at the N-terminus (residues 1–13), a large amino-terminal ectodomain (residues 14–1195), and a short carboxy-terminal endodomain bridged with a transmembrane domain (residues 1196–1255). There is only 20–27% amino acid identity between SARS-CoV S protein and other coronavirus family members [3], [4]. Similar to other coronaviruses, SARS-CoV S glycoprotein contains two non-covalently associated subunits (S1 and S2) functionally. It is believed that binding of S1 subunit to its cellular receptor induces conformational changes in S2, leading to the fusion between viral and cellular membranes, and viral entry [5]. In addition, SARS-CoV S glycoprotein has been proved an important antigen and is able to induce the generation of neutralizing antibody [6].
RNA interference (RNAi) is a cellular process in that double-stranded RNA (dsRNA) molecules of 19–23 nucleotides (nt) can silence targeted genes through sequence-specific cleavage of the cognate RNA transcript. The phenomenon was originally discovered in Caenorhabditis elegans in 1998 [7] but has since been observed in numerous organisms including plant, fungus, Drosophila, seaweed, protozoan, and mammal [8], [9], [10], [11], [12], [13]. The RNAi mechanism appears to be mediated by small dsRNA intermediates. The parent, larger dsRNA is processed by ribonuclease III-like enzymes into smaller fragments in vivo, and the resulting small interfering RNAs (siRNAs) direct posttranscriptional, but pretranslational, degradation of the targeted mRNA. RNAi is becoming a powerful tool to investigate gene function through specific suppression of a particular mRNA and has been employed in therapeutic studies of human diseases including cancer, neurogenerative diseases, and viral infectious diseases [14], [15]. Recently, it has also been used in the study of SARS-CoV [16]. To explore the possibility of silencing SARS-CoV S gene by RNAi technique, we cloned and expressed SARS-CoV spike gene, which encodes N-terminal amino acid residues 1–690 and contains the identified receptor-binding domain (RBD). The in vitro transcribed siRNAs were then introduced into HEK 293T cells expressing SARS-CoV S glycoprotein. Our results demonstrated that the codon-optimized spike gene obviously improved S glycoprotein expression in cultured cells. The specific siRNAs corresponding to SARS-CoV spike gene specifically degraded S mRNA and significantly inhibited S glycoprotein expression.
Materials and methods
Construction of plasmids. Two DNA fragments, corresponding to nt 1–1720 and 1626–2934 of SARS-CoV spike gene, were kindly provided by Prof. Z.H. Yuan (Molecular Virology Laboratory, Fudan University, Shanghai). The DNA fragment encoding N-terminal amino acid residues 1–690 of S glycoprotein, flanked by the ‘Kozak’ consensus sequence, was amplified from the two DNA fragments with the approach of splice overlap extension-polymerase chain reaction (SOE-PCR), and inserted into the multi-cloning site of a eukaryotic expression vector pEGFP-N1 (Clonetech) between its SalI and PstI sites. The resulting plasmid was named pEGFP-S, in which enhanced green fluorescence protein (EGFP) gene was located downstream of S gene. Identity of S gene to the published sequence (GenBank Accession No. AY278554) was confirmed by sequencing (Bioasia, Shanghai). Some of the nucleotides encoding the amino acid residues 1–110 of S glycoprotein in the plasmid pEGFP-S were optimized utilizing mammalian preferred codons, creating the plasmid pEGFP-optS.
Design and transcription of siRNAs. The siRNAs corresponding to SARS-CoV spike gene were designed according to Ambion’s siRNA design guidelines. Scramble siRNA [17] and EGFP siRNA [18] were used as negative and positive controls for silencing, respectively. All sequences of the siRNAs were BLAST searched in the National Center for Biotechnology Information’s (NCBI) “search for short nearly exact matches” mode against all human sequences deposited in the GenBank and RefSeq databases, and were not found to have significant homology to genes other than the targets. The template deoxynucleotides (Table 1 ) used for siRNA transcription were synthesized by Bioasia, Shanghai. The oligonucleotide directed production of siRNAs with T7 RNA polymerase has been described previously [18]. For each transcription reaction, 300 μM of each oligonucleotide template and T7 promoter primer were mixed and denaturalized by heating at 95 °C for 2 min. The mixture was then added with 10× Klenow reaction buffer, dNTP mix (Promega, USA), Exo-Klenow (TaKaRa, Dalian), and nuclease-free water, and incubated at 37 °C for 30 min. The in vitro transcription was performed in 20 μl transcription mix: 6 μl hybridization solution, 4 μl of 5× T7 reaction buffer, 6 μl rNTP mix, 2 μl T7 RNA polymerase, and 2 μl nuclease-free water. After incubation at 37 °C for 2 h, sense and antisense RNAs generated in separate reactions were annealed by mixing both crude transcription reactions and incubating at 37 °C overnight. The concentration of the generated dsRNA was measured by the absorbance at 260 nm in a BioPhotometer (Eppendorf, Germany). S1 nuclease and RNase-free DNase I (TaKaRa, Dalian) were added to final concentrations of 10 and 1 U/μg siRNA, respectively, for the digestion of ssRNA and dsDNA. SiRNAs were assessed by RNA gel electrophoresis on 2% agarose.
Table 1.
Sequences of template deoxynucleotides for siRNAs used for target genes
| Gene targets | Sequences |
|---|---|
| T7 promoter | 5′-TAATACGACTCACTATAGGAGACAGG-3′ |
| EGFP-siRNA | Antisense: 5′-AAGCTGACCCTGAAGTTCATCCCTGTCTC-3′ |
| Sense: 5′-AAGATGAACTTCAGGGTCAGCCCTGTCTC-3′ | |
| Scramble siRNA | Antisense: 5′-AACAAGTCTCGTATGTAGTGGCCTGTCTC-3′ |
| Sense: 5′-AACCACTACATACGAGACTTGCCTGTCTC-3′ | |
| S-siRNA1 | Antisense: 5′-AAGAGCTTTGAGATTGACAAACCTGTCTC-3′ |
| Sense: 5′-AATTTGTCAATCTCAAAGCTCCCTGTCTC-3′ | |
| S-siRNA2 | Antisense: 5′-AACCCTTTCTTTGCTGTTTCTCCTGTCTC-3′ |
| Sense: 5′-AAAGAAACAGCAAAGAAAGGGCCTGTCTC-3′ |
Note. The 8 nucleotide (nt) underlined sequences of template deoxynucleotides for each siRNA are complemented with those of T7 promoter primer. S-siRNA1 and S-siRNA2 target nt 403–423 and nt 871–891 of SARS-CoV spike gene, respectively.
Purification of siRNAs. The in vitro transcribed siRNA was added with one volume of TE-saturated (pH 4.5) phenol:chloroform:isoamyl alcohol (25:24:1), and spun at 13,000 rpm in a microcentrifuge for 10 min. The upper, aqueous phase was collected to a fresh tube, mixed with one volume of chloroform:isoamyl alcohol (24:1), and centrifuged at 13,000 rpm for 10 min. The upper, aqueous phase was then mixed with two volumes of ethanol and 0.1 volume of 3 mmol/L sodium acetate (pH 5.2) and placed on ice for 30 min, followed by centrifugation at 13,000 rpm for 10 min. The pellet was washed with 1 ml of 70% ethanol and suspended in nuclease-free water for further use. The purity and integrity of siRNAs were checked on an agarose gel.
Cell culture and transfection. Human embryonic kidney cell line (HEK) 293T cells were grown at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma) containing 10% heat-inactivated fetal bovine serum (Gibco-BRL, USA) supplemented with l-glutamine (1 mM), streptomycin (100 μg/ml), and penicillin (100 U/ml). Twenty-four hours prior to transfection, the cells were seeded into 24-well plates at a density of 0.5–2 × 105 cells/well with fresh medium (500 μl/well) without antibiotics. For the transfection of adherent HEK 293T cells, a total of 0.8 μg of plasmid DNA or/and 2 μl of siRNA mixed with Lipofectamine 2000 (Invitrogen, CA, USA) were used according to the manufacturer’s instructions. The cells were incubated at 37 °C for various time lengths for gene transcription and expression. The expression of S-EGFP fusion protein was observed directly under an inverted fluorescence microscope.
Fluorescence and flow cytometry analysis. Expression of S-EGFP in transfected cells was examined with fluorescence microscope (Olympus CK40, Japan) at 24, 48, 72, and 96 h after transfection. For flow cytometry analysis of S-EGFP expression, the cells were harvested at 48 h posttransfection and digested with 0.25% trypsin, washed with PBS twice, and then resuspended in PBS to measure the fluorescence using a Becton–Dickinson FACScan flow cytometer with filters (emission, 507 nm; excitation, 488 nm). Samples (about 106 cells each) were counted and analyzed with CellQuest software, using non-transfected HEK 293T cells as control. The values were calculated as the percentage of the cell population that exceeded the fluorescence intensity of the control cells and the mean fluorescence intensity of this population.
Reverse transcription-PCR and real-time quantitative PCR. Total RNA from the transfected or non-transfected cells was extracted at 48 h posttransfection using RNAex Reagent (Watson, Shanghai) and digested with RNase-free DNase I (TaKaRa, Dalian). One microgram of the RNA was then reverse transcribed into cDNA with oligo(dT)15 and the avian myeloblastosis virus (AMV) reverse transcriptase XL (TaKaRa, Dalian) according to manufacturer’s recommendations. Reactions with no reverse transcriptase added were performed in parallel with most experiments and yielded no PCR products. 1 μM of forward primer (5′-CCCATGGGTACACAGACACA-3′) and reverse primer (5′-AATAGGCTGCAGCTGACGTG-3′) for SARS-CoV spike gene was used for semi-quantitative reverse transcription (RT)-PCR with 20 cycles of denaturation (94 °C, 55 s), annealing (56 °C, 55 s), and extension (72 °C, 1 min). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, the primers (forward primer, 5′-TGGGCTACACTGAGCACCAG-3′; reverse primer, 5′-AAGTGGTCGTTGAGGGCAAT-3′) were synthesized based on the human GAPDH mRNA sequence (GenBank Accession No. BC013310). Its reaction conditions were 25 cycles of denaturation (94 °C, 50 s), annealing (60 °C, 30 s), and extension (72 °C, 30 s). Then real-time quantitative PCR (Lightcycler, Roche) was performed as described [19] using SYBR Premix Ex Taq (TaKaRa, Dalian) with the following exceptions. Briefly, reactions were carried out in 20 μl volumes containing 2 μl reverse transcription product. To quantitate SARS-CoV spike gene transcript levels, dilutions of cDNA from the HEK 293T cells transfected with pEGFP-optS were always run in parallel with cDNA from the transfected HEK 293T cells for use as standard curve (dilutions ranged from 106–100 copies of each plasmid). To perform analysis of relative expression of SARS-CoV spike gene using real-time PCR, we adopted the relative quantitative method [20], by normalizing to GAPDH expression levels. Then the percentages of SARS-CoV spike mRNA in siRNAs co-transfected cells relative to that in mock-transfected cells were calculated. The primers for GAPDH were the same as described above. The primers used for the amplification of SARS-CoV spike gene were 5′-TCTGATGCCTTTTCGCTTGA-3′ (forward primer) and 5′-GTGCCCCAAATGTCTTGAGC-3′ (reverse primer).
Western blotting. The transfected or non-transfected cells were harvested and lysed with lysis buffer (50 mM Tris–Cl, pH 8.0, 150 mM NaCl, 0.02% sodium azide, 1% Triton X-100, 1 μg/ml aprotinin, and 100 μg/ml PMSF). Equal amounts of total proteins were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and electrophoretically transferred to nitrocellulose membrane following the protocol suggested by the manufacturer (Bio-Rad, CA). After blocking non-specific-binding sites with 5% non-fat milk, the membrane was incubated with primary antibodies at 4 °C overnight. The primary antibodies used in this experiment were: anti-GFP mouse monoclonal antibody (Santa Cruz, 1:500 dilution) and anti-β-actin goat monoclonal antibody (Santa Cruz, 1:500 dilution). After washing, the blot was incubated with alkaline phosphatase-conjugated goat anti-mouse IgG and horseradish peroxidase-conjugated anti-goat IgG, respectively. Immunoreactive bands were visualized with the 5-bromo-4-chioro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NBT) substrate (Sino-American, Shanghai) and the luminol/enhancer chemiluminescent substrate (ECL) kit (PFBIO, Shanghai), respectively.
Results and discussion
Expression of SARS-CoV S glycoprotein in cultured cells
SARS-CoV spike gene was cloned into pEGFP-N1 with a C-terminal EGFP tag that could be used to monitor protein expression. At 48 h after the transfection, very mild fluorescence was detected in the pEGFP-S transfected HEK 293T cells, while strong fluorescence was observed in the pEGFP-optS transfected cells (Fig. 1 A), which was repeated at least three times but similar results were obtained. To determine the levels of S-EGFP fusion protein expressed in transfected cells, we used convalescent SARS patient sera to detect the fusion protein but obtained very faint bands (data not shown). Therefore, we alternatively employed anti-GFP monoclonal antibody to detect it. Western blot analysis of the cell lysates showed that the S-EGFP fusion protein was expressed at high level in pEGFP-optS transfected cells but at low level in pEGFP-S transfected cells (Fig. 1B). However, the molecular weight of expressed S-EGFP is larger than the predicted one (about 82 kDa) based on amino acid composition. This may represent glycosylated protein which resulted from posttranslational modification by the eukaryotic expression system. The only difference between the two plasmids was that the plasmid pEGFP-optS contained codon-optimized spike gene utilizing mammalian preferred codons. The data indicated that replacement of codons in the spike gene with those frequently used in mammalian increased S-EGFP fusion protein expression in cultured cells, supporting the hypothesis that the observed differences in efficiency of expression of modified and unmodified spike genes were related to codon usage and GC content, as shown in other studies [21], [22]. The underlying mechanisms remain to be elucidated but it is possible that partial optimized codon contained in the spike gene could much more efficiently initiate gene transcription and improve efficiency of gene translation.
Fig. 1.
Differences in the efficiency of expression of modified and unmodified SARS-CoV spike genes. (A) Enhanced fluorescence imaging in codon-optimized SARS-CoV spike gene transfected HEK 293T cells (left panel). (B) Western blot analysis of S-EGFP fusion protein expression in the HEK 293T cells transfected with pEGFP-optS or pEGFP-S.
After all, this transient expression system, transfected with plasmid pEGFP-optS, effectively expresses SARS-CoV S glycoprotein and is suitable for use in the silencing of SARS-CoV S gene expression by siRNA transcribed in vitro.
In vitro transcription of siRNAs
To silence the expression of SARS-CoV S glycoprotein using RNAi technology, we adopted transcription in vitro with T7 RNA polymerase to acquire siRNAs. The sense and antisense of siRNA templates were separately transcribed in vitro with T7 RNA polymerase and annealed to form double-stranded siRNA as described in Materials and methods. The 5′ overhanging leader sequence of the generated dsRNA and DNA template remainders were digested with single-strand specific nuclease (S1 nuclease) and RNase-free DNase I, respectively. The double-stranded siRNAs of 21 nt in length were obtained and found intact as shown in Fig. 2 .
Fig. 2.
Purity and integrity of the siRNAs transcribed in vitro. 1. EGFP-specific siRNA. 2. Scramble siRNA. 3. SARS-CoV S-siRNA1. 4. SARS-CoV S-siRNA2.
In mammalian cells, dsRNA duplexes >30 nt in size lead to the induction of interferon response and global RNA degradation through the activation of RNase L, while duplexes of 21 nt synthesized in vitro induce RNAi after being introduced into human and mouse cell lines. This effect of gene silencing induced by siRNA (<30 nt) was sequence specific without the activation of RNase L [23], [24]. Although siRNAs can be acquired through several methods [25], [26], [27], [28], [29], [30], [31], transcription in vitro by T7 polymerase adopted here is more convenient and reliable than others.
Inhibition of spike gene transcription by siRNAs
To examine RNA levels of SARS-CoV spike gene in siRNAs co-transfected cells, semi-quantitative RT-PCR was performed. The data showed that EGFP-siRNA, S-siRNA1, and S-siRNA2 reduced the accumulation of S mRNA (Fig. 3 A, upper panel, lane 3, 5, and 6 compared to lane 2), and scramble siRNA almost had no effect on S mRNA (Fig. 3A, upper panel, lane 4 compared to lane 2) while GAPDH mRNA was not effected by the four siRNAs (Fig. 3A, lower panel).
Fig. 3.
Reduced SARS-CoV spike transcripts in S-specific siRNA transfected HEK 293T cells. (A) RT-PCR was performed to show the degradation of S mRNA in S-specific siRNA transfected cells. RT-PCR products for the spike gene and the internal control, GAPDH, are shown in the figure. (B) Real-time quantitative PCR was performed for spike and GAPDH RNA content on each sample. The assay shows the percentage of SARS-CoV spike mRNA in siRNA transfected HEK 293T cells relative to that in mock-transfected cells after being normalized to GAPDH mRNA.
To more accurately quantitate RNA levels of SARS-CoV spike gene, we performed real-time quantitative PCR using primers specific to spike gene and GAPDH. Dilutions of cDNA from the pEGFP-optS transfected cells were run for use as standard curve. Based on the standard curve generated, the relative expression of SARS-CoV spike gene in each RNA transcript was obtained by normalizing to GAPDH expression levels, and then the percentages of SARS-CoV spike mRNA in siRNAs co-transfected cells relative to that in mock-transfected cells were calculated. The results of relative quantitative analysis revealed that the transcript levels of SARS-CoV spike gene at 48 h posttransfection were decreased about 10- and 9-fold in cells transfected with SARS-CoV S-siRNA1 and S-siRNA2, respectively. Correspondingly, spike transcript levels were decreased about 16-fold in cells transfected with EGFP-siRNA but not significantly changed in cells transfected with scramble siRNA (Fig. 3B).
RT-PCR and real-time quantitative PCR analysis showed that SARS-CoV spike transcript level in S-siRNA1, S-siRNA2 or EGFP-siRNA transfected HEK 293T cells decreased obviously, whereas no significant inhibition of SARS-CoV spike transcript level was observed in scramble siRNA transfected cells (Fig. 3). Whether RNAi spreading, as demonstrated in plants and C. elegans [32], [33], has occurred in this experiment required further experiments to address. The data above suggested that the effect of gene silencing induced by siRNA should be sequence specific and entire open reading frame (ORF) based. The siRNAs transcribed in vitro can effectively down-regulate SARS-CoV S RNA levels in either EGFP- or S-specific siRNA transfected HEK 293T cells, corresponding to the reported mechanism that siRNAs degraded target mRNA at RNA level instead of at DNA level.
Silencing of SARS-CoV S glycoprotein expression by siRNA
To determine whether the synthesized siRNA could effectively silence SARS-CoV S glycoprotein expression in cultured cells, we co-transfected HEK 293T cells with plasmid pEGFP-optS and the various siRNAs.
The cells were examined microscopically at 48 h posttransfection for green fluorescence. As can be seen in Fig. 4 , the fluorescence imaging was much stronger in the HEK 293T cells transfected with scramble siRNA than in those transfected with SARS-CoV S-siRNA1 and S-siRNA2. Very mild green fluorescence was observed in EGFP-siRNA transfected cells. The fluorescence intensity of HEK 293T cells co-transfected with scramble siRNA showed no significant difference compared with pEGFP-optS transfected cells (Figs. 4, upper panel A and C). The lower panels represent the corresponding image observed by light microscope. Specific silencing of the green fluorescence was confirmed at least in three independent experiments.
Fig. 4.
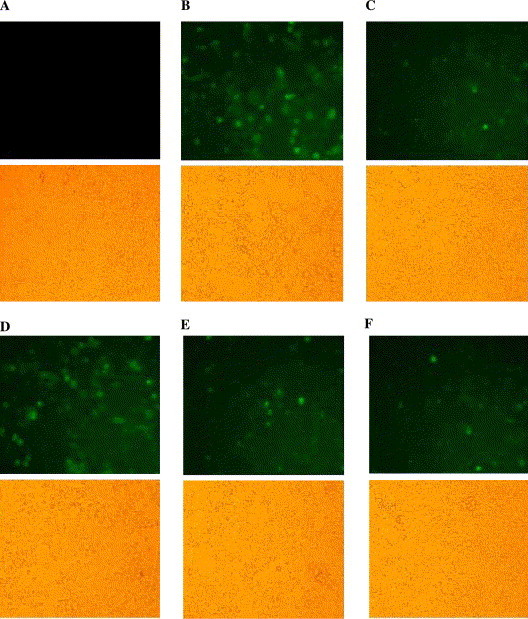
Effect of siRNAs on the expression of S-EGFP in HEK 293T cells. (A) HEK 293T cells. (B) pEGFP-optS transfected. (C) pEGFP-optS and EGFP siRNA co-transfected. (D) pEGFP-optS and scramble siRNA co-transfected. (E) pEGFP-optS and S-siRNA1 co-transfected. (F) pEGFP-optS and S-siRNA2 co-transfected. Upper panels represent the cell fluorescence images recorded at 48 h posttransfection. Lower panels represent the light microscopic view of cells in the same field. Specific silencing of the S-EGFP fusion protein expression was confirmed in three independent experiments.
To further examine the silencing effect of SARS-CoV S-siRNAs, we collected cells and analyzed with fluorescence-activated cell sorting (FACS) 48 h after transfection for EGFP expression with CellQuest software, using non-transfected HEK 293T cells as control. As shown in Fig. 5 , compared with the cells transfected with plasmid pEGFP-optS alone, the cells co-transfected with pEGFP-optS and scramble siRNA gave no significant reduction of EGFP expression, whereas the EGFP-siRNA gave an about 3.7- and 3.8-fold reduction in percentage of fluorescence cell population and mean fluorescence intensity, respectively. Percentage of fluorescence cell population and mean fluorescence intensity reduced about 1.6- and 3.3-fold for S-siRNA1 and 5.8- and 6.1-fold for S-siRNA2. This result was in keeping with fluorescence imaging in Fig. 4.
Fig. 5.
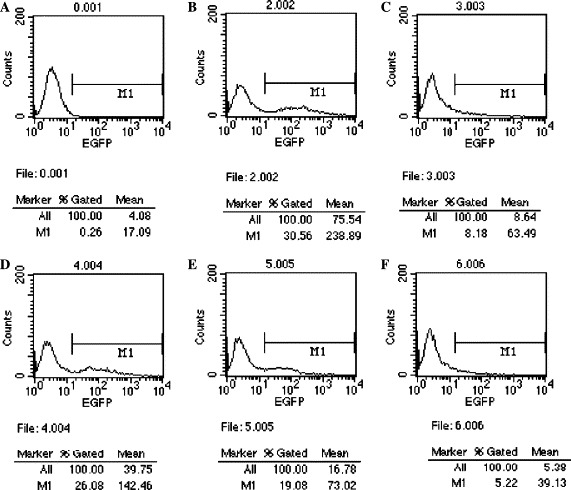
Flow cytometry analysis of EGFP expression in HEK 293T cells. (A–F) show the results from HEK 293T cells control, cells transfected with pEGFP-optS, cells co-transfected with pEGFP-optS and EGFP siRNA, cells co-transfected with pEGFP-optS and scramble siRNA, cells co-transfected with pEGFP-optS and S-siRNA1 and cells co-transfected with pEGFP-optS and S-siRNA2, respectively. At 48 h after transfection, cells were analyzed for EGFP expression by flow cytometry. The percentage of the cell population that exceeded the fluorescence intensity of the control cells and the mean fluorescence intensity of this population were calculated.
To explore the mechanism of siRNA in transfected cells, we examined transfected cells for S-EGFP fusion protein levels in the presence or absence of siRNAs. Western blot analysis showed that compared with the cells transfected with plasmid pEGFP-optS alone, S-EGFP fusion protein expression was reduced in cells co-transfected with EGFP-siRNA, S-siRNA1 or S-siRNA2, but not reduced in cells co-transfected with pEGFP-optS and scramble siRNA (Fig. 6 , upper panel). Interestingly, a little difference in the level of S-EGFP fusion protein expression in S-siRNA1 and S-siRNA2 transfected cells was detected, indicating that the effect of silencing S-EGFP fusion protein expression by S-siRNA2 was better than S-siRNA1, which was consistent with the previous data in Fig. 4, Fig. 5. As a loading control, β-actin was not affected by any of the four siRNAs tested (Fig. 6, lower panel). These data indicated that siRNAs silenced SARS-CoV S glycoprotein expression through blocking the accumulation of S mRNA.
Fig. 6.
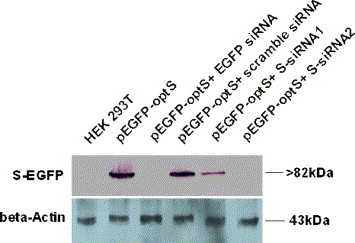
Effect of siRNA on S-EGFP protein expression in HEK 293T cells. Western blot analysis was performed on equal amounts of proteins harvested from mock- or siRNA co-transfected HEK 293T cells at 48 h posttransfection by using GFP- and GAPDH-specific antibodies described in Materials and methods. β-Actin was used as a loading control.
SARS-CoV S glycoprotein initiates entry of SARS-CoV into susceptible cells mediated by combining with cell surface receptors followed by conformational changes leading to membrane fusion [34]. Recently, the angiotensin-converting enzyme 2 (ACE2) was identified as a functional receptor for SARS-CoV [35] and its binding site on the SARS-CoV S glycoprotein was localized between amino acid residues 303 and 537 [36]. Therefore, to block the combination of S glycoprotein with receptor efficiently is crucial to prevent the virus infection. As a cellular defense mechanism, RNAi has been employed in anti-virus infections in viruses such as human immunodeficiency virus and hepatitis C/B virus [37] and inhibition of viral replication in host cells [38]. Zhang et al. [16] introduced vector-mediated RNAi technology into the study of SARS-CoV infection, and found that the siRNAs generated from the DNA vector blocked the expression of SARS-CoV S gene and inhibited the replication of SARS-CoV in the infected cells. Their work enlightened us to explore the possibility of silencing SARS-CoV S glycoprotein expression by siRNA transcribed in vitro by T7 RNA polymerase and blocking its combination with cell surface receptor.
In the present work, we cloned wild and codon-optimized SARS-CoV spike gene encoding N-terminal amino acid residues 1–690, which contained the identified receptor-binding domain (RBD), and found that the codon-optimized spike gene can improve the expression of S glycoprotein in cultured cells. Our results demonstrated that the siRNAs transcribed in vitro were effective, and S-siRNA1 and S-siRNA2 can both silence SARS-CoV S glycoprotein expression, which provided evidence for the further study of SARS-CoV infection of cells mediated by S glycoprotein. Studies on the interaction of SARS-CoV S glycoprotein and its receptor using RNAi technology are in progress. Our findings in this work could help in the development of novel SARS-CoV therapeutic agent and provide a new strategy for the prevention and treatment of SARS-CoV infection in humans.
Acknowledgment
This work was financially supported by the National Key Basic Research and Development Program of China (973 Program No. 2003CB514129).
References
- 1.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. SARS study group. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., vanden Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Zelus B.D., Schickli J.H., Blau D.M., Weiss S.R., Holmes K.V. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37 °C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 2003;77:830–840. doi: 10.1128/JVI.77.2.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruan Y.J., Wei C.L., Ee A.L., Vega V.B., Thoreau H., Su S.T., Chia J.M., Ng P., Chiu K.P., Lim L., Zhang T., Peng C.K., Lin E.O., Lee N.M., Yee S.L., Ng L.F., Chee R.E., Stanton L.W., Long P.M., Liu E.T. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990;8:340–344. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- 9.Romano N., Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein E., Denli A.M., Hannon G.J. The rest is silence. RNA. 2001;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- 11.Misquitta L., Paterson B.M. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc. Natl. Acad. Sci. USA. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plasterk R.H. RNA silencing: the genome’s immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 13.Zamore P.D. Ancient pathways programmed by small RNAs. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y. Mammalian RNAi for the masses. Trends Genet. 2003;19:9–12. doi: 10.1016/s0168-9525(02)00005-7. [DOI] [PubMed] [Google Scholar]
- 15.Dy kxhoorn D.M., Novina C.D., Sharp P.A. Killing the message short RNAs that silence gene expression. Nat. Rev. Mol. Cell. Biol. 2003;4:457–465. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Li T., Fu L., Yu C., Li Y., Xu X., Wang Y., Ning H., Zhang S., Chen W., Babiuk L.A., Chang Z. Silencing SARS-CoV spike protein expression in cultured cells by RNA interference. FEBS Lett. 2004;560:141–146. doi: 10.1016/S0014-5793(04)00087-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Wilson J.A., Jayasena S., Khvorova A., Sabatinos S., Rodrigue-Gervais I.G., Arya S., Sarangi F., Harris-Brandts M., Beaulieu S., Richardson C.D. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao M.M., Ren H., Pan X., Pan W., Qi Z.T. Inhibition of EGFP expression by siRNA in EGFP-stably expressing Huh-7 cells. J. Virol. Methods. 2004;119:189–194. doi: 10.1016/j.jviromet.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Rajeevan M.S., Vernon S.D., Taysavang N., Unger E.R. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J. Mol. Diagn. 2001;3:26–31. doi: 10.1016/S1525-1578(10)60646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heid C.A., Stevens J., Livak K.J., Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J., Liu W.J., Peng S.W., Sun X.Y., Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol. 1999;73:4972–4982. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutton D.W., Havstad P.K., Kemp J.D. Synthetic cryIIIA gene from Bacillus thuringiensis improved for high expression in plants. Transgenic Res. 1992;1:228–236. doi: 10.1007/BF02524753. [DOI] [PubMed] [Google Scholar]
- 23.Stark G.R., Kerr I.M., Williams B.R., Silverman R.H., Schreiber R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 24.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 25.Donze O., Picard D. RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res. 2002;30:e46. doi: 10.1093/nar/30.10.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyagishi M., Taira K. Development and application of siRNA expression vector. Nucleic Acids Res. 2002;(Suppl.):113–114. doi: 10.1093/nass/2.1.113. [DOI] [PubMed] [Google Scholar]
- 27.Yang D., Buchholz F., Huang Z., Goga A., Chen C.Y., Brodsky F.M., Bishop J.M. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svoboda P., Stein P., Schultz R.M. RNAi in mouse oocytes and preimplantation embryos: effectiveness of hairpin dsRNA. Biochem. Biophys. Res. Commun. 2001;87:1099–1104. doi: 10.1006/bbrc.2001.5707. [DOI] [PubMed] [Google Scholar]
- 29.Sui G., Soohoo C., Affar el B., Gay F., Shi Y., Forrester W.C., Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lois C., Hong E.J., Pease S., Brown E.J., Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 31.Rubinson D.A., Dillon C.P., Kwiatkowski A.V., Sievers C., Yang L., Kopinja J., Rooney D.L., Ihrig M.M., McManus M.T., Gertler F.B., Scott M.L., Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 32.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 33.Palauqui J.C., Elmayan T., Pollien J.M., Vaucheret H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimitrov D.S. Cell biology of virus entry. Cell. 2000;101:697–702. doi: 10.1016/s0092-8674(00)80882-x. [DOI] [PubMed] [Google Scholar]
- 35.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S-glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marathe R., Anandalakshmi R., Smith T.H., Pruss G.J., Vance V.B. RNA viruses as inducers, suppressors and targets of posttranscriptional gene silencing. Plant Mol. Biol. 2000;43:295–306. doi: 10.1023/a:1006456000564. [DOI] [PubMed] [Google Scholar]
- 38.Kapadia S.B., Brideau-Andersen A., Chisari F.V. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]


