Abstract
The influenza A virus (IAV) causes seasonal epidemics and occasional but devastating pandemics, which are of a major public health concern. Although several antiviral drugs are currently available, there is an urgent need to develop novel antiviral therapies with different mechanisms of action due to emergence of drug resistance. In this study, two related compounds, chebulagic acid (CHLA) and chebulinic acid (CHLI), were identified as novel inhibitors against IAV replication. A reporter virus-based infection assay demonstrated that CHLA and CHLI exhibit no inhibitory effect on IAV entry or RNA replication during the virus replication cycle. Results of viral release inhibition assay and neuraminidase (NA) inhibition assay indicated that CHLA and CHLI exert their inhibitory effect on the NA-mediated viral release. Moreover, oseltamivir-resistance mutation NA/H274Y of NA is susceptible to CHLA or CHLI, suggesting a different mechanism of action for CHLA and CHLI. In summary, CHLA and CHLI are promising new NA inhibitors that may be further developed as novel antivirals against IAVs.
Keywords: influenza A virus, chebulinic acid, chebulagic acid, neuraminidase, neuraminidase inhibitor
Introduction
Influenza vaccines and antiviral drugs are effective in preventing infection or ameliorating disease severity (Vasileiou et al., 2017). However, influenza infection, as an acute respiratory disease caused by seasonal outbreaks and, periodically, pandemics of influenza viruses and accounting for up to 650,000 annual deaths globally (World Health Organization [WHO], 2018), remains a serious public health concern. This is partly due to the fact that current influenza vaccines can target only selected strains based on annual surveillance and prediction, which does not always match the circulating strains, leading to a sharp drop in vaccine efficacy (de Jong et al., 2000; Nachbagauer and Krammer, 2017). Moreover, emergence of drug resistance strains of influenza A viruses (IAVs) reduces the effectiveness of the current anti-influenza therapies. The three clinically available classes of antivirals for treatment and prevention of influenza infections are the viral ion channel M2 blockers (amantadine and rimantadine), neuraminidase (NA) inhibitors (oseltamivir, zanamivir, peramivir, and laninamivir), and most recently the licensed baloxavir marboxil, which is an oral cap-dependent endonuclease inhibitor of influenza virus polymerase inhibitor (Alves Galvao et al., 2014; Alame et al., 2016; Kashiwagi et al., 2016; Heo, 2018; Noshi et al., 2018). However, high levels of resistance to M2 channel blockers and NA inhibitors have emerged, undermining the efficacy of these drugs (Hata et al., 2007; Samson et al., 2013; van der Vries et al., 2013; Lackenby et al., 2018). Moreover, mutations responsible for reduced susceptibility of influenza A/H3N2 virus to baloxavir have been detected already (Takashita et al., 2019). Therefore, the development of antiviral drugs with different mechanisms of action is urgently needed against influenza viruses.
Natural products contain structurally diversified bioactive chemicals, which are valuable sources for new drug discovery. Among the new medicines approved by the U.S. Food and Drug Administration (FDA) between 1981 and 2010, natural products or their direct derivatives account for 34% (Harvey et al., 2015). In the present study, 352 natural product samples were screened for anti-IAV activity using a phenotypic screening approach based on a recombinant IAV expressing Gaussia luciferase (Zhao et al., 2019). The extracts from both unripe and ripe pods of Terminalia chebula Retz. display potent anti-IAV activity. Further, two constitutes of T. chebula, chebulagic acid (CHLA) and chebulinic acid (CHLI), were identified as novel antivirals against IAV. Detailed studies revealed that both compounds specifically block progeny virus release by inhibiting IAV NA activity. Moreover, CHLA and CHLI showed highly inhibitory efficacy against an oseltamivir-resistant IAV strain, suggesting their potential as novel cost-effective NA inhibitors for controlling influenza virus infections.
Materials and Methods
Library of Natural Product Samples
Dispensing granules of 352 natural product samples were purchased from EFONG Pharmaceutical Company (Foshan, Guangdong, China) and arrayed in 96-well plates at a 20-mg/mL stock concentration in DMSO. All of the sample plates were stored at −80°C until use.
Cells, Viruses, and Compounds
Madin–Darby canine kidney (MDCK) epithelial cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Cellgro, Manassas, VA, United States) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, United States), 1000 units/mL of penicillin, and 100 μg/mL of streptomycin (Invitrogen, Carlsbad, CA, United States). Infections were performed in Opti-MEM containing 2 μg/mL N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)–trypsin (Sigma–Aldrich, St. Louis, MO, United States). All cells were grown at 37°C in 5% CO2.
Recombinant reporter virus PR8-PB2-Gluc and influenza strains A/Puerto Rico/8/1934 (H1N1, A/H1N1/PR8) and A/Wyoming/3/03 (H3N2, A/H3N2/NY) were prepared and stocked in our lab as previously described (Zhao et al., 2018; Wang et al., 2019). Oseltamivir-resistant influenza A/H1N1/pdm(09) virus containing NA/H274Y was provided by Beijing CDC, China, influenza A/Brisbane/10/2007(H3N2) was provided by Chinese Academy of Medical Sciences, and influenza B-Yamagata-like and B-Victoria-like strains were provided by Shandong CDC (Jinan, China).
Fifteen chemical components (> 98% purity) from T. chebula were purchased from the National Institutes for Food and Drug Control (Beijing, China).
High-Throughput Screen
The library of natural product samples was screened using a phenotypic screening approach described previously (Zhao et al., 2019). Briefly, MDCK cells were seeded in 96-well plates at a density of 5000 cells/well 24 h before infection. In the presence of natural product samples (final concentration of 25 μg/mL), MDCK cells were challenged with recombinant reporter virus PR8-PB2-Gluc at an MOI of 0.01. Infection was quantified after 36 h of incubation by measuring the luciferase activity with PierceTM Gaussia Luciferase Glow Assay kit (Thermo Fisher, Hillsboro, OR, United States) according to the manufacturer’s instructions. Data were normalized to signals from the negative controls (virus alone with DMSO), and an average of > 90% inhibition for duplicates was applied for picking hits.
The selected active samples were then reformatted into new 96-well plates and tested against PR8-PB2-Gluc at 25 μg/mL in 0.125% DMSO (v/v) to confirm the primary results. Cell cytotoxicity was examined 48 h post-treatment using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI, United States), treated for the antiviral screen.
The confirmed hit samples were twofold serially diluted, respectively, for dose–response analysis, and the IC50 and CC50 values were determined by fitting dose–response curves with a four-parameter logistic regression to the data in GraphPad Prism software (version 5.02, La Jolla, CA, United States).
One-Cycle Infection Inhibition Assay
To determine whether the hit natural product samples or its derivatives were target to viral entry or genome replication steps, a one-cycle infection inhibition assay was carried out using the reporter virus PR8-PB2-Gluc. Briefly, MDCK cells growing in a 96-well plate were infected with PR8-PB2-Gluc at an MOI of 0.1 in the presence of various concentrations of test samples/compounds. After 1 h of incubation, unabsorbed viruses were removed, and the cells were treated with the tested samples. In order to prevent the second round of infection, DMEM containing 10% FBS, instead of Opti-MEM containing 2 μg/mL of TPCK–trypsin, was used during infection to avoid HA cleavage and infectious virus production. After 24 h, infections were quantified by measuring the luciferase activity with PierceTM Gaussia Luciferase Glow Assay kit (Thermo Fisher, Hillsboro, OR, United States). Meanwhile, in the presence of the reporter virus PR8-PB2-Gluc, cell cytotoxicity for the test compounds was determined.
Virus Release Inhibition Assay
To determine whether the hit natural product samples or the derivatives target to the release of progeny viruses, MDCK cells were infected with the reporter virus PR8-PB2-Gluc at an MOI of 0.1. At 20 h post-infection (p.i.), the culture medium was removed, and the cells were washed with PBS three times. Fresh Opti-MEM containing 2 μg/mL of TPCK–trypsin was added and cultured for 2 h in the presence of various concentrations of test samples/compounds. The culture medium was harvested and titrated using luciferase assay.
Neuraminidase (NA) Inhibition Assay
Neuraminidase inhibition assay was performed using Neuraminidase Inhibitors Screening kit (Beyotime Biotechnology, Shanghai, China) according to manufacturer’s instructions. Briefly, NA and various concentrations of test samples/compounds were added to each well of 96-well plates. In order to fully interact between the compounds and NA, the 96-well plates were mixed for 1 min and incubated at 37°C for 2 min. Then fluorescent substrates were added, mixed, and incubated. After incubation for 1 h, the decrease in fluorescence was monitored to reflect NA inhibition efficacy.
Viral Yield Reduction Assay
Viral yield reduction assay was performed as previously described (Zhu et al., 2011). Briefly, MDCK cells growing in 24-well plates were infected with A/H1N1/PR8, A/H3N2/NY, oseltamivir-resistant influenza A/H1N1/pdm(09), A/H3N2/Brisbane, B-Yamagata-like, and B-Victoria-like strains at an MOI of 0.01 with or without various concentrations of test samples/compounds. The culture supernatants were harvested at 24 h p.i., and virus titers (TCID50/mL) in the culture supernatants were determined using MDCK cells.
Results
Extracts of T. chebula Inhibit IAV Replication
To discover novel antiviral actives against IAV, a library consisting of 352 natural product samples was prepared and screened using a phenotypic screening approach based on a reporter influenza A PR8-PB2-Gluc virus (Zhao et al., 2019) in a primary screen and confirmation screen. As a result, 12 hit samples were identified to inhibit IAV infection (Supplementary Table S1).
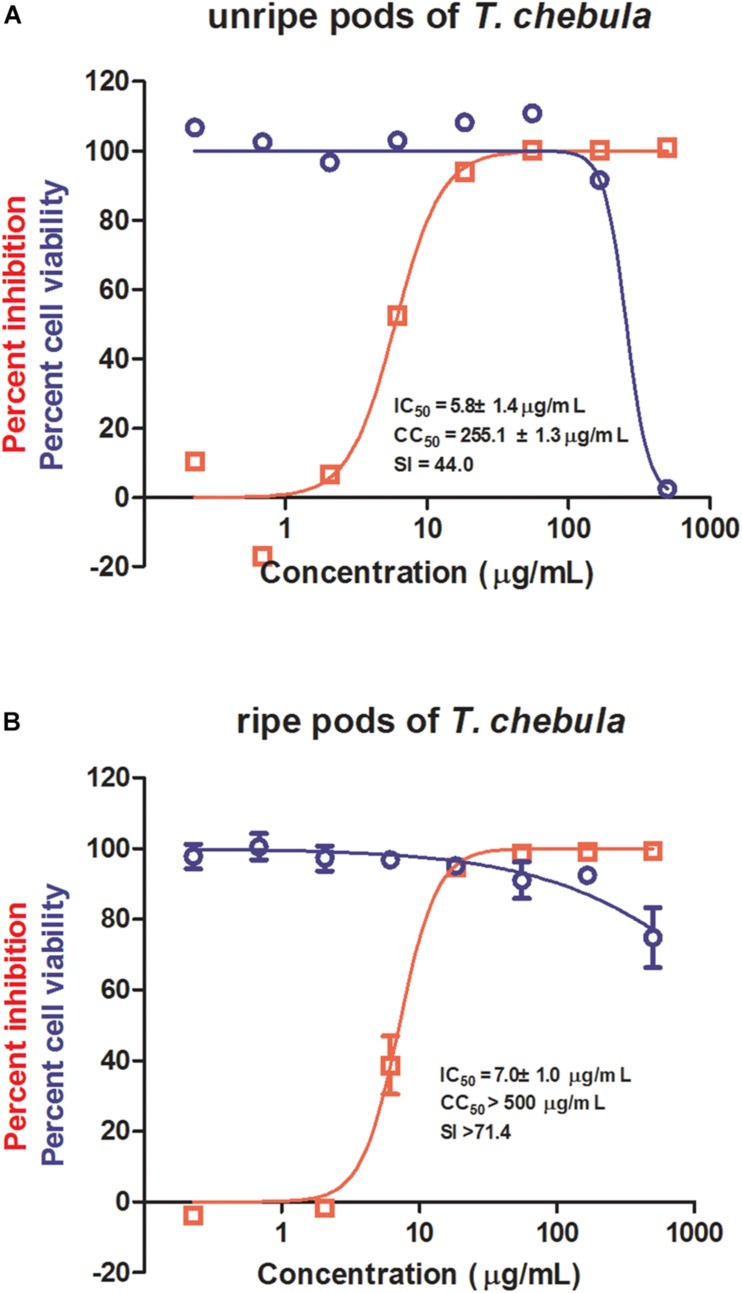
Among the most potent anti-IAV natural product samples, both unripe and ripe pods of T. chebula significantly inhibited IAV replication with IC50s of 5.8 ± 1.4 and 7.0 ± 1.0 μg/mL, respectively (Figures 1A,B). Since unripe pods of T. chebula exhibited a CC50 to MDCK cells of 255.1 ± 1.3 μg/mL, while ripe pods of T. chebula showed no obvious cytotoxicity at a high concentration as 500 μg/mL, it is likely that there is some difference between the compositions of the two natural product sample preparations.
FIGURE 1.
Antiviral activities of unripe and ripe pods of Terminalia chebula on influenza virus. The in vitro dose–response curves against influenza A virus (IAV) (red) and cytotoxicity in Madin–Darby canine kidney (MDCK) cells (blue) of unripe (A) and ripe (B) pods of T. chebula. Data are means ± SD from three independent experiments.
CHLA and CHLI Inhibit IAV Replication
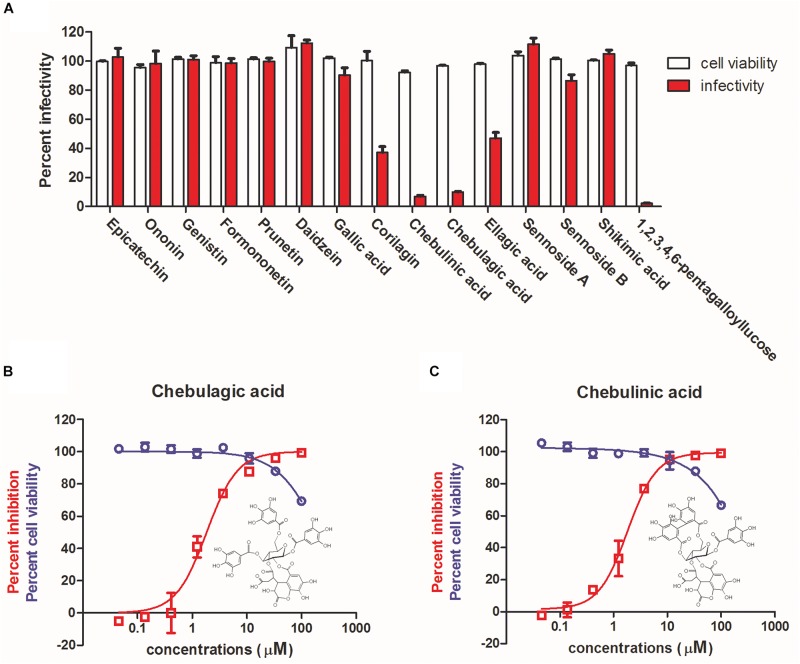
Considering the close relation between unripe and ripe pods of T. chebula, the chemical compositions of the two samples were compared based on spectroscopic data with previously reported literature. Fifteen overlapped constituents were subsequently shortlisted and subjected to antiviral evaluation. As shown in Figure 2A, at a concentration of 12.5 μM, corilagin and ellagic acid showed about 40% inhibition, while CHLA, CHLI, and 1,2,3,4,6-pentagalloyllucose (PGG) showed > 90% inhibition. Since corilagin, ellagic acid, and PGG have been well characterized for their antiviral activity against IAV (Liu et al., 2011; Chen et al., 2015; Zu et al., 2015), CHLA and CHLI, which are structurally related, were chosen for further analysis.
FIGURE 2.
Identification of chebulagic acid (CHLA) and chebulinic acid (CHLI) as novel antiviral actives against influenza virus infection. (A) Antiviral determination of 15 overlapped components of unripe and ripe pods of T. chebula at 12.5 μM. (B) CHLA and (C) CHLI were examined against influenza virus in MDCK cells in vitro for dose–response evaluation (red) and cytotoxicity assay (blue). Insets: molecular structures of (B) CHLA and (C) CHLI. Data are means ± SD from three independent experiments.
High-performance liquid chromatography was performed to further demonstrate that unripe and ripe pods of T. chebula share the constituents of CHLA and CHLI (Supplementary Figure S1), of which the dose–response effects against IAV replication were subsequently determined. As shown in Figures 2B,C, the IC50 values against IAV replication of CHLA and CHLI were 1.36 ± 0.36 and 1.86 ± 0.98 μM, respectively, while the CC50 values of both compounds were greater than 100 μM. These results show that CHLA and CHLI are potent anti-IAV inhibitors.
CHLA and CHLI Do Not Inhibit Virus Entry or RNA Replication of IAV
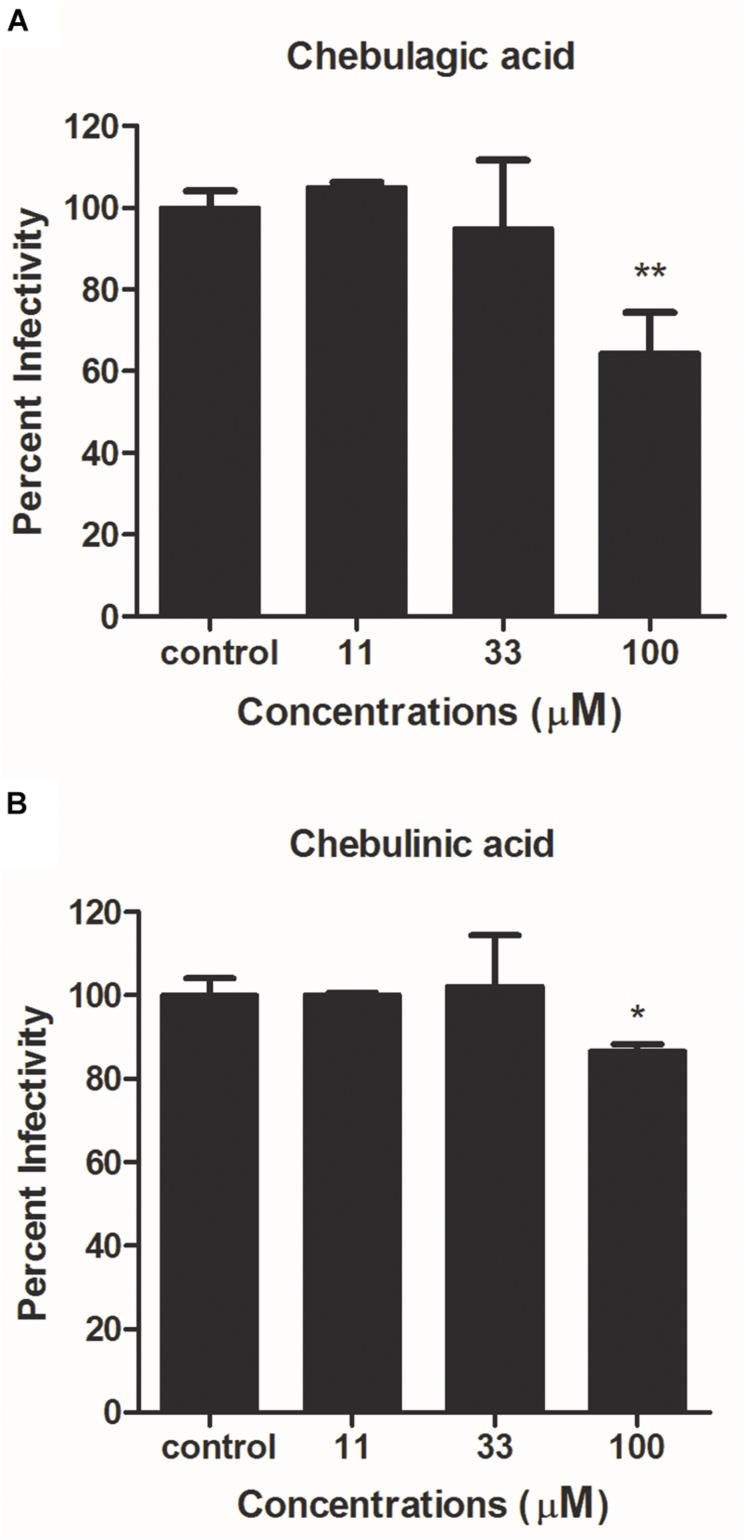
To identify the target(s) and possible mechanism of action for CHLA and CHLI on IAV replication, a reporter IAV-based one-cycle infection assay was performed. Briefly, MDCK cells were infected by the reporter IAV PR8-PB2-Gluc in the absence or presence of test compounds, without the addition of TPCK–trypsin in the culture medium. Without exogenous trypsin-mediated HA cleavage, the progeny viruses are non-infectious and could not initiate a second round of infection. In this context, decreased expression of the reporter luciferase would be expected upon treatment with inhibitors targeting virus entry or RNA replication.
As shown in Figure 3, neither CHLA nor CHLI reduced Gluc expression. Note that the slight decrease in Gluc levels at 100 μM of CHLA and CHLI might be due to cytotoxicity (Supplementary Figure S2). These results suggest that CHLA and CHLI do not interfere with IAV entry or RNA replication.
FIGURE 3.
Reporter luciferase-based one-cycle infection inhibition assay. MDCK cells growing in 96-well plates were infected with reporter influenza PR8-PB2-Gluc virus at an MOI of 0.1, in the absence or presence of increasing concentrations of (A) CHLA and (B) CHLI. Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 4% fetal bovine serum (FBS) were used as culture medium during infection to avoid generation of infectious progeny viruses. At 24 h post infection (p.i.), Gluc expression was determined, and viral infectivities were evaluated. Data are means ± SD from three independent experiments. *p < 0.05; **p < 0.01; Student’s t-test.
CHLA and CHLI Target NA to Block Viral Release
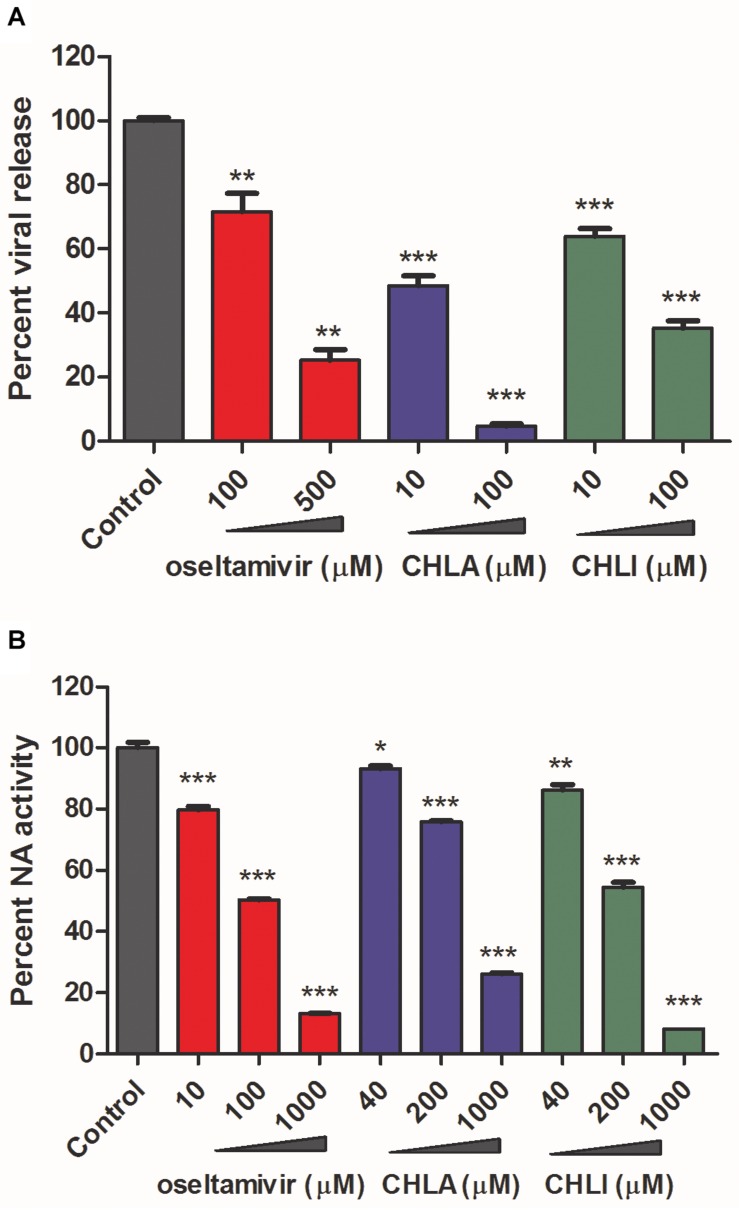
The effects of CHLA and CHLI on viral release were next investigated by a viral release inhibition assay. Briefly, infected cells were washed and cultured in an infection medium for another 2 h, in the absence or presence of different concentrations of test compounds. The progeny viruses were then harvested and titrated. Both CHLA and CHLI inhibited virus release in a dose-dependent manner, like the positive control oseltamivir phosphate (Figure 4A).
FIGURE 4.
CHLA and CHLI block neuraminidase (NA)-mediated virus release. (A) Virus release inhibition assay. MDCK cells were infected with reporter influenza PR8-PB2-Gluc virus at an MOI of 0.1. At 20 h p.i., infected cells were washed three times, and fresh Opti-MEM containing 2 μg/mL of N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)–trypsin was added for another 2-h incubation, in the absence or presence of indicated compounds. The supernatants were harvested, and progeny viruses were titrated by luciferase assay. Data are means ± SD from three independent experiments. **p < 0.01; ***p < 0.001; Student’s t-test. (B) NA inhibition assay. The NA was incubated with fluorimetric substrates in the absence or presence of indicated compounds. The intensity of fluorescence was detected to reflect NA activities. Data are means ± SD from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; Student’s t-test.
It has been well documented that the influenza viral NA activity contributes to progeny virus release from an infected cell surface (Du et al., 2019). We therefore examined whether CHLA and CHLI blocked viral release of IAV by inhibiting viral NA activity. A standard fluorimetric method was used (Li et al., 2016), and the results showed that both CHLA and CHLI inhibited viral NA activity in a dose-dependent manner (Figure 4B).
Together, these results indicate that CHLA and CHLI inhibit IAV replication by preventing the viral NA-mediated progeny virus release.
CHLA and CHLI Are Effective to Inhibit an Oseltamivir-Resistant IAV
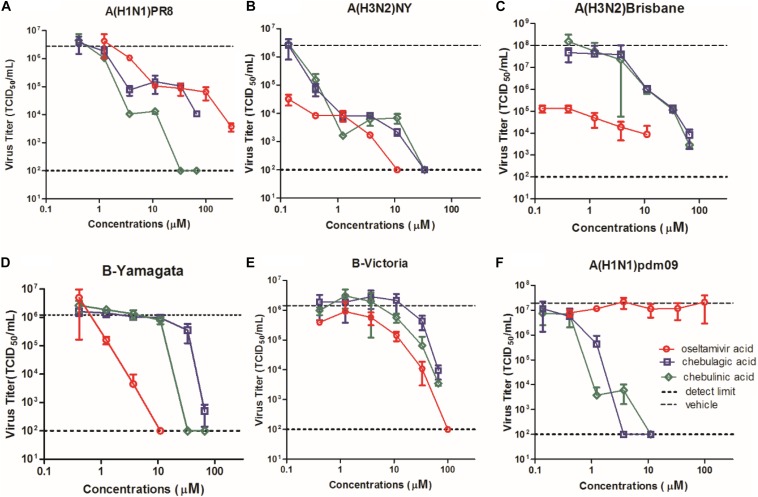
To compare the antiviral potency of CHLA and CHLI with the marketed NA inhibitor oseltamivir carboxylate, a yield reduction assay was performed with six influenza virus strains, including A/H1N1/PR8, A/H3N2/NY, A/H3N2/Brisbane, B-Yamagate, B-Victoria, and an oseltamivir-resistant A/H1N1pdm(09), which contains the NA/H274Y substitution. As a result, CHLA and CHLI showed significant inhibition against all influenza virus strains (Figure 5). Note that the IC90s of CHLA (1.26 μM) and CHLI (1.58 μM) upon A/H1N1/PR8 were lower than that of oseltamivir carboxylate (3.92 μM), suggesting their comparable activities (Figure 5A). While compared to IAVs (Figures 5A–C), IBVs showed less sensitivity to CHLA and CHLI (Figures 5D,E).Interestingly, NA/H274Y in A/H1N1/pdm(09) virus conferred resistance to oseltamivir carboxylate, but the potency of CHLA and CHLI was not affected by the substitution (Figure 5F).
FIGURE 5.
Dose–response curves of CHLA, CHLI, and reference compound oseltamivir acid against virus replication in a yield reduction assay. MDCK cells in 24-well plates were infected with (A) A/H1N1/PR8, (B) A/H3N2/NY, (C) oseltamivir-resistant influenza virus A/H1N1/pdm(09), (D) A/H3N2/Brisbane, (E) B-Yamagata, and (F) B-Victoria at an MOI of 0.01. The culture supernatants were collected at 24 h p.i., and virus titers (TCID50/mL) in the culture supernatants were determined using MDCK cells. Each point represents the mean and standard deviation of three independent experiments.
Discussion
In this study, a phenotypic screening approach was initially used to evaluate 352 natural product samples for activity against IAV infection, and 12 natural product samples were identified as putative hits with high anti-IAV potency. Among these hit natural product samples, unripe and ripe pods of T. chebula displayed the most potent anti-IAV activity.
Unripe and ripe pods of T. chebula are two different medical materials usually prescribed in Traditional Chinese Medicine formula. However, the compositions of unripe pods and ripe pods of T. chebula are highly overlapping, and our study showed that CHLA and CHLI, two overlapped tannin constituents of the two samples, exhibit highly inhibitory effects against IAV replication, with IC50s of 1.36 and 1.86 μM, respectively (Figure 2). Moreover, the SIs of CHLA and CHLI against IAV replication in MDCK cells are > 74 and > 54, respectively, suggesting that the two compounds are good candidates for novel antivirals against IAV (Figure 2).
Previous studies reported that CHLA and CHLI exhibit broad-spectrum antiviral activities by targeting viral glycoprotein–glycosaminoglycan interactions (Lin et al., 2011, 2013; Kesharwani et al., 2017). However, our data demonstrated that CHLA and CHLI do not interfere with influenza virus entry or RNA replication, but act as NA inhibitors targeting virus release.
Neuraminidase is a major surface glycoprotein with a sialidase activity, which contributes to the release of newly formed virions from infected cells and facilitates propagation of the virus (Du et al., 2019). For the purpose of antiviral development, NA is considered one of the major targets (Colman, 1994; Grienke et al., 2012). Currently, four NA inhibitors are available for the prophylaxis and treatment of influenza virus infections, including oseltamivir (Shobugawa et al., 2012), zanamivir (von Itzstein et al., 1993), peramivir (Alame et al., 2016), and laninamivir (Kashiwagi et al., 2016). Among these drugs, oseltamivir remains a first-line therapy since it was approved in 1999. However, resistance to oseltamivir has constantly been reported due to its wide use in clinic, and various mutants with resistance to the other NA inhibitors have also appeared (Samson et al., 2013; van der Vries et al., 2013; Lackenby et al., 2018). Therefore, it is imperative to discover novel NA inhibitors with different structures and mechanisms of action.
There are 11 NA subtypes of IAV, of which N10 and N11 were recently identified in bat IAV genomes (Tong et al., 2012, 2013). The N1–N9 subtypes can be phylogenetically divided into two groups. Group I includes N1, N4, N5, and N8, while group II is comprised of N2, N3, N6, N7, and N9 (Russell et al., 2006). Crystallographic studies show that group 1 NAs (except NA of A/H1N1pdm[09]) possess a large cavity termed 150-cavity adjacent to the catalytic site, but the 150-cavity was not observed in group 2 NAs (Li et al., 2010; Air, 2012). Upon binding of oseltamivir or zanamivir, the open 150-loop of group 1 NAs would adopt a closed conformation (Russell et al., 2006). Besides, inhibitors targeting the 150-cavity may also effectively target group 2 influenza NAs, perhaps by inducing the rigid closed 150-loop of group-2 NAs into a half-open one (Wu et al., 2013). The structure of A/H1N1pdm(09) NA presents a deficient 150-cavity in its crystal structure, conferring resistance to oseltamivir (Li et al., 2010). In addition, another auxiliary binding site adjacent to the sialic acid binding site called “430-cavity” has also been implicated and exploited for the design of new antivirals (Amaro et al., 2007; Swaminathan et al., 2013; Jia et al., 2019).
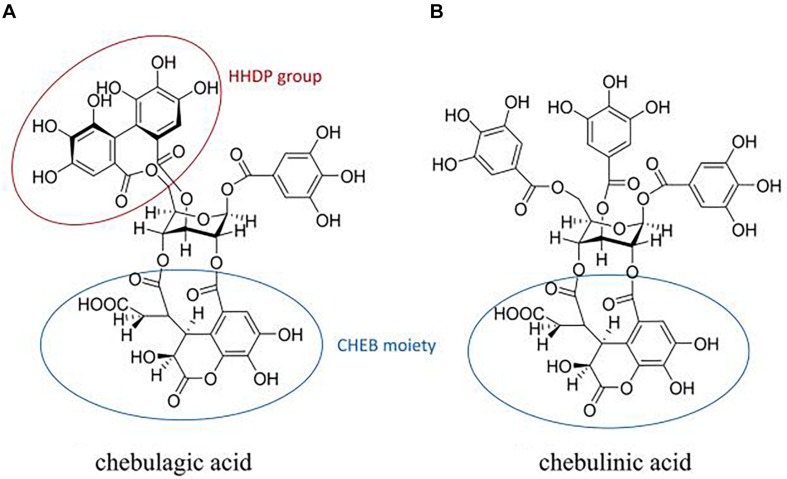
CHLA (1-O-galloyl-2,4-O-chebuloyl-3,6-O-hexahydroxydiph enoyl-β-D-glucose) (Figure 6A) and CHLI (1,3,6-tri-O-galloyl-2,4-O-chebuloyl-β-D-glucopyranoside) (Figure 6B) share the same molecular backbone of a β-D-glucose residue linked to a chebuloyl moiety. The difference is that CHLI has three free galloyl groups attached to the β-D-glucose residue, while CHLA contains only one galloyl group and a hexahydrodiphenoyl (HHDP) group, which is assumed to be a substitution for the two galloyl groups (Olennikov et al., 2015). The presence of the constrained HHDP group in CHLA is considered to result in larger spatial hindrance and less molecular flexibility. Thus, CHLI shows more broad-spectrum and potent biological activities due to the more favorable structure for entering the binding cavities or catalytic pockets of target proteases or enzymes. However, in our screening of anti-influenza activity described above, both of them showed excellent biological activity. The IC90 of CHLI is even better. It can be inferred that the active site is related to its common CHEB structure. Although the mechanism of action of CHLA and CHLI on NA is not clear yet, we predict that they bind to the active site(s) of NA, either one single interaction with the 430-cavity or multiple interactions to both 150- and 430-cavities.
FIGURE 6.
Structures of chebulagic acid (A) and chebulinic acid (B). Circles are framed hexahydrodiphenoyl (HHDP) and chebuloyl (CHEB) moieties.
Conclusion
In summary, we demonstrate that CHLA and CHLI can effectively inhibit IAV replication. These compounds act as NA inhibitors and show antiviral potency to both wild-type and oseltamivir-resistant IAV strains. Therefore, they may be further developed as a potential therapy against IAVs.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
RD, LR, and QC conceived and designed the experiments. PL, YW, XH, XZ, LW, and RD performed the experiments. PL, PZ, and XL analyzed the data. RD, LR, and QC wrote the manuscript. All authors contributed to the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was funded by (1) The Drug Innovation Major Project (Grant No. 2018ZX09711001); (2) the Key Research and Development Projects of Science and Technology Department of Shandong Province (Grant No. 2017CXGC1309); (3) Shandong Provincial Natural Science Foundation, China (Grant No. ZR2019MH078); (4) Project for function implementation of advanced equipment, SDUTCM (Grant No. 2018yq22); and (5) Project for development of TCM Science and Technology in Shandong Province (Grant No. 2019-0031).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00182/full#supplementary-material
References
- Air G. M. (2012). Influenza neuraminidase. Influenza Other Respir. Viruses 6 245–256. 10.1111/j.1750-2659.2011.00304.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alame M. M., Massaad E., Zaraket H. (2016). Peramivir: a novel intravenous neuraminidase inhibitor for treatment of acute influenza infections. Front. Microbiol. 7:450. 10.3389/fmicb.2016.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves Galvao M. G., Rocha Crispino Santos M. A., Alves da Cunha A. J. (2014). Amantadine and rimantadine for influenza A in children and the elderly. Cochrane Database Syst. Rev. 11:CD002745. 10.1002/14651858.CD002745.pub4 [DOI] [PubMed] [Google Scholar]
- Amaro R. E., Minh D. D., Cheng L. S., Lindstrom W. M., Jr., Olson A. J., Lin J. H., et al. (2007). Remarkable loop flexibility in avian influenza N1 and its implications for antiviral drug design. J. Am. Chem. Soc. 129 7764–7765. 10.1021/ja0723535 [DOI] [PubMed] [Google Scholar]
- Chen G. H., Lin Y. L., Hsu W. L., Hsieh S. K., Tzen J. T. C. (2015). Significant elevation of antiviral activity of strictinin from Pu′er tea after thermal degradation to ellagic acid and gallic acid. J. Food Drug Anal. 23 116–123. 10.1016/j.jfda.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman P. M. (1994). Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci. 3 1687–1696. 10.1002/pro.5560031007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J. C., Beyer W. E., Palache A. M., Rimmelzwaan G. F., Osterhaus A. D. (2000). Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J. Med. Virol. 61 94–99. [DOI] [PubMed] [Google Scholar]
- Du R., Cui Q., Rong L. (2019). Competitive cooperation of hemagglutinin and neuraminidase during influenza a virus entry. Viruses 11:E458. 10.3390/v11050458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienke U., Schmidtke M., von Grafenstein S., Kirchmair J., Liedl K. R., Rollinger J. M. (2012). Influenza neuraminidase: a druggable target for natural products. Nat. Prod. Rep. 29 11–36. 10.1039/c1np00053e [DOI] [PubMed] [Google Scholar]
- Harvey A. L., Edrada-Ebel R., Quinn R. J. (2015). The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 14 111–129. 10.1038/nrd4510 [DOI] [PubMed] [Google Scholar]
- Hata M., Tsuzuki M., Goto Y., Kumagai N., Harada M., Hashimoto M., et al. (2007). High frequency of amantadine-resistant influenza A (H3N2) viruses in the 2005-2006 season and rapid detection of amantadine-resistant influenza A (H3N2) viruses by MAMA-PCR. Jpn. J. Infect. Dis. 60 202–204. 10.1097/QAI.0b013e318068d233 [DOI] [PubMed] [Google Scholar]
- Heo Y. A. (2018). Baloxavir: first global approval. Drugs 78 693–697. 10.1007/s40265-018-0899-1 [DOI] [PubMed] [Google Scholar]
- Jia R., Zhang J., Ai W., Ding X., Desta S., Sun L., et al. (2019). Design, synthesis and biological evaluation of “Multi-Site”-binding influenza virus neuraminidase inhibitors. Eur. J. Med. Chem. 178 64–80. 10.1016/j.ejmech.2019.05.076 [DOI] [PubMed] [Google Scholar]
- Kashiwagi S., Watanabe A., Ikematsu H., Uemori M., Awamura S. (2016). Long-acting neuraminidase inhibitor laninamivir octanoate as post-exposure prophylaxis for influenza. Clin. Infect. Dis. 63 330–337. 10.1093/cid/ciw255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesharwani A., Polachira S. K., Nair R., Agarwal A., Mishra N. N., Gupta S. K. (2017). Anti-HSV-2 activity of Terminalia chebula Retz extract and its constituents, chebulagic and chebulinic acids. BMC Complement. Altern. Med. 17:110. 10.1186/s12906-017-1620-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackenby A., Besselaar T. G., Daniels R. S., Fry A., Gregory V., Gubareva L. V., et al. (2018). Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors and status of novel antivirals, 2016-2017. Antiviral Res. 157 38–46. 10.1016/j.antiviral.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang X., Huang L. (2016). Anti-influenza virus activity and constituents. characterization of Paeonia delavayi extracts. Molecules 21:E1133. 10.3390/molecules21091133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Qi J., Zhang W., Vavricka C. J., Shi Y., Wei J., et al. (2010). The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat. Struct. Mol. Biol. 17 1266–1268. 10.1038/nsmb.1909 [DOI] [PubMed] [Google Scholar]
- Lin L. T., Chen T. Y., Chung C. Y., Noyce R. S., Grindley T. B., McCormick C., et al. (2011). Hydrolyzable tannins (chebulagic acid and punicalagin) target viral glycoprotein-glycosaminoglycan interactions to inhibit herpes simplex virus 1 entry and cell-to-cell spread. J. Virol. 85 4386–4398. 10.1128/jvi.01492-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. T., Chen T. Y., Lin S. C., Chung C. Y., Lin T. C., Wang G. H., et al. (2013). Broad-spectrum antiviral activity of chebulagic acid and punicalagin against viruses that use glycosaminoglycans for entry. BMC Microbiol. 13:187. 10.1186/1471-2180-13-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Xiong S., Xiang Y. F., Guo C. W., Ge F., Yang C. R., et al. (2011). Antiviral activity and possible mechanisms of action of pentagalloylglucose (PGG) against influenza A virus. Arch. Virol. 156 1359–1369. 10.1007/s00705-011-0989-9 [DOI] [PubMed] [Google Scholar]
- Nachbagauer R., Krammer F. (2017). Universal influenza virus vaccines and therapeutic antibodies. Clin. Microbiol. Infect. 23 222–228. 10.1016/j.cmi.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noshi T., Kitano M., Taniguchi K., Yamamoto A., Omoto S., Baba K., et al. (2018). In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antivir. Res. 160 109–117. 10.1016/j.antiviral.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Olennikov D., Kashchenko N., Chirikova N. (2015). In vitro bioaccessibility, human gut microbiota metabolites and hepatoprotective potential of chebulic ellagitannins: a case of padma hepaten® formulation. Nutrients 7 8456–8477. 10.3390/nu7105406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. J., Haire L. F., Stevens D. J., Collins P. J., Lin Y. P., Blackburn G. M., et al. (2006). The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443 45–49. 10.1038/nature05114 [DOI] [PubMed] [Google Scholar]
- Samson M., Pizzorno A., Abed Y., Boivin G. (2013). Influenza virus resistance to neuraminidase inhibitors. Antivir. Res. 98 174–185. 10.1016/j.antiviral.2013.03.014 [DOI] [PubMed] [Google Scholar]
- Shobugawa Y., Saito R., Sato I., Kawashima T., Dapat C., Dapat I. C., et al. (2012). Clinical effectiveness of neuraminidase inhibitors—oseltamivir, zanamivir, laninamivir, and peramivir—for treatment of influenza A(H3N2) and A(H1N1)pdm09 infection: an observational study in the 2010–2011 influenza season in Japan. J. Infect. Chemother. 18 858–864. 10.1007/s10156-012-0428-1 [DOI] [PubMed] [Google Scholar]
- Swaminathan K., Dyason J. C., Maggioni A., von Itzstein M., Downard K. M. (2013). Binding of a natural anthocyanin inhibitor to influenza neuraminidase by mass spectrometry. Anal. Bioanal. Chem. 405 6563–6572. 10.1007/s00216-013-7068-x [DOI] [PubMed] [Google Scholar]
- Takashita E., Kawakami C., Ogawa R., Morita H., Fujisaki S., Shirakura M., et al. (2019). Influenza A(H3N2) virus exhibiting reduced susceptibility to baloxavir due to a polymerase acidic subunit I38T substitution detected from a hospitalised child without prior baloxavir treatment, Japan, January 2019. Euro Surveill. 24:1900170. 10.2807/1560-7917.es.2019.24.12.1900170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Li Y., Rivailler P., Conrardy C., Castillo D. A., Chen L. M., et al. (2012). A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U.S.A. 109 4269–4274. 10.1073/pnas.1116200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Zhu X., Li Y., Shi M., Zhang J., Bourgeois M., et al. (2013). New world bats harbor diverse influenza A viruses. PLoS Pathog. 9:e1003657. 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vries E., Schutten M., Fraaij P., Boucher C., Osterhaus A. (2013). Influenza virus resistance to antiviral therapy. Adv. Pharmacol. 67 217–246. 10.1016/b978-0-12-405880-4.00006-8 [DOI] [PubMed] [Google Scholar]
- Vasileiou E., Sheikh A., Butler C., El Ferkh K., von Wissmann B., McMenamin J., et al. (2017). Effectiveness of influenza vaccines in asthma: a systematic review and meta-analysis. Clin. Infect. Dis. 65 1388–1395. 10.1093/cid/cix524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Itzstein M., Wu W. Y., Kok G. B., Pegg M. S., Dyason J. C., Jin B., et al. (1993). Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363 418–423. 10.1038/363418a0 [DOI] [PubMed] [Google Scholar]
- Wang L., Cui Q., Zhao X., Li P., Wang Y., Rong L., et al. (2019). Generation of a reassortant influenza a subtype H3N2 virus expressing gaussia luciferase. Viruses 11:665. 10.3390/v11070665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2018). 2018 Influenza (seasonal) Fact Sheet. Geneva: WHO. [Google Scholar]
- Wu Y., Qin G., Gao F., Liu Y., Vavricka C. J., Qi J., et al. (2013). Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci. Rep. 3:1551. 10.1038/srep01551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wang L., Cui Q., Li P., Wang Y., Zhang Y., et al. (2018). A mechanism underlying attenuation of recombinant influenza a viruses carrying reporter genes. Viruses 10:679. 10.3390/v10120679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wang Y., Cui Q., Li P., Wang L., Chen Z., et al. (2019). A parallel phenotypic versus target-based screening strategy for RNA-dependent RNA polymerase inhibitors of the influenza a virus. Viruses 11:826. 10.3390/v11090826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Li Y., Li S., Li H., Qiu Z., Lee C., et al. (2011). Inhibition of influenza A virus (H1N1) fusion by benzenesulfonamide derivatives targeting viral hemagglutinin. PLoS One. 6:e29120. 10.1371/journal.pone.0029120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu M., Li C., Fang J. S., Lian W. W., Liu A. L., Zheng L. S., et al. (2015). Drug discovery of host CLK1 inhibitors for influenza treatment. Molecules 20 19735–19747. 10.3390/molecules201119653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.