Abstract
Background
Gallbladder cancer (GBC) is the most common cancer of the biliary tract, but molecularly targeted therapies are not available for GBC. Loss of microRNA (miR)-335 expression may be a useful predictor of clinical outcomes and the reversal of its loss of expression may be a useful treatment strategy for GBC. In this study, we investigated whether a long noncoding RNA, nuclear paraspeckle assembly transcript 1 (NEAT1) sponges miR-335 in GBC cells.
Materials and Methods
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR), Western blotting, and immunohistochemistry were used to determine the expression of miR-335; NEAT1; survivin; and Ki67 in GBC cell lines (GBC-SD and SGC-996) and tissue samples from patients (n = 25). Cell Counting Kit-8, colony-formation, and Transwell migration and invasion assays were performed to measure cell proliferation, migration, and invasion. Bioinformatic analysis and dual-luciferase reporter assays were utilized to analyze correlativity.
Results
miR-335 overexpression resulted in inhibition of GBC cell proliferation and invasion. In addition, knockdown of NEAT1 resulted in downregulation of survivin expression. As NEAT1 competitively “sponges” miR-335, NEAT1 knockdown resulted in inhibited GBC cell proliferation and invasion in vitro and GBC tumor growth in vivo. Furthermore, NEAT1 was found to be upregulated in GBC samples, and its expression was inversely correlated with miR-335 levels, but positively correlated with survivin levels.
Conclusion
These findings indicate that NEAT1 promotes survivin expression by functioning as a competitive endogenous RNA for miR-335 in GBC cells; thus, we have identified a potential biomarker and target for GBC diagnosis and therapy.
Keywords: long noncoding RNA, NEAT1, miR-335-5p, competitive endogenous RNA, survivin, gallbladder cancer
Introduction
Globally gallbladder cancer (GBC) is the most common cancer of the biliary tract and the fifth most common digestive tract cancer.1 GBC is uncommon in developed countries, but is more common in certain developing countries. It is characterized by a poor prognosis and usually a late diagnosis, leading to adverse treatment outcomes.2 Currently, complete surgical resection of the tumor is the most effective treatment for GBC, but by the time patients—most—are diagnosed with GBC, the optimal time for surgery has passed.3 In recent years, with rapid developments in tumor biology and sequencing technologies, the molecular pathogenesis of GBC has been elucidated in detail. Therefore, identification of new molecular targets and the development of new therapeutic strategies against GBC are urgently needed.
MicroRNAs (miRs, miRNAs) are small noncoding single-stranded RNAs that bind to the 3′ untranslated regions of their target mRNAs to suppress protein expression.4 Increasing evidence indicates that miRNAs play key roles in the progression of many human cancers, by acting as oncogenic RNAs or tumor suppressors.5 miR-335 is reported to be a tumor suppressor that is downregulated in many cancers, including thyroid cancer, lung cancer, osteosarcoma, and neuroblastoma.6–9 Recently, the expression of miR-335 was found to be significantly lower in GBC tissues, but its involvement in GBC and the associated molecular mechanisms remain unclear.
Long noncoding RNAs (lncRNAs) are noncoding RNAs that are longer than 200 nucleotides in length; they often show dysregulated expression in cancers and play an important role in the initiation and/or progression of cancers.10 In GBC, the lncRNA TUG1 promotes cell proliferation and metastasis by downregulating miR-300, and the lncRNA MINCR promotes GBC progression by stimulating EZH2 expression.11,12 The lncRNA, MALAT1, facilitates growth and inhibits senescence by antagonizing ABI3BP in GBC cells.13 Another lncRNA, PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in GBC.3 Accumulating evidence suggests that the expression of the lncRNA, nuclear paraspeckle assembly transcript 1 (NEAT1), is dysregulated in many human cancers, and that this closely correlates with poor prognosis.14 NEAT1 promotes the progression of colon cancer by sponging miR-495-3p and activating CDK6.15 It performs an oncogenic function in triple-negative breast cancer, by promoting the development of chemoresistance and cancer stemness.14 Nevertheless, it is not known whether the aberrant expression of NEAT1 in GBC is associated with the progression of malignancy. Importantly, the mechanism by which NEAT1 exerts its oncogenic effect remains to be identified.
In this study, we first showed that overexpression of miR-335 inhibited GBC cell proliferation and invasion. Next, we demonstrated that NEAT1 upregulated survivin by competitively “sponging” miR-335, and this enhanced GBC cell proliferation and invasion in vitro and promoted GBC tumor growth in vivo; thus, we have identified a potential biomarker and/or target for GBC diagnosis and therapy.
Materials and Methods
Clinical Samples and Cell Lines
A total of 25 human GBC samples (Supplementary Table 1) and corresponding normal gallbladder tissue samples were obtained from the Department of Biliary II, Shanghai Eastern Hepatobiliary Surgery Hospital, Naval Military Medical University (Shanghai, China), after obtaining written informed consent. This study was performed according to an established protocol approved by the Ethics Committee of the Shanghai Eastern Hepatobiliary Surgery Hospital, Naval Military Medical University. Tissue samples were collected during surgical resection and were immediately frozen in liquid nitrogen. GBC cell lines, GBC-SD and SGC-996 were purchased from Guangzhou Gino Bio Co., Ltd. (Guangzhou, China), and the human gallbladder epithelium cell line, H69, was acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin.
RNA Extraction and qRT-PCR Assays
Total RNA was extracted using TRIzol reagent (Life Technologies Inc., Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using the Superscript III Reverse Transcription Reagent (Life Technologies). qRT-PCR analysis was performed using SYBR Green chemistry on an ABI7500 real-time PCR detection system (Applied Biosystems, Foster City, CA, USA). β-Actin or U6 small nuclear RNA served as the endogenous control. Each experiment was performed in triplicate, and the 2−ΔΔCt method was used to calculate relative expression.
Western Blotting
For Western blotting, proteins in cell lysates were separated on SDS-polyacrylamide gel by electrophoresis, and electrotransferred onto a polyvinylidene difluoride membrane, which was then sequentially probed with primary and secondary antibodies. Signals were detected by chemiluminescence reagents (ECL Kit, Pierce Biotechnology, Waltham, MA, USA) and imaged on a Tanon-5200 Chemiluminescent Imaging System (Bio-tanon, Shanghai, China). Anti-survivin (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and anti-β-actin (Proteintech, Wuhan, China) antibodies were employed for Western blotting.
Oligonucleotides and Transfection
An miR-335 mimic, an miR-335 inhibitor, and their negative controls (NC mimic and NC inhibitor, respectively) were purchased from Ribobio (Guangzhou, China). siRNAs specifically targeting NEAT1 (siNEAT1) and a scrambled control siRNA (siNC) were synthesized by GenePharma (Shanghai, China). All transfections were performed using Lipofectamine 2000 (Life Technologies).
Lentivirus Vector Construction and Infection
A lentiviral vector encoding a short hairpin RNA (shRNA) specific for NEAT1 (designated as shNEAT1) and its negative control (designated as shNC), as well as a lentivirus expressing anti-miR-335 (designated anti-miR-335) and its negative control (anti-NC) were purchased from Hanbio (Shanghai, China). All transfections were performed using Lipofectamine 2000 (Life Technologies).
Luciferase Reporter Assay
The plasmids, pmirGLO-NEAT1-wt or pmirGLO-NEAT1-mt, carrying wild-type or mutant binding sites for miR-335, respectively, were cotransfected with either the miR-335 or NC mimic, into HEK293T cells via Lipofectamine-mediated gene transfer. Survivin-wt and -mt 3′-UTRs were constructed and transfected into cells along with the miR-335 or NC mimic, respectively. Firefly and Renilla luciferase activities were measured 48 h after transfection, according to the manufacturer’s instructions. The ratio of the luminescence from the firefly luciferase to that of Renilla luciferase was calculated as the relative luciferase activity.
Cell Counting Kit-8 Assay
Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8) assay at 1, 2, 3, and 4 days after transfection. Cells were seeded in a 96-well plate at a density of 1500 cells/well and 10 μL of CCK-8 was added to 90 μL of cell culture medium per well. The cells were then incubated for 2 h and cell viability was determined by measuring the absorbance at 450 nm.
Colony-Formation Assay
For colony-formation assays, 1000 cells were seeded in each well of a 6-well plate and incubated at 37°C for 2 weeks. The colonies were fixed and stained with a solution containing 0.1% crystal violet and 20% methanol and were then counted. All stainings were performed in triplicate.
Cell Migration and Invasion Assays
Cell migration was assessed in a 24-well Transwell chemotaxis chamber (BD Biosciences, Franklin Lakes, NJ). Cells were seeded at a density of 4 × 104 in the upper chamber a Transwell plate containing a non-coated membrane for the migration assay. For the invasion assays, cells were seeded at a density of 4 × 104 in the upper chamber of a Transwell plate containing a Matrigel-coated membrane. After several hours of incubation at 37°C, cells that migrated or invaded were fixed with 20% methanol and stained with crystal violet (0.1%). These cells were then counted and imaged using an inverted microscope (Olympus, Tokyo, Japan).
Tumor Xenograft Experiments
GBC-SD cells stably expressing a control shRNA, shNEAT1, an miR-335 inhibitor, or shNEAT1 + an miR-335 inhibitor were subcutaneously injected into either side of the flank of 6-week-old nude mice (n = 4 mice per group). Tumor volumes were calculated (1/2 × length × width2) on a weekly basis. After 5 weeks, the mice were euthanized, and the tumors were excised and subjected to immunohistochemistry (IHC). All animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committee of the Naval Military Medical University.
IHC
After heat-induced epitope retrieval, paraffin-embedded sections were incubated with 3% H2O2 and blocked for another 60 min with 3% normal serum. Sections were then incubated with primary antibodies overnight at 4°C. An Elite ABC Staining Kit and DAB Peroxidase Substrate Kit (Vector Laboratories, Inc., Burlingame, CA, USA) were used to visualize antibody binding, according to the manufacturer’s instructions. Anti-survivin (Abcam, Cambridge, UK) and anti-Ki67 (Cell Signaling Technology, Beverly, MA, USA) antibodies were used for IHC. ImageJ software was used to analyze IHC staining in randomly selected slides from four mice in each group (more than six representative fields in each slide were analyzed). Positive staining in each slide was scored and presented as relative expression level (the protein expression level in shNC mice was arbitrarily set at 1). Data are expressed as means ± SD.
Statistical Analysis
Results are presented as means ± SD. Statistical analysis was performed with Prism, version 5.0 (GraphPad, San Diego, CA, USA), using the Mann–Whitney test to compare two groups and a one-way ANOVA, followed by Tukey’s post hoc test to compare two or more groups. Correlation analysis was performed using Spearman correlation coefficient rank test. P values <0.05 were considered statistically significant.
Results
miR-335 Decreased GBC Cell Proliferation and Invasion
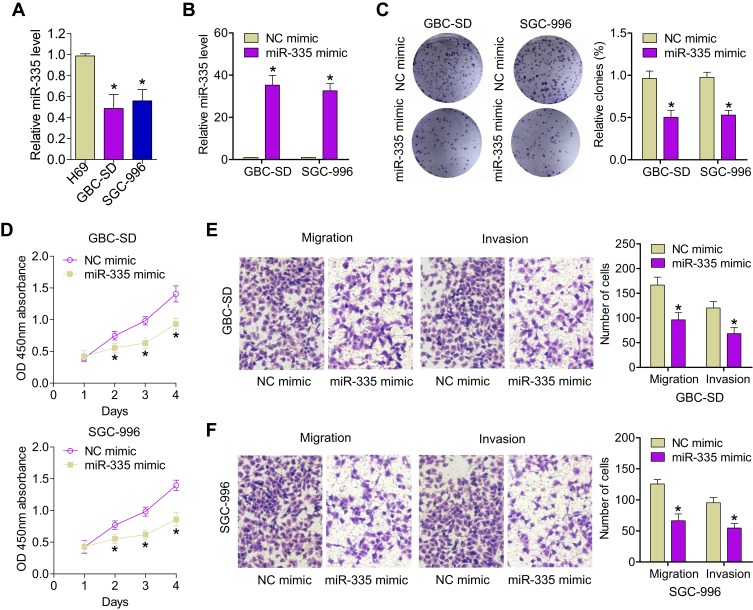
It has been demonstrated that miR-335 levels are lower in GBC tumors than in that in nondysplastic gallbladder epithelia.16 Nonetheless, the function of miR-335 and its mechanism of action in GBC remain unclear. Here, we determined the expression of miR-335 in the human nontumorigenic biliary epithelial cell line, H69 and in GBC cell lines, SGC-996 and GBC-SD, by qRT-PCR. We found that miR-335 was significantly underexpressed in GBC cell lines with respect to that in the normal biliary epithelial cell line (Figure 1A). Then, we overexpressed miR-335 in SGC-996 and GBC-SD cells by transiently transfecting a miR-335 mimic. qRT-PCR was performed to confirm miR-335 overexpression in these cells (Figure 1B). A colony-formation assay showed that miR-335 mimic-transfected cells formed significantly fewer and significantly smaller colonies than NC mimic-transfected cells (Figure 1C). CCK-8 assay showed that the viability of GBC cells transfected with the miR-335 mimic was markedly lower than the viability of GBC cells transfected with the NC mimic (Figure 1D). Transwell migration and invasion assay demonstrated that transfection of the miR-335 mimic into GBC cells clearly reduced their migratory and invasive abilities (Figure 1E). These results suggest that overexpression of miR-335 markedly decreases the viability and the migration and invasiveness of GBC cells.
Figure 1.
Overexpression of miR-335 inhibits GBC cell proliferation and invasion. (A) The relative expression of miR-335 in GBC cells (GBC-SD and SGC-996) and in the human nontumorigenic biliary epithelial cell line, H69, was determined by qRT-PCR. (B) The transfection efficiency of the miR-335 mimic was evaluated by qRT-PCR. (C) Representative images and quantification of colony-formation assay data. (D) Changes in GBC cell proliferation after transfection with the miR-335 mimic or the NC mimic were determined by CCK-8 assay. (E) Representative images and quantification of data from Transwell migration and invasion assays of GBC-SD cells transfected with the miR-335 or NC mimics. (F) Representative images and quantification of data from Transwell migration and invasion assays of SGC-996 cells transfected with the miR-335 or NC mimics. *P < 0.05.
NEAT1 Upregulated Survivin Expression by Sponging miR-335
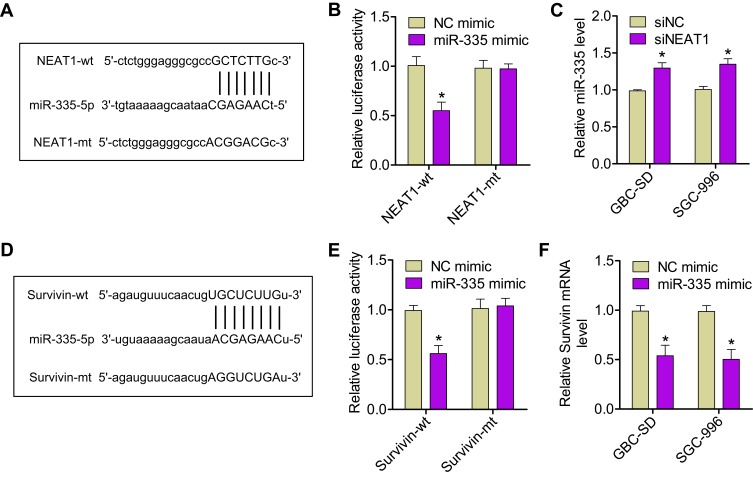
We next aimed to identify the molecular mechanisms responsible underlying miR-335 downregulation in GBC. It has recently been reported that, in gastric cancer, miR-335 expression is subject to post-transcriptional regulation by a competing endogenous RNA (ceRNA).17 In this study, NEAT1 was found to contain miR-335-binding elements and to function as a sponge for miR-335. We hypothesized that NEAT1 could regulate GBC progression by binding to miR-335. We first confirmed the binding of miR-335 to the sites predicted by the online bioinformatic database, starBase 2.0 (Figure 2A). Dual reporter luciferase assays—performed in HEK293T cells—demonstrated that the miR-335 mimic significantly decreased the luciferase activity of NEAT1-wt but not that of NEAT1-mt (Figure 2B), indicating the existence of miR-335-binding site(s) in NEAT1. Moreover, we found that NEAT1 knockdown significantly increased miR-335 levels in GBC-SD and SGC-996 cells (Figure 2C). Taken together, these results suggest that, in GBC cells, NEAT1 can act as a sponge and reduce miR-335 levels.
Figure 2.
NEAT1 promoted survivin expression by sponging miR-335. (A) The miR-335 response element in NEAT1, as predicted by the starBase 2.0 algorithm. (B) Relative luciferase activity of HEK293T cells after cotransfection with either the NEAT1-wt or -mt reporter construct, along with either the miR-335 or NC mimic. (C) Expression of miR-335 was assessed by qRT-PCR in GBC-SD and SGC-996 cells transfected with either siNEAT1 or siNC. (D) Bioinformatic analysis indicated that the 3′-UTR sequence of survivin was complementary to the seed sequence of miR-335. (E) Relative luciferase activity of HEK293T cells after cotransfection with either the survivin-wt or -mt 3′-UTR reporter gene, along with either the miR-335 or NC mimic. (F) Expression of survivin mRNA was measured by qRT-PCR in GBC cells transfected with either the miR-335 or NC mimic. *P < 0.05.
In addition, to elucidate the molecular mechanisms underlying the regulation of miR-335 expression, we examined miR-335 targets that were computationally predicted by the TargetScan algorithm. Among the high-scoring mRNA targets predicted for miR-335, survivin mRNA was selected for further research. Our previous study had shown that survivin is implicated in GBC, as a critical regulator of tumor cell apoptosis.18 Bioinformatic analysis showed that a sequence within the 3′-UTR of survivin mRNA was complementary to the seed sequence of miR-335 (Figure 2D). The dual-luciferase reporter assay showed that cotransfection with the miR-335 mimic and survivin-wt resulted in marked decrease in luciferase activity; however, cotransfection with the miR-335 mimic and survivin-mt did not have any effect on luciferase activity (Figure 2E). Overexpression of miR-335 in GBC cells significantly reduced the expression of survivin at mRNA and protein levels (Figure 2F and S1A). An online bioinformatic database predicted that NEAT1 and survivin had the same putative binding site in miR-335 (Figure 2A and D). Furthermore, NEAT1 knockdown resulted in decreased luciferase activity in cells transfected with survivin-wt (Figure S1B). We next tested whether NEAT1 regulates the expression of survivin in GBC cells in an miR-335-dependent manner. We found that NEAT1 knockdown results in decreased expression of survivin at mRNA and protein levels in GBC cells (Figure S1C and D). These results indicated that NEAT1 positively regulated survivin expression by sponging miR-335 in GBC cells.
The NEAT1/miR-335/survivin axis affects GBC cell proliferation and invasion in vitro.
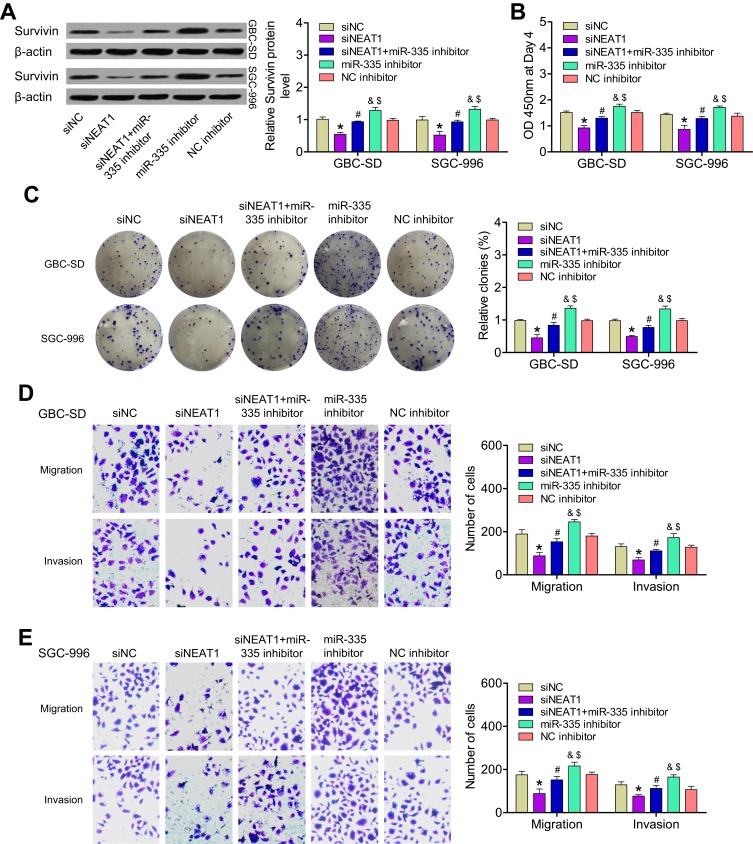
We next investigated whether NEAT1 promoted the expression of survivin by targeting miR-335 in GBC cells. Compared to its expression in control cells, survivin expression (mRNA and protein levels) was significantly lower in GBC-SD and SGC-996 cells transfected with a NEAT1-specific siRNA and was higher in cells transfected with an miR-335 inhibitor. By contrast, in cells cotransfected with an miR-335 inhibitor and siNEAT1, the upregulation of survivin by NEAT1 was attenuated (Figure 3A). We continued to investigate the effects of the NEAT1/miR-335/survivin axis on the proliferation and invasiveness of GBC cells. CCK-8 and colony-formation assays showed that the viability and colony-formation ability of GBC cells were clearly reduced upon siNEAT1 transfection and were increased by miR-335 inhibitor transfection. These effects were attenuated by simultaneous cotransfection of the miR-335 inhibitor and siNEAT1 (Figure 3B and C). The knockdown of NEAT1 decreased the number of migratory and invasive GBC cells, but this effect was reversed by cotransfection of the miR-335 inhibitor (Figure 3D and E). These results showed that NEAT1 promoted GBC proliferation and invasion by regulating the miR-335/survivin pathway.
Figure 3.
Influence of the NEAT1/miR-335/survivin axis on GBC cell proliferation and invasion in vitro. (A) Expression of survivin protein was determined in GBC-SD and SGC-996 cells transfected with siNC, siNEAT1, or an miR-335 inhibitor or cotransfected with an miR-335 inhibitor and siNEAT1. (B) Changes in cell proliferation in the above-mentioned GBC cell groups were determined by CCK-8 assay. (C) Quantification of colony-formation assay data from different groups of GBC cells. (D, E) Quantification of data from Transwell migration and invasion assays of different groups of GBC cells. *P < 0.05 vs the siNC group; #P < 0.05 vs the siNEAT1 group; &P < 0.05 vs the siNEAT1 + miR-335 inhibitor group; $P < 0.05 vs the NC inhibitor group.
The Oncogenic Activity of NEAT1 Was, in Part, Mediated by Downregulation of miR-335 in vivo
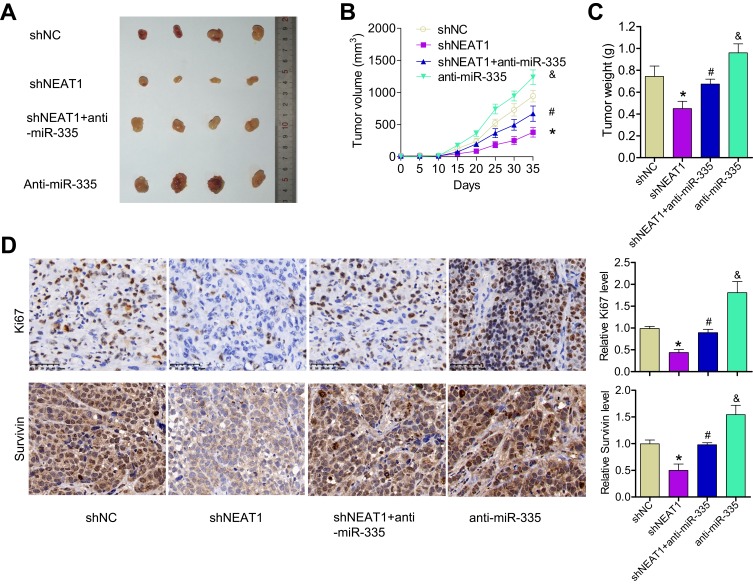
To verify our in vitro findings, we established an in vivo xenograft model in nude mice. ShNEAT1, anti-miR-335, or both shNEAT1 and anti-miR-335 were transfected into GBC-SD cells, which were then injected into nude mice. Tumors were allowed to form and grow for 5 weeks, during which tumor size and weight were measured at regular intervals. The results showed that NEAT1 knockdown resulted in markedly reduced tumor volume, whereas anti-miR-335 clearly promoted tumor growth (Figure 4A and B). However, double knockdown of NEAT1 and miR-335 had no significant impact on tumor formation (Figure 4A and B). Meanwhile, smaller tumors were observed in mice treated with shNEAT1 than in mice treated with shNC. Larger tumors were observed in mice treated with anti-miR-335 than in those treated with shNC, but this effect was attenuated by simultaneous cotreatment with the miR-335 inhibitor and shNEAT1 (Figure 4C). IHC showed lower expression of survivin and fewer Ki67-positive cells in tumors derived from shNEAT1-transfected cells; this effect was significantly attenuated by anti-miR-335 treatment (Figure 4D).
Figure 4.
Knockdown of NEAT1 inhibited tumor growth, in part through miR-335 downregulation in vivo. (A) Representative image of xenograft tumor formation in nude mice injected with shNC, shNEAT1, anti-miR-335, and shNEAT1 + anti-miR-335 transfected cells (n = 4 per group). (B) Tumor volume growth curves. (C) Tumor weight. (D) IHC showing Ki67 and survivin levels in tumor. *P < 0.05 vs the siNC group; #P < 0.05 vs the siNEAT1 group; &P < 0.05 vs the siNEAT1 + miR-335 inhibitor group.
NEAT1 Was Upregulated in GBC Samples and Was Inversely Correlated with miR-335 Levels
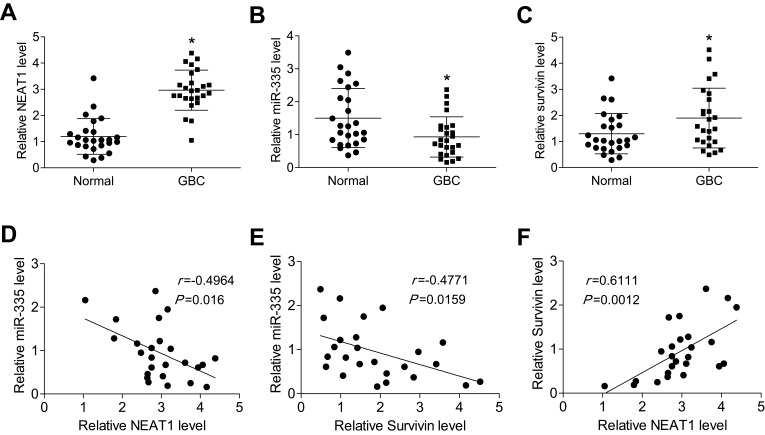
We next measured the levels of NEAT1, miR-335, and survivin in GBC tissue samples (n = 25) and normal gallbladder tissue samples (n = 25) by qRT-PCR. We found that NEAT1 and survivin levels were significantly higher, and miR-335 levels were significantly lower in GBC samples, compared with those in normal gallbladder tissue samples (Figure 5A–C). Moreover, we evaluated the correlations between NEAT1 and miR-335 levels, between NEAT1 and survivin levels, and between miR-335 and survivin levels in GBC samples. As shown in Figure 5D–F, NEAT1 levels negatively correlated with miR-335 levels, but positively correlated with survivin levels in GBC samples. Further, miR-335 levels negatively correlated with survivin levels.
Figure 5.
NEAT1 was upregulated in GBC tissue samples, and its levels inversely correlated with miR-335 levels, but positively correlated with survivin levels. (A) The relative expression of NEAT1 in human GBC samples and normal gallbladder tissue samples. The relative expression of miR-335 (B) and survivin (C) in human GBC samples and normal gallbladder tissue samples (D) A significant inverse correlation was observed between NEAT1 and miR-335 levels in GBC samples. (E) A significant inverse correlation was observed between miR-335 and survivin mRNA levels in GBC samples. (F) A significant positive correlation was observed between NEAT1 and survivin mRNA levels in GBC samples. *P < 0.05.
Discussion
A lot of evidence suggests that certain miRNAs target specific mRNAs related to the initiation and progression of GBC and are involved in regulating the proliferation and invasiveness of GBC cells.19,20 For example, miR-139-5p is associated with a poor prognosis and it controls glycolysis by downregulating PKM2 in GBC,21 whereas miRNA-30a-5p inhibits GBC cell proliferation, migration, and metastasis by targeting E2F7 mRNA.22 In the present study, we first identified miR-335 as a key miRNA involved in GBC cell proliferation and invasion. We then explored the potential effects of lncRNAs on the regulatory functions of miR-335 in GBC. We showed that NEAT1 significantly reduced the activity of miR-335 by sequestering miR-335 molecules. Therefore, the following possible mechanism is proposed: NEAT1 sponges miR-335 to promote survivin expression, and by extension, GBC cell proliferation and invasion in vitro and tumor growth in vivo, thereby facilitating GBC progression.
LncRNAs are a newly identified class of noncoding RNAs that perform important functions, such as regulation of cell cycle, cell growth, migration, and apoptosis, and are involved in almost all physiological and pathological processes.23,24 Several studies have uncovered the effectiveness of lncRNAs as miRNA sponges, affecting GBC initiation and progression as tumor suppressors or oncogenic RNAs. For example, H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in GBC;25 MALAT1 functions as a ceRNA by sponging miR-363-3p, thereby regulating MCL-1 expression in GBC;26 and the MINCR/miR-26a-5p/EZH2 axis is reported to be involved in the proliferation, invasion, and apoptosis of GBC cells.11 In our study, we observed that NEAT1 contained miR-335-binding elements, and an online bioinformatic database identified NEAT1 as a potential sponge of miR-335. Our dual-luciferase reporter assay confirmed that miR-335 binds to a site(s) within NEAT1. Further, NEAT1 has been shown to be enriched in miRNA-binding sites and to function as a ceRNA that interacts with miRNAs; it is also known to play an essential role in tumor initiation and progression. NEAT1 facilitates melanoma cell proliferation, migration, and invasion by regulating miR-495-3p and E2F327 and promotes colorectal cancer progression by competing with SIRT1 mRNA for binding to miR-34a and by activating the Wnt/β-catenin signaling pathway.28 In our study, we found that NEAT1 knockdown resulted in a significant increase in miR-335 levels in GBC-SD and SGC-996 cells. Thus, we can speculate that NEAT1 serves as a ceRNA that interacts with miR-335, thereby influencing the expression of miR-335 target mRNAs in GBC.
We examined computationally predicted targets of miR-335 and found that survivin was a possible target. Survivin—also known as baculoviral inhibitor of apoptosis repeat-containing 5—is the newest member of a family of proteins that is involved in inhibiting apoptosis; it was recently suggested as a novel biomarker of cancer.29,30 Overexpression of survivin in tumors is generally associated with poor prognosis and high drug resistance.31 Our previous study implicated survivin as a critical regulator of tumor cell apoptosis in GBC.18 In the present study, bioinformatic analysis showed that the 3′-UTR of survivin mRNA is complementary to the seed sequence of miR-335; dual-luciferase reporter assays confirmed the interaction between miR-335 and survivin mRNA. Overexpression of miR-335 resulted in significantly reduced survivin expression in GBC cells. Online bioinformatic database searches predicted that NEAT1 and survivin shared almost the same putative binding site in miR-335. Survivin expression significantly decreased in GBC-SD and SGC-996 cells after transfection with an NEAT1-specific shRNA, but increased after transfection with an miR-335 inhibitor. By contrast, in cells cotransfected with an miR-335 inhibitor and shNEAT1, the stimulatory effect of NEAT1 on survivin expression was attenuated. We demonstrated that the oncogenic activity of NEAT1 was, in part, mediated by the downregulation of miR-335 in vitro and in vivo. Furthermore, NEAT1 was found to be upregulated in GBC samples, and its expression was inversely correlated with miR-335 levels, but positively correlated with survivin levels. These data indicated that NEAT1 positively regulated survivin expression by sponging miR-335 in GBC cells.
Several studies also have shown that plasma NEAT1 and miR-335 levels are abnormal in some cancers. For example, the plasma levels of NEAT1 are higher in lymph node-negative patients, lymph node-positive patients, and particularly, in triple-negative patients, indicating that NEAT1 dysregulation is associated with more aggressive tumors.32 Downregulation of miR-335 has also been demonstrated in samples from gastric cancer patients, and this phenomenon is inversely correlated with methylation of the promoter region, supporting the hypothesis that methylation of miR-335 may serve as a promising non-invasive treatment strategy for gastric cancer.33 Plasma NEAT1 levels may also be abnormal in GBC patients. In future experiments, we will collect multiple GBC plasma samples to measure NEAT1 levels and analyze the correlation between NEAT1 expression and GBC.
In conclusion, we found that miR-335 overexpression results in inhibition of proliferation and invasion of GBC cells. In addition, we demonstrated that NEAT1 promotes survivin expression by competitively sponging miR-335, thereby promoting GBC cell proliferation and invasion in vitro and GBC tumor growth in vivo. Moreover, NEAT1 was found to be upregulated in GBC tissue samples, and to inversely correlate with miR-335 levels, but positively correlate with survivin levels. These findings suggest that NEAT1 is a potential biomarker and a target for GBC diagnosis and therapy.
Acknowledgments
This study was supported by the National Natural Science Foundation (81772528).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Misra S, Chaturvedi A, Misra NC, Sharma ID. Carcinoma of the gallbladder. Lancet Oncol. 2003;4(3):167–176. doi: 10.1016/S1470-2045(03)01021-0 [DOI] [PubMed] [Google Scholar]
- 2.Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: recent update. World J Gastroenterol. 2017;23(22):3978–3998. doi: 10.3748/wjg.v23.i22.3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Yu Y, Li H, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(1):33. doi: 10.1186/s12943-019-0947-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800 [DOI] [PubMed] [Google Scholar]
- 6.Luo L, Xia L, Zha B, et al. miR-335-5p targeting ICAM-1 inhibits invasion and metastasis of thyroid cancer cells. Biomed Pharmacother. 2018;106:983–990. doi: 10.1016/j.biopha.2018.07.046 [DOI] [PubMed] [Google Scholar]
- 7.Tang H, Zhu J, Du W, et al. CPNE1 is a target of miR-335-5p and plays an important role in the pathogenesis of non-small cell lung cancer. J Exp Clin Cancer Res. 2018;37(1):131. doi: 10.1186/s13046-018-0811-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89. doi: 10.1186/s12943-018-0837-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch J, Fay J, Meehan M, et al. MiRNA-335 suppresses neuroblastoma cell invasiveness by direct targeting of multiple genes from the non-canonical TGF-beta signalling pathway. Carcinogenesis. 2012;33(5):976–985. doi: 10.1093/carcin/bgs114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunch H. Gene regulation of mammalian long non-coding RNA. Mol Genet Genomics. 2018;293(1):1–15. doi: 10.1007/s00438-017-1370-9 [DOI] [PubMed] [Google Scholar]
- 11.Wang SH, Yang Y, Wu XC, et al. Long non-coding RNA MINCR promotes gallbladder cancer progression through stimulating EZH2 expression. Cancer Lett. 2016;380(1):122–133. doi: 10.1016/j.canlet.2016.06.019 [DOI] [PubMed] [Google Scholar]
- 12.Ma F, Wang SH, Cai Q, et al. Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma. Biomed Pharmacother. 2017;88:863–869. doi: 10.1016/j.biopha.2017.01.150 [DOI] [PubMed] [Google Scholar]
- 13.Lin N, Yao Z, Xu M, et al. Long noncoding RNA MALAT1 potentiates growth and inhibits senescence by antagonizing ABI3BP in gallbladder cancer cells. J Exp Clin Cancer Res. 2019;38(1):244. doi: 10.1186/s13046-019-1237-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin VY, Chen J, Cheuk IW, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10(4):270. doi: 10.1038/s41419-019-1513-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z, Dang J, Song A, Cui X, Ma Z, Zhang Z. NEAT1 promotes colon cancer progression through sponging miR-495-3p and activating CDK6 in vitro and in vivo. J Cell Physiol. 2019;234(11):19582–19591. doi: 10.1002/jcp.v234.11 [DOI] [PubMed] [Google Scholar]
- 16.Peng HH, Zhang YD, Gong LS, Liu WD, Zhang Y. Increased expression of microRNA-335 predicts a favorable prognosis in primary gallbladder carcinoma. Onco Targets Ther. 2013;6:1625–1630. doi: 10.2147/OTT.S53030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Zhang M, Sun G. Long non-coding RNA NEAT1 regulates the proliferation, migration and invasion of gastric cancer cells via targeting miR-335-5p/ROCK1 axis. Pharmazie. 2018;73(3):150–155. doi: 10.1691/ph.2018.7877 [DOI] [PubMed] [Google Scholar]
- 18.Qiu Y, Li X, Yi B, et al. Protein phosphatase PHLPP induces cell apoptosis and exerts anticancer activity by inhibiting Survivin phosphorylation and nuclear export in gallbladder cancer. Oncotarget. 2015;6(22):19148–19162. doi: 10.18632/oncotarget.v6i22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin YP, Hu YP, Wu XS, et al. miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell Death Dis. 2018;9(2):182. doi: 10.1038/s41419-017-0258-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Zhan M, Xu SW, et al. miR-218-5p restores sensitivity to gemcitabine through PRKCE/MDR1 axis in gallbladder cancer. Cell Death Dis. 2017;8(5):e2770. doi: 10.1038/cddis.2017.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Yu Y, Chen X, et al. MiR-139-5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Prolif. 2018;51(6):e12510. doi: 10.1111/cpr.2018.51.issue-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye YY, Mei JW, Xiang SS, et al. MicroRNA-30a-5p inhibits gallbladder cancer cell proliferation, migration and metastasis by targeting E2F7. Cell Death Dis. 2018;9(3):410. doi: 10.1038/s41419-018-0444-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guenzl PM, Barlow DP. Macro lncRNAs: a new layer of cis-regulatory information in the mammalian genome. RNA Biol. 2012;9(6):731–741. doi: 10.4161/rna.19985 [DOI] [PubMed] [Google Scholar]
- 24.Xu MD, Qi P, Du X. Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Mod Pathol. 2014;27(10):1310–1320. doi: 10.1038/modpathol.2014.33 [DOI] [PubMed] [Google Scholar]
- 25.Wang SH, Ma F, Tang ZH, et al. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res. 2016;35(1):160. doi: 10.1186/s13046-016-0436-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S-H, Zhang W-J, Wu X-C, et al. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J Cell Mol Med. 2016;20(12):2299–2308. doi: 10.1111/jcmm.2016.20.issue-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Y, Zhou Y, Han H, Li P, Wei W, Lin N. lncRNA NEAT1 facilitates melanoma cell proliferation, migration, and invasion via regulating miR-495-3p and E2F3. J Cell Physiol. 2019;234(11):19592–19601. doi: 10.1002/jcp.v234.11 [DOI] [PubMed] [Google Scholar]
- 28.Luo Y, Chen JJ, Lv Q, et al. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/beta-catenin signaling pathway. Cancer Lett. 2019;440–441:11–22. doi: 10.1016/j.canlet.2018.10.002 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Lu P, Du H, Zhang L. LINK-A lncRNA promotes proliferation and inhibits apoptosis of mantle cell lymphoma cell by upregulating survivin. Med Sci Monit. 2019;25:365–370. doi: 10.12659/MSM.912141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahmoudian-Sani MR, Alghasi A, Saeedi-Boroujeni A, Jalali A, Jamshidi M, Khodadadi A. Survivin as a diagnostic and therapeutic marker for thyroid cancer. Pathol Res Pract. 2019;215(4):619–625. doi: 10.1016/j.prp.2019.01.025 [DOI] [PubMed] [Google Scholar]
- 31.Kim YH, Kim SM, Kim YK, Hong SP, Kim MJ, Myoung H. Evaluation of survivin as a prognostic marker in oral squamous cell carcinoma. J Oral Pathol Med. 2010;39(5):368–375. [DOI] [PubMed] [Google Scholar]
- 32.Muller V, Oliveira-Ferrer L, Steinbach B, Pantel K, Schwarzenbach H. Interplay of lncRNA H19/miR-675 and lncRNA NEAT1/miR-204 in breast cancer. Mol Oncol. 2019;13(5):1137–1149. doi: 10.1002/mol2.2019.13.issue-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandoval-Borquez A, Polakovicova I, Carrasco-Veliz N, et al. MicroRNA-335-5p is a potential suppressor of metastasis and invasion in gastric cancer. Clin Epigenetics. 2017;9:114. doi: 10.1186/s13148-017-0413-8 [DOI] [PMC free article] [PubMed] [Google Scholar]