Nontyphoidal Salmonella species are globally disseminated pathogens and are the predominant cause of gastroenteritis. The pathogenesis of salmonellosis has been extensively studied using in vivo murine models and cell lines, typically challenged with Salmonella enterica serovar Typhimurium. Although S. enterica serovars Enteritidis and Typhimurium are responsible for most of the human infections reported to the Centers for Disease Control and Prevention (CDC), several other serovars also contribute to clinical cases of salmonellosis.
KEYWORDS: Salmonella enterica, virulence, macrophages, pathogenicity, cytokines, infection, Salmonella, whole-genome sequencing
ABSTRACT
Nontyphoidal Salmonella species are globally disseminated pathogens and are the predominant cause of gastroenteritis. The pathogenesis of salmonellosis has been extensively studied using in vivo murine models and cell lines, typically challenged with Salmonella enterica serovar Typhimurium. Although S. enterica serovars Enteritidis and Typhimurium are responsible for most of the human infections reported to the Centers for Disease Control and Prevention (CDC), several other serovars also contribute to clinical cases of salmonellosis. Despite their epidemiological importance, little is known about their infection phenotypes. Here, we report the virulence characteristics and genomes of 10 atypical S. enterica serovars linked to multistate foodborne outbreaks in the United States. We show that the murine RAW 264.7 macrophage model of infection is unsuitable for inferring human-relevant differences in nontyphoidal Salmonella infections, whereas differentiated human THP-1 macrophages allowed these isolates to be further characterized in a more human-relevant context.
INTRODUCTION
Salmonella is a zoonotic pathogen that is responsible for illnesses on a global scale and poses a significant burden to public health (1). Invasive salmonellae, such as the host-restricted Salmonella enterica serovars Typhi and Paratyphi, cause fever in humans, killing nearly 217,000 people in 2000 (2). Infection with nontyphoidal Salmonella (NTS), such as S. enterica serovars Enteritidis and Typhimurium, results in gastroenteritis and is estimated to cause 155,000 deaths annually (3). Approximately 5% of individuals presenting with gastroenteritis following infection by NTS will develop bacteremia, with increased risk among immunologically compromised individuals (4). These estimates do not consider the emergence of invasive NTS (iNTS), which, instead of being associated with diarrhea, is linked with many host risk factors that influence the epidemiology of iNTS disease in Africa, including malaria, malnutrition among infants and children, and HIV infection (5).
Salmonella species are nonfastidious bacteria that can survive outside the host in a range of food matrices, under low-moisture conditions, and in food processing environments. In humans, infection commonly occurs following the ingestion of contaminated food or water. Sources for contamination vary and can range from the presence of the bacterium on the surface of raw produce to the shedding of Salmonella bacteria in the fecal and urinary excretions of reservoir animals (6, 7).
Upon entering the host, salmonellae are challenged by a series of adverse conditions, which include the low-pH environment of the stomach, the membrane-disrupting properties of bile in the small intestine, and a battery of phagocytic host immune cells, such as macrophages (8, 9). Salmonella has adapted to efficiently use mucosal carbohydrates and has an intrinsic resistance to inducible antimicrobials produced by epithelial host cells during inflammation. This provides a competitive growth advantage that can lead to intestinal colonization whereby S. enterica outgrows host microbiota in the lumen of the inflamed intestine. This virulence strategy suppresses competing microbiota by host inflammatory responses and increases the numbers of intestinal S. enterica bacteria (10). Deployment of a type III secretion system (T3SS) apparatus is fundamental to the pathogenesis of S. enterica and enables the bacterium to translocate effector proteins into the host cell cytoplasm (11). The acquisition of Salmonella pathogenicity island (SPI)-encoded virulence factors via horizontal gene transfer followed by evolution has enabled this microorganism to exploit a privileged replicative niche, avoiding the host innate immune system within intracellular vesicles called the Salmonella-containing vacuole (SCV) (12, 13). The protection afforded by the SCV allows Salmonella to thrive and sets in motion a cycle of infection whereby the bacterium can proliferate and basolaterally reinfect epithelial cells, as well as become engulfed by additional, localized phagocytic cells.
The importance of Salmonella in both clinical and public health settings has fueled research into the virulence mechanisms and associated pathogenicity of this bacterium (14, 15). Many of these studies use S. Typhimurium infection in mice as a model for typhoid fever due to the similar pathology observed, which included intestinal and extraintestinal lesions that frequently occur in human hosts infected with typhoidal serovars (16). The in vitro transcriptome of S. Typhimurium has been characterized extensively by sequencing-based methods (17, 18). Similarly, the ex vivo response of S. Typhi and S. Typhimurium during infection of macrophages has also been elucidated (19–21).
Although S. Typhimurium infection of mice results in symptoms that mimic human typhoid fever, this serovar, along with S. Enteritidis, is predominantly associated with gastroenteritis in humans. In mice pretreated with streptomycin, S. Typhimurium causes acute colitis. This is similar to human intestinal disease induced by S. Typhimurium in that it is predominantly colitic, with little or no inflammation in the ileum of the lower intestines. Previous studies have shown that S. Typhimurium mutants lacking a functional T3SS are unable to mount complete virulence in murine models of gastroenteritis (22–24). Despite the fact that more than 2,610 serovars of Salmonella have been reported to date, few studies are available that describe the infections arising from other S. enterica serovars (25–28). A large focus has been placed on two serovars, S. Enteritidis and S. Typhimurium. Both are commonly associated with food poisoning episodes and are frequently reported as the cause of most human infections reported by the Centers for Disease Control and Prevention (CDC) in the United States.
In this study, we selected 10 isolates representing atypical Salmonella serovars with confirmed links to foodborne outbreaks and evaluated the ability of these bacteria to survive within macrophages using murine and human models of infection. We found that the established murine macrophage model of infection was less suitable than the human macrophage model to discern human-relevant differences between the isolates. By investigating the whole genomes of all 10 atypical serovar isolates and comparing regions known to be involved in the pathogenicity of Salmonella with those identified in the S. Typhimurium serovar, we could begin to identify differences between these isolates that could potentially explain their ability to cause a foodborne outbreak. Together, these results are of significance to public health and highlight the diversity encountered in environmental Salmonella serovars, which may not be fully appreciated in studies focusing on reference isolates.
RESULTS
In vitro characterization of the isolates.
To simulate the host-specific challenges ingested salmonellae encounter along the alimentary canal during infection in an in vitro laboratory environment, isolates (Table 1) were characterized for their acid tolerance and swim/swarm motility (see Fig. S3 in the supplemental material). In addition, differences in susceptibility to salts of the main acid constituents of bile were determined by MIC and minimum bactericidal concentration (MBC) assays.
TABLE 1.
Isolates used in this study
| S. enterica serovar | Strain | Outbreak | Location | Yr of isolation | Isolation source |
|---|---|---|---|---|---|
| Anatum | CFSAN003959 | Papaya | Mexico | 2012 | Papaya |
| Bareilly | CFSAN001111 | Tuna | India | 2012 | Tuna scrape, frozen |
| Cubana | CFSAN002050 | Alfalfa sprouts | Arizona, USA | 2012 | Alfalfa sprouts |
| Heidelberg | CFSAN002063 | Chicken | Washington, USA | 2012 | Clinical |
| Javiana | CFSAN000905 | Green onion | Mexico | 2009 | Canal water |
| Montevideo | CFSAN000264 | Black pepper | Rhode Island, USA | 2010 | Black pepper |
| Newport | CFSAN003345 | Eastern shore sampling | Virginia, USA | 2011 | Goose feces |
| Saintpaul | CFSAN004090 | Jalapeño/serrano pepper | Mexico | 2008 | Jalapeño pepper |
| Tennessee | CFSAN001387 | Peanut butter | Georgia, USA | 2007 | Peanut butter |
| Typhimurium | 14028S | ||||
| Typhimurium | ST4/74 | ||||
| Weltevreden | CFSAN001415 | Prison tuna | Virginia, USA | 2005 | Tuna |
The acid tolerance response of Salmonella is a complex defense mechanism employed by the pathogen to defend against the acid shock experienced in the stomach (8). Increased numbers of cells for most isolates were recorded following 1 and 2 h of growth at pH 2.5 before significant reductions in viable numbers occurred at 4 h (see Fig. S1 in the supplemental material). Motility has been suggested as a virulence determinant with respect to invasion, with notable nonmotile and/or host-adapted exceptions (29–32). After 24 h of incubation at 21°C, S. enterica serovar Bareilly CFSAN001111 and S. enterica Javiana CFSAN000905 exhibited a reduced swim phenotype compared to that of S. Typhimurium ST4/74 (P < 0.001) whereas S. enterica Newport CFSAN003345 and S. enterica Saintpaul CFSAN004090 exhibited an increased swim phenotype (P < 0.001). At 37°C, S. enterica Anatum CFSAN003959 and S. enterica Tennessee CFSAN001387 exhibited a reduced swim phenotype after 8 and 24 h compared to that of S. Typhimurium ST4/74 (P < 0.001) (see Fig. S2 in the supplemental material). In the absence of glucose as a carbon source, no swarm motility was observed for any of the isolates, regardless of temperature or incubation time (Fig. S2). During digestion, contraction of the gallbladder releases bile into the small intestine. Salmonella enterica Weltevreden CFSAN001415 showed a 2-fold difference in susceptibility to sodium deoxycholate (DOC) in comparison to S. Typhimurium ST4/74, although the MBC was comparable to those of the other isolates (see Table S1 in the supplemental material).
Intracellular survival of atypical S. enterica serovars in murine and human macrophages.
Salmonella Typhimurium pathogenesis has been extensively studied in vivo using murine models and ex vivo using murine cell lines (such as J774.2 and RAW 264.7). However, differences in the ability of Salmonella serovars to survive and replicate within human macrophages are currently not well described (33). Differentiated human monocyte cell lines (including THP-1 and U937) have been used to explore the replication of S. enterica serovars Enteritidis and Typhimurium. Few studies describing the bacterial replication in human macrophages of atypical NTS serovars have been reported, despite their epidemiological importance and their contributions to clinical cases of salmonellosis (33, 34).
To study the ability of the isolates to survive phagocytosis, infections were performed using murine RAW 264.7 and differentiated human THP-1 macrophages by the gentamicin protection assay adapted from previously described protocols (35–38). Salmonella Typhimurium 14028S and ST4/74 were included as reference strains in all infection assays. Infections were carried out at a multiplicity of infection (MOI) of 10:1. Viable internalized bacteria were enumerated at 2, 4, 8, and 24 hours postinfection (hpi) in RAW 264.7 macrophages and at 2, 4, 8, 24, and 168 hpi in THP-1 macrophages.
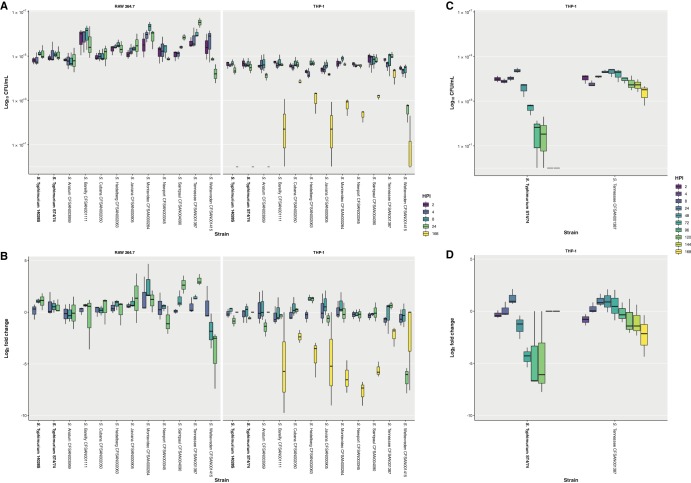
Of the 10 atypical serovars tested in this study, all isolates were found to persist within RAW 264.7 macrophages for 24 hpi, with many of these serovar increasing in number over the course of the infection. In the case of S. Weltevreden CFSAN001415, a 1-log10 decrease in intracellular bacteria between 2 and 24 hpi was recorded, although this isolate was still recoverable. In contrast, S. Tennessee CFSAN001387 exhibited a 1-log10 increase in intracellular bacteria between 2 and 24 hpi. Similar numbers of viable bacteria were recorded for S. Typhimurium 14028S and ST4/74 (Fig. 1A).
FIG 1.
Survival of isolates following phagocytosis by RAW 264.7 and THP-1 macrophages, assessed by gentamicin protection assay. (A) Survival of isolates at 2, 4, 8, and 24 hours postinfection (hpi) following phagocytosis by RAW 264.7 macrophages and at 2, 4, 8, 24, and 168 hpi following phagocytosis by THP-1 macrophages, reported as log10 CFU/ml. (B) Survival of isolates at 4, 8, and 24 hpi following phagocytosis by RAW 264.7 macrophages and at 4, 8, 24, and 168 hpi following phagocytosis by THP-1 macrophages, reported as log2 fold change compared to survival at 2 hpi. Results for isolates in the study correspond to the mean of three independent assays (n = 3) with duplicate technical replicates. Results for reference Salmonella enterica serovar Typhimurium strains 14028S and ST4/74 correspond to the mean of six independent assays (n = 6) with duplicate technical replicates. (C) Survival at 2, 4, 8, 24, 48, 72, 96, 120, 144, and 168 hpi of S. enterica Tennessee CFSAN001387, in comparison with S. Typhimurium ST4/74, following phagocytosis by THP-1 macrophages, reported as log10 CFU/ml. (D) Survival at 4, 8, 24, 48, 72, 96, 120, 144, and 168 hpi of S. Tennessee CFSAN001387, in comparison with S. Typhimurium ST4/74, following phagocytosis by THP-1 macrophages, reported as log2 fold change compared to survival at 2 hpi. Results correspond to the mean of three independent assays (n = 3) with triplicate technical replicates. The lower and upper hinges correspond to the 25th and 75th percentiles, with the whiskers extending ±1.5 times the range between first and third quartiles.
The highest and lowest mean CFU/ml values for all 10 atypical serovars are shown (see Table S2 in the supplemental material). When the log2 fold change was calculated, the largest decreases were recorded for S. Weltevreden CFSAN001415, at 8 and 24 hpi (Fig. 1B). Of note, S. Bareilly CFSAN001111 showed an unusual infection profile in that it had the highest mean CFU/ml at 2 hpi, possibly indicative of increased invasiveness, despite normalization of the infections by centrifugation of the cell culture plates. The latter observation was maintained for all subsequent time points. However, the mean CFU/ml values at 4, 8, and 24 hpi for S. Bareilly CFSAN001111 were lower than those for other isolates.
Compared to S. Typhimurium ST4/74, significant differences were observed in the infection profile of specific isolates at individual time points (see Table S3 in the supplemental material). Time courses longer than 24 h were not possible in RAW 264.7 macrophages, as this resulted in cell death or proliferation of the macrophages themselves that would skew the MOI. Although differences between these isolates were observed in RAW 264.7 macrophages, the inability to consistently extend the time course of the assay beyond 24 hpi limited the utility of this ex vivo murine model to discern any further differences in the ability of the isolates to survive and persist following phagocytosis. To mitigate this limitation and differentiate the isolates in a human-relevant context, additional assays were carried out in differentiated THP-1 macrophages.
Of the 10 atypical serovars tested in this study, all isolates persisted within THP-1 macrophages for 24 hpi, with the majority exhibiting no significant changes in viable numbers up to this time point. As with RAW 264.7 macrophages, the only exception noted when infecting THP-1 macrophages was recorded for S. Weltevreden CFSAN001415, which exhibited a 1-log10 decrease in mean CFU/ml between 2 and 24 hpi but remained recoverable. Upon extending this assay beyond 24 h to 168 hpi, equivalent to 7 days, all strains except for S. Anatum CFSAN003959 and the S. Typhimurium 14028S and ST4/74 reference strains were recoverable, with the majority exhibiting a 2-log10 decrease in intracellular bacteria. Exceptions to this were noted for S. enterica Cubana CFSAN002050, S. enterica Heidelberg CFSAN002063, and S. Tennessee CFSAN001387, all of which showed a 1-log10 decrease in intracellular bacteria between 2 and 168 hpi, with S. Tennessee CFSAN001387 displaying the smallest decrease of all the isolates. Conversely, S. Weltevreden CFSAN001415 demonstrated the largest decrease in bacterial cell numbers at 168 dpi, but unlike S. Anatum CFSAN003959 and both S. Typhimurium 14028S and ST4/74 reference strains, it could still be recovered at the end of the assay (Fig. 1A).
The highest and lowest mean CFU/ml values among the 10 study isolates are shown (Table S2). In THP-1 macrophages, S. Weltevreden CFSAN001415 had the lowest mean CFU/ml values at 24 and 168 hpi of the recoverable isolates, as observed by the log2 fold change (Fig. 1B). As observed in RAW 264.7 macrophages, S. Tennessee CFSAN001387 had the highest mean CFU/ml value at 24 hpi, as well as at 168 hpi, in THP-1 macrophages. Compared to S. Typhimurium ST4/74, significant differences were observed in the infection profiles of specific isolates at individual time points (Table S2).
Fewer significant differences were observed between isolates compared to S. Typhimurium ST4/74 infection in human THP-1 macrophages versus murine RAW 264.7 macrophages, further highlighting the potential unsuitability of the murine model for inferring human-relevant differences between isolates in NTS infection. Overall, the viable intracellular bacteria recorded at later time points, including at 24 hpi, was significantly higher in RAW 264.7 than THP-1 macrophages. Bacterial cell numbers reached as high as 1 × 107 mean CFU/ml in RAW 264.7 macrophages for some atypical serovars, compared with values that did not exceed 1 × 105 mean CFU/ml in THP-1 macrophages for all isolates. This 2-log10 difference supports our observations showing the inability of RAW 264.7 macrophages, compared with that of THP-1 macrophages, to clear infecting bacteria.
The viability of both murine and human macrophages following infection with each of the selected bacterial isolates was measured using colorimetric assays to measure extracellular glucose 6-phosphate (G6P) and lactate dehydrogenase (LDH) activities compared with those of uninfected control macrophages. No significant differences were observed in host cell viability following an MOI of 10:1 (see Fig. S4 and S5 in the supplemental material). This is in agreement with recent studies that used flow cytometry-based techniques to quantify apoptosis in macrophages infected with different Salmonella strains (33, 39).
As S. Tennessee CFSAN001387 was noted to be the most proliferative isolate in THP-1 macrophages, exhibiting the lowest reduction in viable intracellular bacteria between 2 and 168 hpi, further assays were performed to directly compare to S. Typhimurium ST4/74, with CFU being enumerated at 2, 4, 8, 24, 48, 72, 96, 120, 144, and 168 hpi. This was done to determine when S. Typhimurium ST4/74 was no longer recoverable compared with S. Tennessee CFSAN001387. Salmonella Typhimurium ST4/74 exhibited an overall 1-log10 reduction between 2 and 48 hpi, with an additional 2-log10 reduction in viable bacteria by 72 h. After 96 and 120 hpi, S. Typhimurium ST4/74 was barely detected and was unrecoverable at 144 and 168 hpi. In comparison, S. Tennessee CFSAN001387 exhibited an overall 1-log10 reduction between 2 and 168 hpi, similar to that in the previous infections (Fig. 1C and D). Differences between these two isolates were significant at multiple individual time points (see Table S4 in the supplemental material).
Host response to atypical S. enterica infection by murine and human macrophages.
As there were notable differences exhibited by these Salmonella serovars in their ability to replicate and survive within murine and human macrophages, the host response to infection was investigated by quantifying proinflammatory cytokine and infection-relevant chemokine release using standard immunoassay protocols.
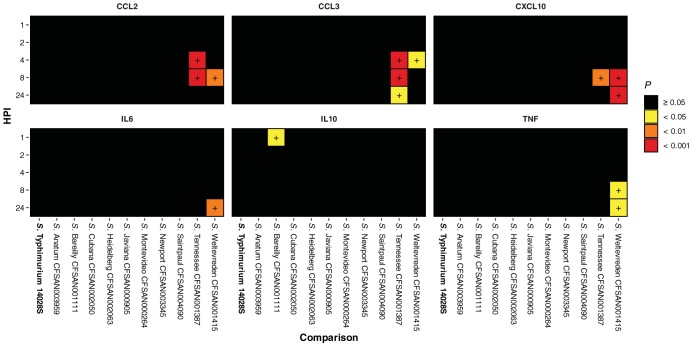
In RAW 264.7, increased CCL2 and CCL3 chemokine release was observed at 4, 8, and 24 hpi with S. Tennessee CFSAN001387 and S. Weltevreden CFSAN001415 compared to that in uninfected control macrophages. When comparing infection with these 10 atypical serovars (Table 1) to that with S. Typhimurium ST4/74, increased proinflammatory cytokine (including interleukin 6 [IL-6], IL-10, and tumor necrosis factor [TNF]) and chemokine (including CCL2, CCL3, and CXCL10) release was observed (Fig. 2; see also Table S5 and Fig. S6 in the supplemental material).
FIG 2.
Significant RAW 264.7 chemokine and proinflammatory cytokine release following infection with isolates compared to that following S. Typhimurium ST4/74 infection. Increased release of chemokine and proinflammatory cytokine proteins by RAW 264.7 macrophages at 1, 2, 4, 8, and 24 hpi with isolates compared to that following infection with S. Typhimurium ST4/74, as determined by one-way analysis of variance (ANOVA). Values correspond to adjusted probability (P) as determined by post hoc analysis of significance using Tukey’s range test. Differences were deemed significant by arbitrary cutoffs at P < 0.05, 0.01, and 0.001. Quantification was determined using a multiplex magnetic bead-based enzyme-linked immunosorbent assay (ELISA) and the Luminex 200 xMAP platform (see Table S7 in the supplemental material).
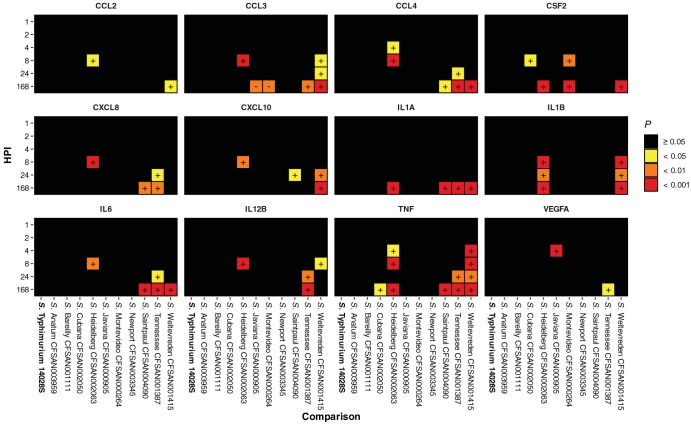
In THP-1, increased proinflammatory cytokine (including CXCL8, IL-1B, IL-6, and TNF), cytokine (including CSF2, IL-1A, IL-12B, and VEGFA), and chemokine (including CCL2, CCL3, CCL4, and CXCL10) release was recorded at 8, 24, and 168 hpi across a selection of these isolates in comparison with that in uninfected control macrophages. As with RAW 264.7 macrophages, when comparing infection with the 10 atypical serovars in this study to S. Typhimurium ST4/74 in THP-1 macrophages, significant increased proinflammatory cytokine and chemokine release was observed, further differentiating the isolates by the innate host response (Fig. 3 and Table S5).
FIG 3.
Significant THP-1 chemokine, cytokine, and proinflammatory cytokine release following infection with isolates compared to that following S. Typhimurium ST4/74 infection. Increased release of chemokine, cytokine, and proinflammatory cytokine proteins by THP-1 macrophages at 1, 2, 4, 8, 24, and 168 hpi with isolates compared to that following infection with S. Typhimurium ST4/74, as determined by one-way ANOVA. Values correspond to adjusted probability (P) as determined by post hoc analysis of significance using Tukey’s range test. Differences were deemed significant by arbitrary cutoffs at P < 0.05, 0.01, and 0.001. Quantification was determined using an electrochemiluminescence-based ELISA method and the Sector Imager 2400 platform (Meso Scale Discovery) (Table S7).
Salmonella serovar Heidelberg CFSAN002063 stimulated the release of CCL3, CSF2, CXCL8, CXCL10, IL-1A, IL-1B, IL-6, IL-12, and TNF to levels in excess of those observed for S. Typhimurium ST4/74, particularly at 8 hpi and from 4 to 168 hpi with respect to TNF (see Fig. S7 in the supplemental material). In the gentamicin protection assays, it was noted that although both S. Tennessee CFSAN001387 and S. Weltevreden CFSAN001415 could be recovered at 168 hpi from THP-1 macrophages, these two isolates exhibited different infection profiles. The former displayed the smallest reduction in viable intracellular bacteria over the time course of the infection, whereas the latter displayed the largest reduction in viable numbers. This can be accounted for in the overlapping yet contrasting cytokine profile of THP-1 macrophages following infection with these isolates. Salmonella Tennessee CFSAN001387 and S. Weltevreden CFSAN001415 stimulated the release of CCL3, CCL4, IL-1A, IL-6, IL-12B, and TNF to levels higher than those observed for S. Typhimurium ST4/74 (Fig. 4). In addition, S. Weltevreden CFSAN001415 stimulated the release of CCL2, CXCL10, CSF2, and IL-1B, triggering a broader response than that observed for S. Tennessee CFSAN001387 (Fig. 4).
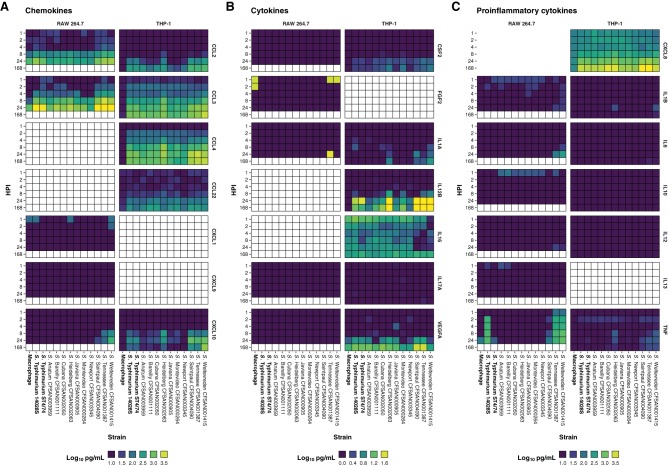
FIG 4.
Chemokine, cytokine, and proinflammatory cytokine release from infected RAW 264.7 and THP-1 macrophages. Release of chemokine (A), cytokine (B), and proinflammatory cytokine (C) targets by RAW 264.7 and THP-1 macrophages following infection with isolates, reported as log10 pg/ml. Cells colored white correspond to time points not assayed with respect to protein or cell line. Quantification was determined as described in the “Cytokine quantification” section of Materials and Methods (Table S7).
Overall, the human macrophages mounted a much greater proinflammatory response to infection in comparison to that in the murine model (Fig. 4). A homolog of the human CXCL8 gene is absent in mice. However, the murine Cxcl1 gene codes for a functionally homologous protein (40). In this study, the latter was not released from infected RAW 264.7 macrophages at the levels observed for CXCL8 in THP-1 macrophages. As all infections were carried out in pure macrophage cultures, the true effect of the observed chemokine release cannot be fully appreciated in this experimental model, as the activation and/or recruitment of other phagocytic cells in a coculture population or in an in vivo environment would have an impact on bacterial survival.
Distribution and similarity of SPI proteins from atypical S. enterica serovars in comparison to S. Typhimurium ST4/74.
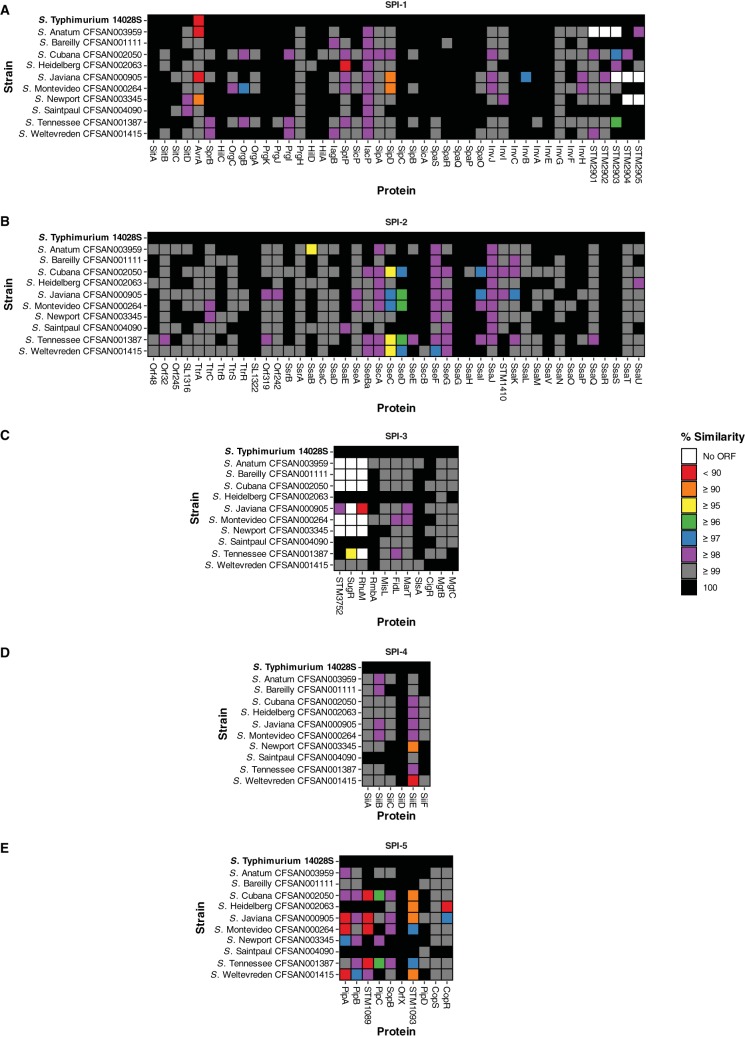
Whole-genome sequencing was used to facilitate a comparative analysis of all 10 atypical serovar isolates to elucidate the genetic variation among them, with particular focus on virulence determinants and SPI locus gene content (41, 42). Sequencing was performed using the Illumina MiSeq and Pacific Biosciences RS II sequencing platforms. De novo assemblies were performed on these data, and the N50 length for Illumina-sequenced assemblies ranged from 403 to 763 kbp, with an average N50 length of 586 kbp (see Table S6 in the supplemental material). These genome sequences were used to determine the relationships between the isolates to identify key differences that may explain the phenotypes observed in the previous experiments. Genetic diversity was characterized by amino acid sequence similarity, which was used to identify SPI regions that were highly conserved between these isolates despite the broad range of serovars, in addition to distinct variable regions and/or the absence of key effector proteins in specific isolates (Fig. 5; see also Fig. S8 in the supplemental material).
FIG 5.
Sequence variability within SPI-1 to SPI-5. The amino acid sequence similarity for the SPI gene content of each isolate included in this study was compared with that of S. Typhimurium ST4/74.
SPI-1 and its associated T3SS have been extensively implicated in Salmonella virulence and in the ability of this pathogen to invade host eukaryotic cells, trigger inflammation, and transport effector proteins (43, 44). Sequence variation at the amino acid level in comparison to S. Typhimurium ST4/74 was greatest in the AvrA, OrgB, SptP, SipD, InvB, and SL2883 proteins. Several SPI-1encoded genes were absent in many of the atypical serovars but present in S. Typhimurium ST4/74, although not yet fully characterized (Fig. 5).
The AvrA protein has been previously shown to be crucial, playing a role in the inhibition of the antiapoptotic NF-κB pathway (45). In S. Typhimurium 14028S, S. Anatum CFSAN003959, S. Javiana CFSAN000905, and S. Newport CFSAN003345, AvrA showed differences at the amino acid sequence level that may affect its ability to function as a protease (46). SptP is a tyrosine phosphatase involved in the inhibition of Raf activation and the subsequent MAP kinase pathway (47). Its identification here is consistent with the phenotype observed for S. Heidelberg CFSAN002063 that stimulated high levels of TNF release at 4, 8, 24, and 168 hpi in THP-1 macrophages. Many proteins that are SPI-1 associated in S. Typhimurium ST4/74, including STM2901, STM2902, STM2903, SL2883, STM2904, and STM2905, showed mixed conservation, being either highly similar in the majority of isolates or unidentifiable (in the cases of S. Anatum CFSAN003959, S. Javiana CFSAN000905, and S. Newport CFSAN003345).
SPI-2 and its associated T3SS contributes to the ability of Salmonella to translocate effectors across the membrane of the SCV when the bacterium is internalized in epithelial cells and macrophages (48, 49). The integral function of SPI-2 for intracellular survival can be observed by the degree of amino acid sequence conservation across all atypical serovars (Fig. 5). As these isolates were implicated in multistate foodborne outbreaks and, as demonstrated above, are capable of surviving within both RAW 264.7 and THP-1 macrophages, this observation is consistent with the expressed phenotype and raises the question as to whether potential differences in the expression of some or all of these genes may further explain the differences shown in intracellular survival. Sequence variation at the amino acid level in comparison to S. Typhimurium ST4/74 was greatest in SsaB, SseB, SseC, and SseD. Loss of the effector protein SsaB (SpiC) has been shown to promote defective virulence phenotypes due to an inability to translocate all SPI-2 effectors (50). This may explain the observed infection phenotype in S. Anatum CFSAN003959, the only isolate among the 10 atypical serovars studied that was unrecoverable at 168 hpi in THP-1 macrophages, as it had the lowest amino acid sequence similarity for SsaB.
SseBCD proteins function as a translocon that facilitates the secretion of effector proteins by intracellular Salmonella (51). Both S. Cubana CFSAN002050 and S. Tennessee CFSAN001387 display differences in all three SseBCD proteins, with S. Tennessee CFSAN001387 being least similar at the amino acid sequence level compared to S. Typhimurium ST4/74. Salmonella serovar Tennessee CFSAN001387 was shown in this study to survive within THP-1 macrophages significantly better than S. Typhimurium ST4/74.
High levels of similarity to S. Typhimurium ST4/74 were observed for SPI-3 across the isolates, with the exception of the complete loss or major differences in STM3752, SugR, and RhuM, as reported previously (41). SPI-4 and its associated type I secretion system (T1SS) have been implicated in adhesion, contributing to intestinal inflammation in animal models (52, 53). S. Newport CFSAN003345 and S. Weltevreden CFSAN001415 exhibited low levels of amino acid sequence similarity to S. Typhimurium ST4/74 with respect to SiiE, which has been shown to be important for persistent infection in macrophages (54). SPI-5 encodes many SPI-1 and SPI-2 T3SS targeted effector proteins. Sequence variation was observed for CopR, PipA, STM1089, and STM1093 in multiple isolates, with mutations in PipA having previously been implicated in enteric salmonellosis (55).
DISCUSSION
Studies aimed at elucidating the host response to NTS serovars, including S. Enteritidis and S. Typhimurium, have been largely facilitated by the availability of ex vivo cell culture and in vivo murine infection models. This study focused on atypical serovars of this genus, utilizing isolates cultured from foodborne outbreaks, the majority of which are frequently listed in the top 20 serovars reported annually by the CDC as responsible for laboratory-confirmed human cases of salmonellosis (https://www.cdc.gov/nationalsurveillance/salmonella-surveillance.html).
A number of in vitro experiments were performed that were designed to mimic the immediate challenges faced by Salmonella enterica upon entering a human host following the consumption of contaminated food sources, namely, tolerance to low pH and to bile, which are encountered in the stomach and lower intestine, respectively. Across the board for the panel of atypical Salmonella serovar isolates that were assayed, minimal differences were observed in the ability to survive low pH, with significant reductions in viable CFU recorded at 4 h (Fig. S1). In the context of an in vivo infection, this represents sufficient acid tolerance in the majority of cases, as food will begin to pass to the lower intestines, where the remaining Salmonella bacteria that have survived will focus on outgrowing the host microbiota. With respect to bile tolerance, the only significant difference that was observed among the study isolates was the increased susceptibility to DOC of S. Weltevreden CFSAN001415 compared to that of other isolates (Table S1). Salmonella serovar Weltevreden has begun to emerge as a dominant foodborne pathogen with global distribution (28, 57). However, this bile susceptibility, coupled with characteristically minimal antibiotic resistance characteristics for this serovar, may explain the limited number of outbreaks observed compared to those caused by more virulent S. Heidelberg isolates.
The ability of these atypical S. enterica serovar isolates to survive within macrophages was assayed using both murine 264.7 and human THP-1 cell line models of infection. In the murine model of infection, the most notable difference observed for bacterial viability was a 1-log10 reduction in CFU/ml for S. Weltevreden CFSAN001415 between 2 and 24 hpi (Fig. 1A). This was in contrast to results for S. Tennessee CFSAN001387, which represented the largest increase in the number of viable internalized bacteria; these two isolates exhibited the extremes of variability among the panel of isolates in this model of infection (Fig. 1A). It was not possible to consistently perform infections with a time course longer than 24 hpi for the RAW 264.7 cell line, which limited the utility of this model to study long-term intracellular survival of Salmonella isolates.
To remedy this limitation with a view to gaining a more relevant appreciation of any potential phenotypic differences, further gentamicin protections assays were performed using the human THP-1 differentiated macrophage cell line. In this ex vivo model of infection, it was possible to extend the time course beyond 24 hpi to 168 hpi (7 days postinfection), allowing for the viable intracellular CFU to be tracked closely for both S. Typhimurium ST4/74 and S. Tennessee CFSAN001387 (Fig. 1C). The environmental isolate S. Tennessee CFSAN001387 survived with the human THP-1 macrophages for up to 168 hpi, and an overall 1-log10 reduction in CFU relative to 2 hpi was recorded. This was not the case for S. Typhimurium ST4/74, which could only be recovered at decreased numbers for the 120 hpi time point and was no longer detected following the lysis of viable THP-1 macrophages at 144 and 168 hpi, indicating that the infection had been cleared in this closed system (Fig. 1D).
An aspect of the host response to infection that was further investigated as part of this study was the release of proinflammatory cytokines and infection-relevant chemokines from infected macrophages. When assaying these targets from infected murine RAW 264.7 macrophages, statistically significant increased release of CCL2 and CCL3 chemokines along with several proinflammatory cytokine targets was observed following infection with S. Typhimurium ST4/74 relative to uninfected macrophage controls, congruent with existing literature. Of interest from this study was that similar increases were observed to a much greater degree from both S. Tennessee CFSAN001387 and S. Weltevreden CFSAN001415 compared with those observed in infection by the reference strain S. Typhimurium ST4/74 (Fig. 2 and Table S5).
Taking into consideration that a homolog for the important macrophage chemokine CXCL8 is absent from the innate immune system of mice, further experiments were performed to quantify the proinflammatory cytokine and infection-relevant chemokine release from infected human THP-1 macrophages using supernatants from the cell culture infections carried out in this study (Fig. 3). It was observed that a much broader response to infection with the panel of atypical Salmonella enterica serovar isolates was mounted by the human macrophages, especially with respect to a number of proinflammatory cytokine targets (Fig. 4). Of note, S. Heidelberg CFSAN002063 stimulated the release of the most targets, representing a statistically significant increase relative to that in infection with S. Typhimurium ST4/74, especially at 4 and 8 hpi. This may begin to explain the much larger number of foodborne outbreaks by this serovar that result in hospitalizations compared with the other atypical serovars included in this study, as well as the clinical manifestation, which is reported from these cases as being a more violent illness rather than the gastroenteritis caused by other Salmonella enterica serovars. Whereas other isolates from the atypical serovars included in this study managed to fly under the radar of the host innate immune system, causing minimal or no difference in proinflammatory cytokine or infection-relevant chemokine release compared to that in uninfected macrophage controls or S. Typhimurium ST4/74 infection, S. Weltevreden CFSAN001415 triggered a broader response among infected human THP-1 macrophages. This led to a statistically significant increase in the release of several proinflammatory cytokines, especially at later time points in the infection course, such as 168 hpi. This is notable because S. Weltevreden CFSAN001415 exhibited the largest decrease in the number of viable intracellular bacteria between 2 and 168 hpi (of the isolates that could still be recovered) compared with S. Tennessee CFSAN001387, which exhibited the smallest decrease in viable intracellular CFU, suggesting that a much greater immune response is mounted against S. Weltevreden, leading to the infection being cleared at a greater rate (Fig. 1B, 3, and 4).
One limitation of the experimental infections performed as part of this study was that pure cultures of macrophage cell lines, both RAW 264.7 and THP-1, were used. It would be prudent to include coculture infection models with multiple cell lines as future work in this area, given that a number of chemokine signals were released from the infected macrophages, which have been shown in previous literature to be involved in recruiting other host cell types to the site of infection. This limitation may also influence the results obtained for the proinflammatory cytokine and infection-relevant chemokine data, as this closed cell culture system does not represent the true turnover of these signal markers in an in vivo system. For longer incubation periods, to limit the manipulation of the infected macrophages, this could lead to a local accumulation of a given cytokine target beyond what is typically observed in an in vivo infection.
A comparative analysis of the underlying genetic differences that may be harbored by these isolates was performed following whole-genome sequencing. These analyses focused on known virulence determinants of the Salmonella genus, namely, a panel of SPIs. Both SPI-1 and SPI-2 harbor a T3SS, and several differences in these loci were identified among the genomes of the atypical S. enterica serovar isolates compared with the S. Typhimurium ST4/74 reference genome (Fig. 5A and B). In SPI-1, the protease AvrA showed amino acid-level differences, with <90% sequence similarity compared with S. Typhimurium ST4/74, which could explain why this isolate, along with the reference strains S. Typhimurium 14028S and ST4/74, could not be recovered from long-term infections in human THP-1 macrophages (Fig. 1C and 5A). A number of isolates, which included S. Cubana CFSAN002050, S. Javiana CFSAN000905, S. enterica serovar Montevideo CFSAN000264, S. Tennessee CFSAN001387, and S. Weltevreden CFSAN001415, showed the greatest genetic diversity among the SseBCD proteins, which encode a translocon facilitating the secretion of effector proteins by phagocytosed Salmonella. These isolates were frequently among those that exhibited interesting infection phenotypes by way of long-term survival and/or an increased response to infection by the human macrophages, as determined by proinflammatory cytokine and infection-relevant chemokine release. These loci and the observed phenotypes suggest a number of hypotheses that would warrant further investigation as to their potential relatedness.
These data allowed identification of a number of statistically significant phenotypic differences, as well as of the underlying genetic diversity, among the isolates relative to the S. Typhimurium reference strains that could potentially explain their differing and increased ability to survive within macrophage infection models or to cause an increased innate immune response. These experiments have highlighted the potential unsuitability of the widely used murine RAW 264.7 macrophage infection model to infer human-relevant distinctions between isolates. We have shown that for NTS serovars, differences in the inflammatory response of human macrophages can further differentiate these microorganisms in a manner that was not possible with murine macrophages (Fig. 3). In the case of specific proinflammatory cytokines such as TNF, RAW 264.7 macrophages responded to infection with S. Typhimurium 14028S and ST4/74, S. Tennessee CFSAN001387, and S. Weltevreden CFSAN001415, whereas THP-1 macrophages displayed a broader proinflammatory response to the range of serovars studied (Fig. 4).
Furthermore, the reference strains S. Typhimurium 14028S and ST4/74 that are often included in in vitro research emerged as the biological outliers in many respects with regard to their infection phenotype (Fig. 1). It is possible that these “outlier” characteristics explain why S. Typhimurium and S. Enteritidis are so successful at infecting individuals and causing foodborne outbreaks, as together these serovars represent the majority of NTS illnesses. It would be advantageous for Salmonella to not cause a self-limiting illness in its host, as can be seen with commensal serovars of Salmonella enterica found in poultry that do not necessarily lead to illness. In contrast with this “under the radar” strategy, we saw many examples in this study where S. Heidelberg CFSAN002063 induced such a flagrant proinflammatory response that the prospects for a prolonged illness where the bacteria can engage in a long-term intracellular lifestyle among host epithelial cells is limited. These can be seen from the clinical manifestations of S. Heidelberg, which typically represent severe gastroenteritis over a shorter timespan than that caused by more “benign” serovars of Salmonella. Similarly, the advantages of the biological “outlier” characteristics observed in S. Typhimurium can be seen to a lesser extent in both S. Tennessee CFSAN001387 and S. Weltevreden CFSAN001415 when considering their infection profiles and associated host responses. Both isolates could be recovered following long-term infection past the point that S. Typhimurium 14028S and ST4/74 were recoverable. In the case of S. Tennessee CFSAN001387, this isolate represented the lowest decrease in the number of viable intracellular bacteria between 2 and 168 hpi and induced a lesser proinflammatory cytokine and infection-relevant chemokine response than that of S. Weltevreden CFSAN001415, which presented a much broader host immune response and exhibited the greatest decrease in the number of viable intracellular bacteria between 2 and 168 hpi.
These data suggest more differences than previously acknowledged for S. enterica serovars, with broad implications for public health. In agreement with this notion, potential key differences were identified in established virulence determinants of Salmonella, such as SPI gene content, which may begin to bridge the gap between the observed phenotypes and the underlying genotypes following further study into these generated hypotheses. Further work will be required to understand the full scope of other potential targets within the genomes of these isolates, such as the accessory gene content unique to individual strains, many of which are currently poorly characterized.
MATERIALS AND METHODS
Bacterial isolates and culture methods.
All Salmonella isolates were stored at −80°C in lysogeny broth (LB) broth (Sigma-Aldrich) supplemented with 15% (vol/vol) glycerol. Working cultures were prepared by streaking isolates and restreaking individual colonies onto Mueller-Hinton (MH) agar (Sigma-Aldrich). Individual colonies from restreaked isolates were used to inoculate 5 ml MH broth (Sigma-Aldrich) and grown overnight at 37°C with orbital shaking at 200 rpm. Overnight cultures were then used in the subsequent experiments as detailed below.
Acid resistance.
The ability to survive in a low-pH culture medium was assessed for isolates as described previously (58). Briefly, bacterial cultures were individually grown overnight (18 h) without shaking in 5 ml buffered MH broth (morpholineethanesulfonic acid [MES] hydrate, 2% [wt/vol], pH 5) containing 0.4% (wt/vol) glucose at 37°C. A volume of 333 μl of overnight culture was centrifuged at 10,000 rpm for 10 min. The supernatant was removed, and the cell pellet was recovered and resuspended in 2 ml prewarmed, buffered MH broth (MES hydrate, 2% [wt/vol], pH 2.5) containing 0.4% (wt/vol) glucose and incubated at 37°C in a 24-well plate. Time point measurements were taken at 0 h (before acidification), 1, 2, and 4 h postacidification. At each time point, 10 μl of the culture was diluted in 1,990 μl of maximum recovery diluent (MRD) medium (Oxoid) in a fresh 24-well plate and incubated for 30 min at room temperature to allow the bacteria to recover. Samples were then decimally diluted in phosphate-buffered saline (PBS; Sigma-Aldrich), and 100-μl aliquots of these dilutions were plated directly onto LB agar. Agar plates were incubated for 18 h at 37°C before enumeration of the CFU.
Motility assays.
Swim and swarm motility were assessed for isolates as described previously (58). Briefly, swim motility plates (MH broth containing 0.3% [wt/vol] agar) were stab inoculated. Swarm motility plates (MH broth containing 0.6% [wt/vol] agar) were inoculated by spotting 1 μl of overnight culture. Inoculated plates were then incubated at 21°C (ambient room temperature) for 8 and 24 h and at 37°C for 8 h. The diameter of visible colony spread was measured in mm from three directions and the average value recorded.
Tolerance to bile salts.
The MIC and MBC values for sodium deoxycholate (DOC) and sodium cholate was determined for isolates using the broth dilution method according to Clinical and Laboratory Standard Institute (CLSI) guidelines adapted from the protocol as described previously (59).
Bacterial growth curves.
The growth of isolates in LB broth was assessed using a Multiskan FC microplate photometer (Thermo Fisher Scientific). Measurements were taken every 15 min over 24 h of the optical density at 620 nm (OD620). The instrument was kept at 37°C with shaking during kinetic intervals.
Preparation of bacterial inoculum for infection.
The MIC and MBC for gentamicin (CN) were determined for isolates using the broth dilution method according to CLSI guidelines. Isolates were classified as susceptible or resistant according to the lower working concentration of 20 μg/ml used in the gentamicin protection assays detailed below.
Inoculum stocks for each isolate were prepared by streaking and restreaking individual bacterial isolates onto LB agar. Individual colonies from restreaked isolates were taken and used to inoculate 5 ml LB broth before growth overnight at 37°C with orbital shaking at 200 rpm. Overnight cultures were centrifuged at 5,500 relative centrifugal force (RCF) for 10 min. The supernatant was discarded, and the bacterial cell pellet was resuspended in 5 ml PBS before centrifugation again at 5,500 RCF for 10 min. Finally, the pellet was resuspended in 5 ml of PBS solution (15% [vol/vol] glycerol) before aliquoting (250 μl/microcentrifuge tube) and freezing at −80°C. Representative inoculum stocks for each isolate were decimally diluted in PBS, and 100-μl aliquots of the dilutions were plated onto LB agar. Agar plates were incubated for 18 h at 37°C before enumeration of CFU.
Ex vivo gentamicin protection assay.
The ability to survive and proliferate following phagocytosis by murine RAW 264.7 (Sigma-Aldrich) and human THP-1 (Sigma-Aldrich) macrophages was assessed for gentamicin-susceptible isolates adapted from protocols described previously using S. Typhimurium 14028S and ST4/74 as reference strains (36, 60).
RAW 264.7 macrophages were grown in antibiotic-free Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich) and incubated at 37°C in a humidified atmosphere with 5% CO2. THP-1 monocytes were grown in antibiotic-free RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% (vol/vol) heat-inactivated FBS and incubated at 37°C in a humidified atmosphere with 5% CO2. Cell viability was assessed using trypan blue and a TC20 automated cell counter (Bio-Rad).
Cells were subcultured, and 1 ml was directly seeded into 24-well plates at a density of 1 × 105 cells/ml per well. THP-1 monocytes were differentiated to adherent macrophages by supplementing media with 20 ng/ml phorbol 12-myristate 13-acetate (PMA) for 5 days.
Prior to infection, inoculum stocks for each bacterial isolate to be assessed were diluted in complete medium to 1 × 106 bacteria/ml for an MOI of 10:1 and incubated at 37°C for 1 h. Macrophages were washed 3 times with 1 ml Hanks’ balanced salt solution (HBSS) before 1 ml of the bacterial suspension, prepared as outlined above, was added to each well; 1 ml of complete medium was added to uninfected control wells. These 24-well plates were centrifuged at 300 RCF for 5 min at room temperature (21°C) before incubation at 37°C with 5% CO2 for 1 h to allow for phagocytosis.
Following phagocytosis, the cells were washed 3 times with 1 ml HBSS. A volume of 1 ml complete medium supplemented with 100 μg/ml gentamicin was added to each well before incubation at 37°C with 5% CO2 for 1 h to kill external bacteria. After 1 h, cells were washed 3 times with 1 ml HBSS. Another volume of 1 ml of complete medium supplemented with 20 μg/ml gentamicin was then added to each well before incubation at 37°C with 5% CO2 until the desired time points.
Cultures were processed at each time point by washing the cells 3 times with 1 ml HBSS before 1 ml 1% (vol/vol) Triton X-100/PBS solution was added to the infected cells prior to incubation at room temperature for 10 min. Lysed supernatants were decimally diluted in PBS, and 100-μl aliquots of the dilutions were plated onto LB agar. Agar plates were incubated for 18 h at 37°C before enumeration of the CFU.
Relative viability assay for RAW 264.7 and THP-1 macrophages by glucose-6-phosphate dehydrogenase and lactate dehydrogenase.
Relative viability of infected macrophages was determined by comparison to uninfected control cells using the Vybrant cytotoxicity assay kit (Life Technologies) and Pierce LDH cytotoxicity assay kit (Life Technologies) to measure extracellular glucose-6-phosphate dehydrogenase (G6PD; EC 1.1.1.49) and lactate dehydrogenase (LDH; EC 1.1.1.27) activity in cell culture supernatants according to manufacturer’s instructions. All samples and standards were assayed in duplicate.
Cytokine quantification.
A panel of proinflammatory cytokines and infection-relevant chemokines were quantified from the supernatants of infected RAW 264.7 and THP-1 macrophages. For RAW 264.7 supernatants, targets were quantified at 0 h (before infection) and at 1, 2, 4, 8, and 24 hpi. For THP-1 supernatants, targets were quantified at 0 h (before infection) and at 1, 2, 4, 8, 24, and 168 hpi.
RAW 264.7 cytokine/chemokine release was measured using a multiplex magnetic bead-based kit (Life Technologies) and the 200 xMAP platform (Luminex). Similarly, THP-1 cytokine/chemokine release was measured using an electrochemiluminescence-based V-PLEX kit (Meso Scale Discovery) and the Sector Imager 2400 platform (Meso Scale Discovery). The measured levels of many targets were below the range of detection in RAW 264.7 supernatant samples and for the purposes of this study were recorded at the lower level of detection for the assay used when comparing to S. Typhimurium ST4/74 infection or infected THP-1 macrophages. Assays were performed according to the manufacturer’s protocol for cell culture supernatant samples. All samples and standards were assayed in duplicate.
Bacterial whole-genome sequencing.
Whole-genome sequencing was carried out as described previously (61). Briefly, genomic DNA (gDNA) was purified from overnight cultures of bacterial isolates grown in Trypticase soy broth (TSB; Becton, Dickinson) incubated at 37°C using the DNeasy blood and tissue kit (Qiagen). Libraries were prepared using 1 ng gDNA with the Nextera XT kit (Illumina) and sequenced using the MiSeq platform (Illumina) with a v2 kit (2 × 250 bp).
For single-molecule real-time (SMRT) sequencing on the PacBio RS II platform, libraries using 6 μg gDNA were sheared to a size of 10 kb using g-Tubes (Covaris Inc., Woburn, MA) according to the manufacturer’s instructions. The SMRTbell 10-kb template libraries were constructed using a DNA template prep kit 1.0 with the 10-kb insert library protocol (Pacific Biosciences, Menlo Park, CA) and sequenced using P4-C2 chemistry on three SMRT cells with a 240-minute collection protocol along with Stage Start.
Whole-genome assembly and annotation.
For Illumina MiSeq data, Jellyfish (version 2.2.6) was used to generate a k-mer spectrum before inspection of the quality of the reads using FastQC (version 0.11.5) (62, 63). Error correction was performed using BFC (version r181) (64). A relaxed sliding window trim for an average Phred quality score of 10 was performed using Trimmomatic (version 0.36) before the genomes were de novo assembled with SPAdes (version 3.9.0) using the default k-mer size selection and the automatic coverage cutoff threshold (65, 66). The quality of the subsequent assemblies was assessed using Bandage (version 0.8.0) and QUAST (version 4.3) (67, 68). Contigs were excluded from the assembly if they were shorter than 200 bp.
Analysis of the PacBio data was implemented using SMRT Analysis version 2.3.0. The best de novo assembly was established with the PacBio Hierarchical Genome Assembly Process (HGAP) version 3.0 program using the continuous long reads from the three SMRT cells. The assembly outputs from HGAP produced circular contiguous sequences with overlapping regions at the end that can be identified using dot plots in Gepard (version 1.40) (69). Genomes were checked manually for even sequencing coverage. Afterwards the interim consensus sequence was used to determine the final consensus and accuracy scores using the Quiver consensus algorithm (70).
All sequences and assemblies are publicly available, with accession numbers provided where appropriate, and were submitted for annotation using the NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAP) (71).
Sequence analysis.
Salmonella pathogenicity island (SPI) gene content for isolates was compared to that of S. Typhimurium ST4/74. Homologous amino acid sequences for each protein were identified and generated using BLAST+ (version 2.4.0) and Biopython (version 1.68) (72–74). Amino acid sequence similarity was assessed using a Needleman-Wunsch global alignment through the EMBOSS analysis software (version 6.6.0) (75).
Pangenome analysis.
To limit bias among different annotation tools for downstream analyses, additional annotation of all isolates included in this study was performed using Prokka (version 1.11) (76–83). Presence or absence of protein sequences from all strains was determined using the pangenome pipeline Roary (version 3.6.6) (84–87) (see Fig. S9 in the supplemental material). Visualization of the pangenome data was performed using Anvi’o (version 2.0.2) (88, 89).
Statistics.
The R statistical computing environment was used for all statistical analyses (90, 91). Multiple comparisons for normally distributed data were performed by one-way analysis of variance (ANOVA) where appropriate, and post hoc analysis of significance was assessed by Tukey’s range test.
Data availability.
Accession numbers for raw sequencing data are in Table S6 in the supplemental material. Whole-genome sequencing data have been deposited in the Sequence Read Archive (SRA). Individual run accession numbers (SRR) for demultiplexed isolate data are listed in Table S6.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Research IT HPC Service at University College Dublin for providing computational facilities and support that contributed to the research results reported within this paper.
D.H. is supported by the Food Safety Authority of Ireland Newman Fellowship in Food Safety.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crump JA, Luby SP, Mintz ED. 2004. The global burden of typhoid fever. Bull World Health Organ 82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 3.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 4.Acheson D, Hohmann EL. 2001. Nontyphoidal salmonellosis. Clin Infect Dis 32:263–269. doi: 10.1086/318457. [DOI] [PubMed] [Google Scholar]
- 5.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. 2015. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg Infect Dis 21:941–949. doi: 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäumler AJ, Tsolis RM, Ficht TA, Adams LG. 1998. Evolution of host adaptation in Salmonella enterica. Infect Immun 66:4579–4587. doi: 10.1128/IAI.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton Behravesh C, Mody RK, Jungk J, Gaul L, Redd JT, Chen S, Cosgrove S, Hedican E, Sweat D, Chávez-Hauser L, Snow SL, Hanson H, Nguyen T-A, Sodha SV, Boore AL, Russo E, Mikoleit M, Theobald L, Gerner-Smidt P, Hoekstra RM, Angulo FJ, Swerdlow DL, Tauxe RV, Griffin PM, Williams IT, Salmonella Saintpaul Outbreak Investigation Team. 2011. 2008 outbreak of Salmonella Saintpaul infections associated with raw produce. N Engl J Med 364:918–927. doi: 10.1056/NEJMoa1005741. [DOI] [PubMed] [Google Scholar]
- 8.Riesenberg-Wilmes MR, Bearson B, Foster JW, Curtis R. 1996. Role of the acid tolerance response in virulence of Salmonella Typhimurium. Infect Immun 64:1085–1092. doi: 10.1128/IAI.64.4.1085-1092.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella Typhimurium. Microbiology 150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 10.Santos RL, Raffatellu M, Bevins CL, Adams LG, Tükel Ç, Tsolis RM, Bäumler AJ. 2009. Life in the inflamed intestine, Salmonella style. Trends Microbiol 17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat Rev Microbiol 6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 12.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steele-Mortimer O. 2008. The Salmonella-containing vacuole – moving with the times. Curr Opin Microbiol 11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coburn B, Grassl GA, Finlay BB. 2007. Salmonella, the host and disease: a brief review. Immunol Cell Biol 85:112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- 15.Hurley D, McCusker MP, Fanning S, Martins M. 2014. Salmonella-host interactions – modulation of the host innate immune system. Front Immunol 5:481. doi: 10.3389/fimmu.2014.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Bäumler AJ. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect Immun 71:1–12. doi: 10.1128/iai.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kröger C, Dillon SC, Cameron ADS, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hébrard M, Händler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton J. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton J. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Sabbagh SC, Lepage C, McClelland M, Daigle F. 2012. Selection of Salmonella enterica serovar Typhi genes involved during interaction with human macrophages by screening of a transposon mutant library. PLoS One 7:e36643. doi: 10.1371/journal.pone.0036643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srikumar S, Kröger C, Hébrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron ADS, Hokamp K, Hinton J. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 22.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt W-D. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/iai.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hapfelmeier S, Hardt W-D. 2005. A mouse model for S. Typhimurium-induced enterocolitis. Trends Microbiol 13:497–503. doi: 10.1016/j.tim.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun 73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen SJ, Bishop R, Brenner FW, Roels TH, Bean N, Tauxe RV, Slutsker L. 2001. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987–1997. J Infect Dis 183:753–761. doi: 10.1086/318832. [DOI] [PubMed] [Google Scholar]
- 26.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PAD, Weill F-X. 2010. Supplement 2003–2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S, the S. enterica MLST study group. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makendi C, Page AJ, Wren BW, Le Thi Phuong T, Clare S, Hale C, Goulding D, Klemm EJ, Pickard D, Okoro C, Hunt M, Thompson CN, Phu Huong Lan N, Tran Do Hoang N, Thwaites GE, Le Hello S, Brisabois A, Weill F-X, Baker S, Dougan G. 2016. A phylogenetic and phenotypic analysis of Salmonella enterica serovar Weltevreden, an emerging agent of diarrheal disease in tropical regions. PLoS Negl Trop Dis 10:e0004446. doi: 10.1371/journal.pntd.0004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockman HA, Curtiss R. 1990. Salmonella Typhimurium mutants lacking flagella or motility remain virulent in BALB/c mice. Infect Immun 58:137–143. doi: 10.1128/IAI.58.1.137-143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Smith NH, Nelson K, Crichton PB, Old DC, Whittam TS, Selander RK. 1993. Evolutionary origin and radiation of the avian-adapted non-motile salmonellae. J Med Microbiol 38:129–139. doi: 10.1099/00222615-38-2-129. [DOI] [PubMed] [Google Scholar]
- 31.Toguchi A, Siano M, Burkart M, Harshey RM. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol 182:6308–6321. doi: 10.1128/jb.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 33.Lathrop SK, Binder KA, Starr T, Cooper KG, Chong A, Carmody AB, Steele-Mortimer O. 2015. Replication of Salmonella Typhimurium in human monocyte-derived macrophages. Infect Immun 83:2661–2671. doi: 10.1128/IAI.00033-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwan WR, Huang X-Z, Hu L, Kopecko DJ. 2000. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect Immun 68:1005–1013. doi: 10.1128/iai.68.3.1005-1013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaudaux P, Waldvogel FA. 1979. Gentamicin antibacterial activity in the presence of human polymorphonuclear leukocytes. Antimicrob Agents Chemother 16:743–749. doi: 10.1128/aac.16.6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steele-Mortimer O, Méresse S, Gorvel J-P, Toh B-H, Finlay BB. 1999. Biogenesis of Salmonella Typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell Microbiol 1:33–49. doi: 10.1046/j.1462-5822.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- 37.Steele-Mortimer O, Brumell JH, Knodler LA, Méresse S, Lopez A, Finlay BB. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol 4:43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- 38.Malik-Kale P, Winfree S, Steele-Mortimer O. 2012. The bimodal lifestyle of intracellular Salmonella in epithelial cells: replication in the cytosol obscures defects in vacuolar replication. PLoS One 7:e38732. doi: 10.1371/journal.pone.0038732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braukmann M, Methner U, Berndt A. 2015. Immune reaction and survivability of Salmonella Typhimurium and Salmonella Infantis after infection of primary avian macrophages. PLoS One 10:e0122540. doi: 10.1371/journal.pone.0122540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modi WS, Yoshimura T. 1999. Isolation of novel GRO genes and a phylogenetic analysis of the CXC chemokine subfamily in mammals. Mol Biol Evol 16:180–193. doi: 10.1093/oxfordjournals.molbev.a026101. [DOI] [PubMed] [Google Scholar]
- 41.McWhorter AR, Chousalkar KK. 2015. Comparative phenotypic and genotypic virulence of Salmonella strains isolated from Australian layer farms. Front Microbiol 6:12. doi: 10.3389/fmicb.2015.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhanani AS, Block G, Dewar K, Forgetta V, Topp E, Beiko RG, Diarra MS. 2015. Genomic comparison of non-typhoidal Salmonella enterica serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky isolates from broiler chickens. PLoS One 10:e0128773. doi: 10.1371/journal.pone.0128773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehrbar K, Friebel A, Miller SI, Hardt W-D. 2003. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J Bacteriol 185:6950–6967. doi: 10.1128/jb.185.23.6950-6967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hapfelmeier S, Stecher B, Barthel M, Kremer M, Müller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt W-D. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- 45.Collier-Hyams LS, Zeng H, Sun J, Tomlinson AD, Bao ZQ, Chen H, Madara JL, Orth K, Neish AS. 2002. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-κB pathway. J Immunol 169:2846–2850. doi: 10.4049/jimmunol.169.6.2846. [DOI] [PubMed] [Google Scholar]
- 46.Liao AP, Petrof EO, Kuppireddi S, Zhao Y, Xia Y, Claud EC, Sun J. 2008. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLoS One 3:e2369. doi: 10.1371/journal.pone.0002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin SL, Le TX, Cowen DS. 2003. SptP, a Salmonella Typhimurium type III-secreted protein, inhibits the mitogen-activated protein kinase pathway by inhibiting Raf activation. Cell Microbiol 5:267–275. doi: 10.1046/j.1462-5822.2003.t01-1-00274.x. [DOI] [PubMed] [Google Scholar]
- 48.Waterman SR, Holden DW. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol 5:501–511. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- 49.Figueira R, Holden DW. 2012. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158:1147–1161. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 50.Freeman JA, Rappl C, Kuhle V, Hensel M, Miller SI. 2002. SpiC Is required for translocation of Salmonella pathogenicity island 2 effectors and secretion of translocon proteins SseB and SseC. J Bacteriol 184:4971–4980. doi: 10.1128/jb.184.18.4971-4980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikolaus T, Deiwick J, Rappl C, Freeman JA, Schröder W, Miller SI, Hensel M. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J Bacteriol 183:6036–6045. doi: 10.1128/JB.183.20.6036-6045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerlach RG, Jäckel D, Stecher B, Wagner C, Lupas A, Hardt W-D, Hensel M. 2007. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell Microbiol 9:1834–1850. doi: 10.1111/j.1462-5822.2007.00919.x. [DOI] [PubMed] [Google Scholar]
- 53.Gerlach RG, Jäckel D, Geymeier N, Hensel M. 2007. Salmonella pathogenicity island 4-mediated adhesion is coregulated with invasion genes in Salmonella enterica. Infect Immun 75:4697–4709. doi: 10.1128/IAI.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgan E, Bowen AJ, Carnell SC, Wallis TS, Stevens MP. 2007. SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect Immun 75:1524–1533. doi: 10.1128/IAI.01438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood MW, Jones MA, Watson PR, Hedges S, Wallis TS, Galyov EE. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol Microbiol 29:883–891. doi: 10.1046/j.1365-2958.1998.00984.x. [DOI] [PubMed] [Google Scholar]
- 56. Reference deleted.
- 57.Jain P, Nandy S, Bharadwaj R, Niyogi SK, Dutta S. 2015. Salmonella enterica serovar Weltevreden ST1500 associated foodborne outbreak in Pune, India. Indian J Med Res 141:239–241. doi: 10.4103/0971-5916.155595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martins M, McCusker MP, McCabe EM, O’Leary D, Duffy G, Fanning S. 2013. Evidence of metabolic switching and implications for food safety from the phenome(s) of Salmonella enterica serovar Typhimurium DT104 cultured at selected points across the pork production food chain. Appl Environ Microbiol 79:5437–5449. doi: 10.1128/AEM.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hernández SB, Cota I, Ducret A, Aussel L, Casadesús J. 2012. Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet 8:e1002459. doi: 10.1371/journal.pgen.1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drecktrah D, Knodler LA, Ireland R, Steele-Mortimer O. 2006. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic 7:39–51. doi: 10.1111/j.1600-0854.2005.00360.x. [DOI] [PubMed] [Google Scholar]
- 61.Toro M, Retamal P, Allard M, Brown EW, Evans P, Gonzalez-Escalona N. 2015. Draft genome sequences of 33 Salmonella enterica clinical and wildlife isolates from Chile. Genome Announc 3:e00054-15. doi: 10.1128/genomeA.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon A. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Cambridge, United Kingdom. [Google Scholar]
- 63.Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H. 2015. BFC: correcting Illumina sequencing errors. Bioinformatics 31:2885–2887. doi: 10.1093/bioinformatics/btv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 70.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 71.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 73.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 76.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 80.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 82.Seemann T. 2018. Barrnap: BAsic Rapid Ribosomal RNA Predictor. https://github.com/tseemann/barrnap.
- 83.Skennerton C. 2019. MinCED – Mining CRISPRs in Environmental Datasets. https://github.com/ctSkennerton/minced.
- 84.Enright AJ, Dongen SV, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 86.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 91.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers for raw sequencing data are in Table S6 in the supplemental material. Whole-genome sequencing data have been deposited in the Sequence Read Archive (SRA). Individual run accession numbers (SRR) for demultiplexed isolate data are listed in Table S6.