Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections, and Chlamydia pneumoniae causes community-acquired respiratory infections. In vivo, the host immune system will release gamma interferon (IFN-γ) to combat infection. IFN-γ activates human cells to produce the tryptophan (Trp)-catabolizing enzyme indoleamine 2,3-dioxygenase (IDO). Consequently, there is a reduction in cytosolic Trp in IFN-γ-activated host cells.
KEYWORDS: Chlamydia, persistence, IFN-γ, amino acid starvation, tRNA synthetases, tryptophan, leucine, amino acid limitation, codon-dependent transcription, tRNA synthetase inhibitors
ABSTRACT
Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections, and Chlamydia pneumoniae causes community-acquired respiratory infections. In vivo, the host immune system will release gamma interferon (IFN-γ) to combat infection. IFN-γ activates human cells to produce the tryptophan (Trp)-catabolizing enzyme indoleamine 2,3-dioxygenase (IDO). Consequently, there is a reduction in cytosolic Trp in IFN-γ-activated host cells. In evolving to obligate intracellular dependence, Chlamydia has significantly reduced its genome size and content, as it relies on the host cell for various nutrients. Importantly, C. trachomatis and C. pneumoniae are Trp auxotrophs and are starved for this essential nutrient when the human host cell is exposed to IFN-γ. To survive this, chlamydiae enter an alternative developmental state referred to as persistence. Chlamydial persistence is characterized by a halt in the division cycle, aberrant morphology, and, in the case of IFN-γ-induced persistence, Trp codon-dependent changes in transcription. We hypothesize that these changes in transcription are dependent on the particular amino acid starvation state. To investigate the chlamydial response mechanisms acting when other amino acids become limiting, we tested the efficacy of prokaryote-specific tRNA synthetase inhibitors, indolmycin and AN3365, to mimic starvation of Trp and leucine, respectively. We show that these drugs block chlamydial growth and induce changes in morphology and transcription consistent with persistence. Importantly, growth inhibition was reversed when the compounds were removed from the medium. With these data, we find that indolmycin and AN3365 are valid tools that can be used to mimic the persistent state independently of IFN-γ.
INTRODUCTION
Chlamydial diseases are significant causes of morbidity in humans. Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections in the world. In 2017, the U.S. Centers for Disease Control and Prevention received over 1.7 million reports of chlamydial infections (1). This number is likely an underestimate, as most infections are asymptomatic and, therefore, undetected (2). The strains responsible for these infections are primarily confined to the urogenital serovars, D to K, but can also contain those of the invasive serovars, L1 to L3. Untreated C. trachomatis urogenital infections can ascend the genital tract, potentially leading to pelvic inflammatory disease and tubal factor infertility (3). Chlamydia pneumoniae is a respiratory pathogen responsible for approximately 10% of community-acquired cases of pneumonia. The presence of antibodies in >50% of adults in the United States, as well as several other countries, suggests infection with C. pneumoniae is relatively common (see reference 4 for an extended review). Additionally, long-term sequelae such as atherosclerosis and adult-onset asthma have been associated with C. pneumoniae infection (5, 6).
Chlamydiae are obligate intracellular bacteria that require a host cell to complete their developmental cycle. Chlamydial development involves interconversion between two distinct developmental forms: the elementary body (EB) and the reticulate body (RB). EBs are infectious, metabolically quiescent, environmentally stable, and compact in size (0.3 μm). RBs are the noninfectious, metabolically active replicative form that measure approximately 0.8 μm in diameter (as reviewed in reference 7). After initial attachment, EBs are internalized into an endocytic vesicle of the host cell and begin primary differentiation into RBs. Soon thereafter, chlamydial proteins are secreted into the vesicle membrane and host cell cytosol to prevent targeting of the chlamydia-containing vacuole to the lysosome. This modified endosome is known as the chlamydial inclusion and is a protective vacuole that masks the invading organisms from host cell defenses for the entirety of their development (8). Following the establishment of the inclusion and primary differentiation into an RB, the organism rapidly multiplies by a polarized budding mechanism (9). Chlamydia asynchronously undergoes secondary differentiation into EBs until the organisms are released from the cell through lysis or extrusion. The duration of this developmental process is approximately 48 h for C. trachomatis or 96 h for the slower growing C. pneumoniae.
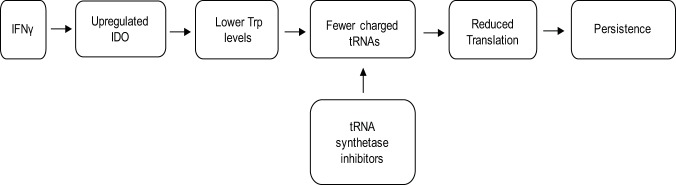
During infection, host immune cells respond to Chlamydia by releasing the cytokine gamma interferon (IFN-γ) (10). IFN-γ will bind its receptor and activate multiple signaling pathways. The major IFN-γ-induced antichlamydial effector in human cells is indoleamine 2,3-dioxygenase (IDO) (11). IDO will catabolize cytosolic tryptophan (Trp) into N′-formylkynurenine, a metabolite that cannot be used by C. trachomatis or C. pneumoniae (11–14). Although IFN-γ regulates over 200 human genes (15), IDO expression, with the resulting depletion of available Trp (16, 17) and decrease in translation (18), is the driving factor for inhibiting chlamydial growth (Fig. 1). This is supported by the ability to restore growth in cell culture by adding additional Trp to the medium, by pharmacologically inhibiting IDO in the presence of IFN-γ, or by using IDO mutant cells (19–21).
FIG 1.
A flowchart illustrating the events leading to IFN-γ-mediated persistence. By using tRNA synthetase inhibitors to affect translation, a more direct route to persistence is hypothesized.
Because C. trachomatis and C. pneumoniae are Trp auxotrophs and depend on host Trp to grow, they must respond to this starvation condition to maintain viability (22). Interestingly, Chlamydia has eliminated the stringent response (relA/spoT), which is used by most eubacteria to respond to amino acid starvation (22, 23). This raises the intriguing question of how they respond to amino acid limitation. Phenotypically, chlamydial RBs transition into an alternative developmental state termed persistence to cope with this stress (16). Persistent chlamydiae are thought to be associated with the chronic sequelae linked to chlamydial diseases. Importantly, various stressors can trigger persistence, and not all persistent transcriptomic and proteomic responses are the same (as reviewed in reference 24). Nevertheless, persistence models are characterized by bacteria (i) remaining viable yet noninfectious, (ii) being nonreplicative, (iii) exhibiting an aberrant morphology, and (iv) reactivating to resume the development cycle after the stress is removed (18, 25). Importantly, and as it relates to IFN-γ-induced persistence, transcriptomic and proteomic changes also occur, and these changes are predictable based on the Trp content of the transcript or protein (26).
The mechanisms for entry into, maintenance of, and exit from persistence are not known, and investigation of these mechanisms is made difficult by the nature of inducing persistence through IFN-γ exposure. Therefore, it is important to study Chlamydia’s transition into persistence while simultaneously controlling for as many variables as possible. This requires an amino acid starvation model without the confounding variables that come with using IFN-γ. In an effort to reproduce and model IFN-γ-induced persistence in the absence of IFN-γ, the most straightforward approach is to remove Trp from the culture medium. However, this will induce host cell autophagy and lysosomal degradation of proteins, which can regenerate amino acid pools that Chlamydia can scavenge (27).
We hypothesized that bacterial tRNA synthetase inhibitors would offer an alternative pathway to decrease translation in Chlamydia in an amino acid-dependent manner (Fig. 1). These inhibitors would, therefore, afford an opportunity to specifically mimic starvation for different amino acids to compare and contrast amino acid starvation responses in Chlamydia. We show that combining Trp-depleted media with the Trp analog indolmycin is sufficient to induce persistence in Chlamydia in cell culture. Indolmycin is a prokaryotic tryptophanyl-tRNA synthetase inhibitor that acts through competitive inhibition as a Trp analog (28). Additionally, AN3365, a prokaryotic leucyl-tRNA synthetase inhibitor, was used in this study to investigate the possibility of inducing persistence through the starvation of an amino acid other than Trp. AN3365 is an antibiotic in the aminomethyl benzoxaborole class shown to be active against Gram-negative bacteria (29). Unlike indolmycin, AN3365 is a noncompetitive inhibitor of bacterial leucyl-tRNA synthetases that locks the protein in its editing conformation, preventing release of charged leucyl-tRNAs (29). These tools will facilitate the modeling of specific amino acid starvation responses in Chlamydia without affecting the host cells. Here, we validate the use of these compounds, indolmycin and AN3365, as tools that can be used within the chlamydial field to investigate mechanisms engaged by Chlamydia to enter, maintain, and exit persistence in response to amino acid starvation.
RESULTS
The bacterial tRNA synthetase inhibitors indolmycin and AN3365 block chlamydial growth.
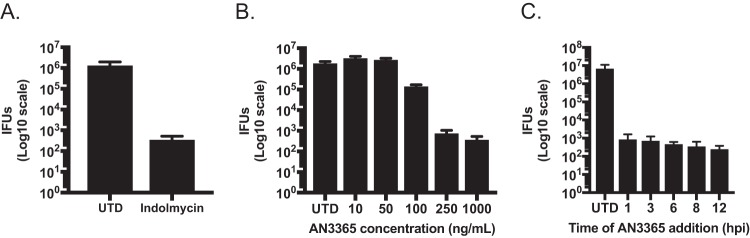
To determine whether the bacterial tRNA synthetase inhibitors indolmycin and AN3365 were effective against Chlamydia, we measured inclusion-forming units (IFUs) as a metric for chlamydial growth in the presence and absence of the inhibitors. Initial empirical experiments with indolmycin, a competitive inhibitor for tryptophanyl-tRNA charging, revealed that it had the most consistently reproducible effects at 120 μM or higher when used in Trp-deplete (0 mg/liter) Dulbecco’s modified Eagle medium (DMEM) (see Fig. S1 in the supplemental material). We chose to perform this and all subsequent experiments with indolmycin by adding it at a 120 μM concentration in the absence of Trp at 10 h postinfection (hpi). Under these conditions, indolmycin treatment reduced the generation of IFUs to the limit of detection of the assay (Fig. 2A). To determine the effective concentration for AN3365, we performed a dose-response assay and observed that concentrations in excess of 250 ng/ml (added at 10 hpi) were sufficient to reduce chlamydial growth to basal levels (i.e., near the limit of detection) (Fig. 2B). Importantly, as a noncompetitive inhibitor of leucyl-tRNA charging, AN3365 was effective in the presence of normal medium levels of leucine (105 mg/liter). AN3365 was also effective at blocking chlamydial growth when added at various time points up to 12 hpi during the developmental cycle (Fig. 2C). From these data, we conclude that indolmycin and AN3365 effectively blocked chlamydial growth. Nevertheless, a decrease in recoverable IFUs suggests one of three outcomes: (i) complete loss of viability, (ii) decrease in the rate of development (i.e., prolonged RB-only phase), or (iii) entry into persistence.
FIG 2.
Indolmycin and AN3365 effectively reduce recoverable infectious elementary bodies. For all experiments, HEp-2 cells were infected with C. trachomatis L2, and inclusion-forming units (IFUs) were collected at 24 hpi and titrated on a fresh monolayer of HEp-2 cells in the absence of antibiotics. (A) Indolmycin was added at 120 μM at 10 hpi. (B) Effect of various doses of AN3365 on IFU production when added at 10 hpi. (C) AN3365 was added at different times postinfection at 1 μg·ml−1 as indicated. Data are representative of three separate biological replicates. Error bars represent the standard deviations between biological replicates.
Indolmycin and AN3365 induce morphological aberrance in C. trachomatis.
To help distinguish between the possible reasons for a decrease in recoverable IFUs, immunofluorescence microscopy was used to analyze chlamydial morphology (Fig. 3). HEp-2 cells were infected with C. trachomatis serovar L2, treated or not with the indicated compounds at 10 hpi or pretreated with IFN-γ as described in Materials and Methods, and fixed and processed for imaging at 24 hpi. Both indolmycin and AN3365 treatment resulted in noticeably larger individual organisms, similar to that observed during IFN-γ treatment (16). However, there was no additive effect of both IFN-γ treatment and inhibitors (see Fig. S2). Electron microscopic analysis of organisms treated with inhibitors also revealed aberrant morphology (see Fig. S3). Interestingly, the labeling of major outer membrane protein (MOMP) was not uniform around the membrane of the organisms in the treated cells. The ompA gene encodes 30 Leu (L) and 7 Trp (W) codons. Decreased availability of L or W may therefore negatively impact translation of MOMP that in turn may lead to nonuniform localization along the organism’s membrane (see also reference 16). Conversely, antibodies targeting Hsp60_1, which contains 0 W codons, labeled both indolmycin- and IFN-γ-treated samples. Interestingly, Hsp60_1 labeling in the AN3365-treated sample was observed despite the presence of 45 L codons. Whether this represents continued synthesis of Hsp60_1 or a stable pool of the protein that was present prior to starvation remains unknown at this time. These observations are consistent with what was previously described during IFN-γ-induced persistence (12).
FIG 3.
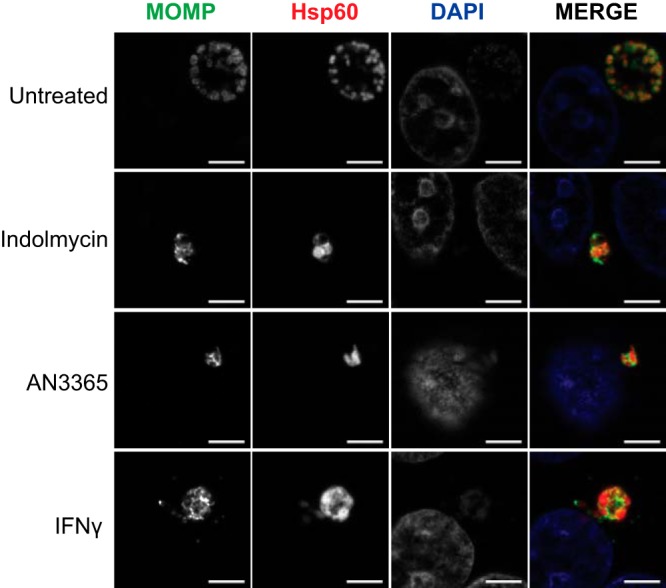
Inhibition of tRNA synthetase results in morphological aberrance in C. trachomatis L2. Representative images of HEp-2 cells infected with C. trachomatis and treated or not as indicated. Cells were fixed and stained 24 hpi using primary antibodies to major outer membrane protein (MOMP) and chlamydial Hsp60_1. Treatments of 120 μM indolmycin and 1 μg·ml−1 AN3365 given at 10 hpi resulted in smaller inclusions and morphologically aberrant organisms similar to IFN-γ-treated organisms. All images were acquired on Zeiss LSM 800 confocal microscope with Airyscan at 63× optical magnification with 2× digital zoom. Bars, 5 μm.
We also measured host cell activity (a proxy for cell numbers) under various treatment conditions to ensure that the inhibitors had no impact on the host cell. During treatment with cycloheximide, a eukaryotic protein synthesis inhibitor, or IFN-γ, which depletes host Trp pools, cell activity was decreased as expected and compared to that of untreated controls (see Fig. S4). However, treatment with inhibitors or growth in Trp-free medium did not impact cell activity. Taken together, these data in conjunction with the IFU and microscopy data presented above support the conclusion that the inhibitors are specific to Chlamydia and that the organisms are nonreplicative, display an aberrant morphology, and are not proceeding through the normal developmental cycle.
Removal of indolmycin and AN3365 rescues C. trachomatis growth and morphology.
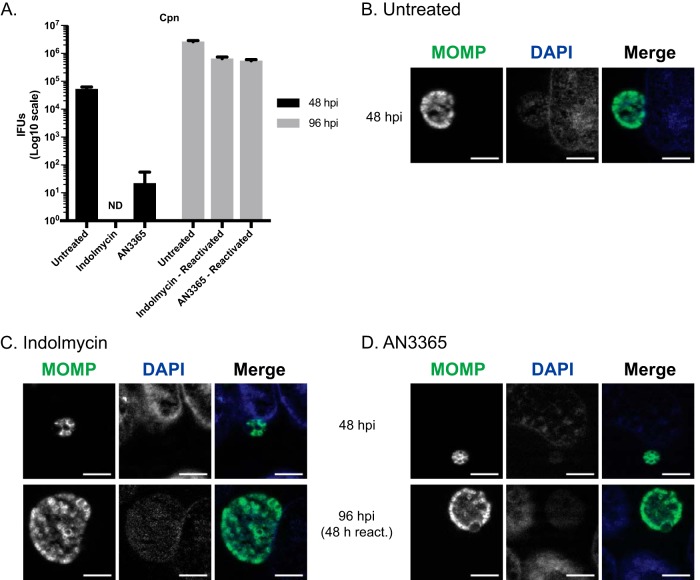
A key characteristic of persistence is its reversibility, such that the organism can revert back to a developmentally competent RB. To determine whether or not C. trachomatis remains viable under indolmycin or AN3365 treatment, we attempted to rescue the organisms by removing each treatment (Fig. 4). HEp-2 cells were infected with C. trachomatis serovar L2 and treated or not with the designated compounds at 10 hpi. Treatment medium was aspirated at 24 hpi, and samples were washed 3× with Dulbecco's phosphate-buffered saline (DPBS) before replenishing with normal medium. After 24- and 48-h recovery periods following drug removal, IFUs were quantified and organism morphology was assessed. In the presence of indolmycin or AN3365, chlamydial growth was inhibited up to 72 hpi (Fig. 4A). After removal of these compounds from the medium, chlamydial growth was restored, as represented by a logarithmic increase in IFUs at 48 (24 h postrecovery) and 72 (48 h postrecovery) hpi. These IFU data were corroborated by the detection of RBs 24 h postrecovery that were indistinguishable from those at 24 hpi for untreated organisms (Fig. 4B to D). Moreover, recoverable IFUs were intermediate between that observed at the 24- and 48-hpi untreated groups. This is expected when considering treatment time at 10 hpi essentially pauses the developmental cycle. After 24 h of recovery, the organisms have undergone approximately 34 h of development. We conclude that the effects of indolmycin and AN3365 are reversible, consistent with what is observed during the removal of IFN-γ from persistent cultures (18, 25, 30). Collectively, the data presented in Fig. 2, 3, and 4 indicate that the tRNA synthetase inhibitors induce a persistent growth state that is reversible upon removal of the compounds.
FIG 4.
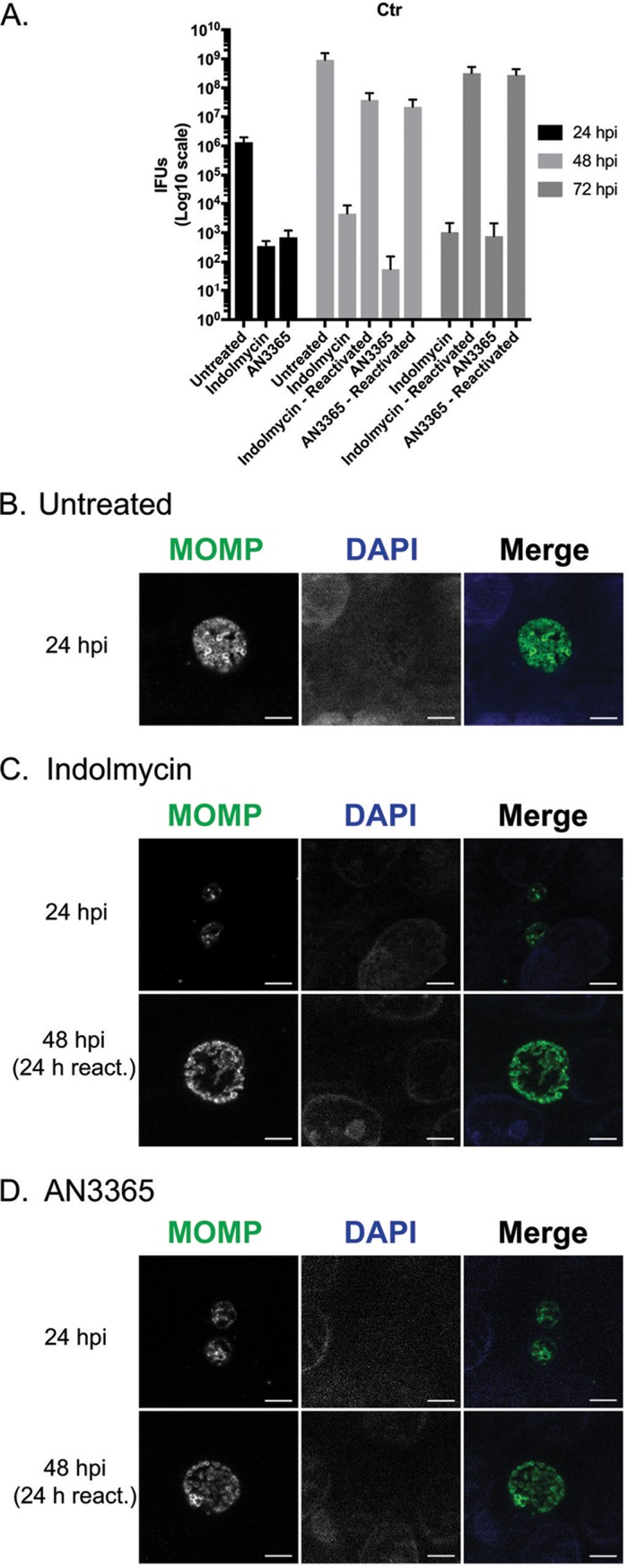
Removal of indolmycin and AN3365 allows reactivation from persistence. HEp-2 cells were infected with C. trachomatis and treated or not with the designated tRNA synthetase inhibitor at 10 hpi. In untreated and reactivated samples, DMEM was removed at 24 hpi followed by three DPBS washes and replenishment with unmodified DMEM. Cultures were allowed to recover for an additional 24 or 48 h before fixation. (A) IFU samples were collected from replicate wells at designated points throughout the experiment. Error bars represent variability between three biological replicates. (B) Representative images of untreated C. trachomatis infected HEp-2 cells at 24 or 48 hpi. Representative images of 120 μM indolmycin- (C) or 1 μg·ml−1 AN3365-treated (D) C. trachomatis-infected HEp-2 cells at 24, 48 (24 h reactivation [react]), or 72 (48 h react) hpi. All images were acquired on an AXIO Imager.Z2 with ApoTome.2 at ×100 magnification. Bars, 5 μm.
Indolmycin and AN3365 induce persistence in C. pneumoniae.
Although the underlying mechanisms of persistence are unknown, we hypothesize that they are conserved between C. trachomatis and C. pneumoniae. C. pneumoniae is a slower growing species than C. trachomatis, particularly the L2 serovar, suggesting it would be equally, if not more, sensitive to tRNA synthetase inhibitors. Therefore, we suspected the use of indolmycin and AN3365 on C. pneumoniae would produce a phenotype similar to that observed during IFN-γ exposure. To test this, we infected HEp-2 cells with C. pneumoniae AR39. Samples were treated or not with the designated compound at 24 hpi or with IFN-γ at time of infection. As seen in Fig. 5, the morphologies of organisms treated with indolmycin or AN3365 closely resemble those treated with IFN-γ.
FIG 5.
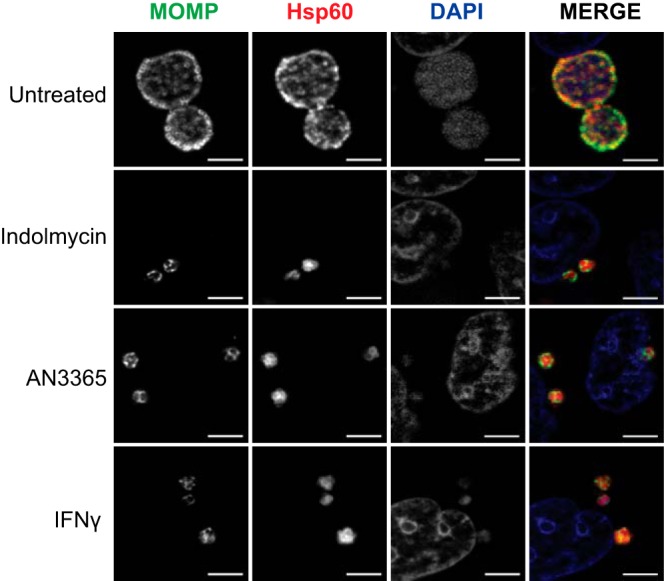
Immunofluorescence images of HEp-2 cultures infected with C. pneumoniae at 48 hpi show morphological similarity between 120 μM indolmycin and 1 μg·ml−1 AN3365 treatments with IFN-γ-induced persistent organisms. Indolmycin and AN3365 were added at 24 hpi. IFN-γ was added at the time of infection. All images were acquired on an AXIO Imager.Z2 with ApoTome.2 at ×100 magnification. Bars, 5 μm.
Furthermore, treatment using indolmycin or AN3365 reduced recoverable IFUs below or to the limit of detection, respectively (Fig. 6A). Following the removal of indolmycin or AN3365, recoverable IFUs approached levels between those from the 48- and 96-hpi untreated samples (Fig. 6A). Additionally, morphological aberrance observed under the effects of indolmycin or AN3365 was reversed following the removal of the respective compound (Fig. 6B to D). We conclude from these data that tRNA synthetase inhibitors are broadly applicable to study persistence in Chlamydia species.
FIG 6.
Removal of indolmycin and AN3365 allows reactivation from persistence in C. pneumoniae. HEp-2 cells were infected with C. pneumoniae and treated or not with the designated tRNA synthetase inhibitor at 24 hpi. DMEM was removed at 48 hpi followed by three DPBS washes and replenishment with unmodified DMEM. Cultures were allowed to recover for an additional 48 h before processing. (A) IFU samples were collected from replicate wells at designated points throughout the experiment. Error bars represent variability between three biological replicates. ND, not detectable. (B) Representative images of untreated C. pneumoniae-infected HEp-2 cells at 48 hpi. Representative images of 120 μM indolmycin- (C) or 1 μg·ml−1 AN3365-treated (D) C. pneumoniae-infected HEp-2 cells at 48 hpi or 96 (48 h reactivation) hpi. All images were acquired on an AXIO Imager.Z2 with ApoTome.2 at ×100 magnification. Bars, 5 μm.
The bacterial tRNA synthetase inhibitors induce transcriptional changes consistent with IFN-γ-mediated persistence.
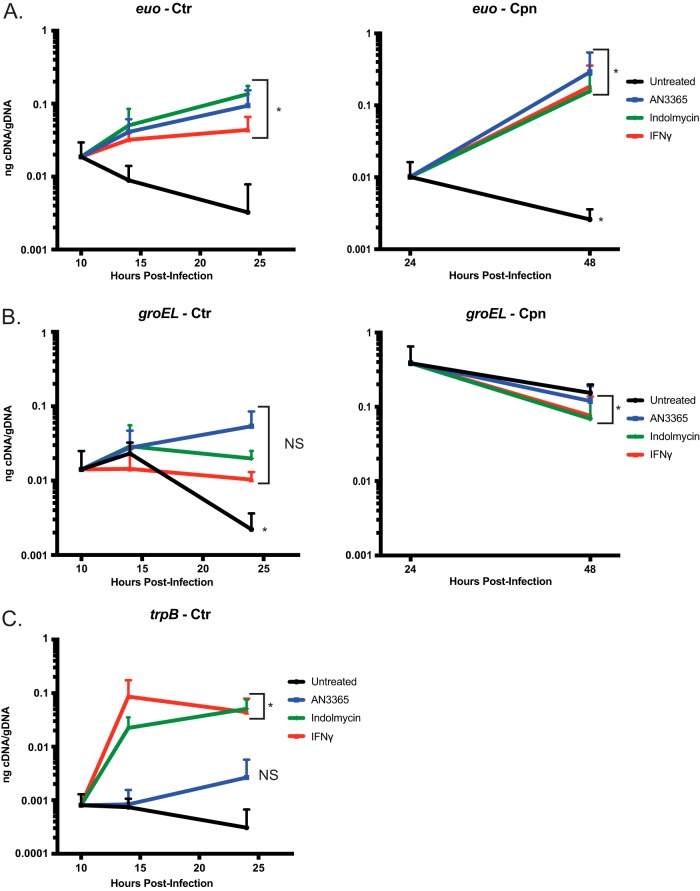
To determine whether the tRNA synthetase inhibitors induce transcriptional changes consistent with persistence, nucleic acids were isolated from infected cultures, and the abundance of selected transcripts was measured and normalized to genomic DNA content. Increased transcription of euo has been previously associated with IFN-γ-mediated persistence (18, 30). This elevated transcriptional response was observed in both C. trachomatis (beginning at 4 h posttreatment) and C. pneumoniae when exposed to either indolmycin or AN3365 treatments and resembled IFN-γ-induced persistence (Fig. 7A). In addition to euo, groEL_1 has also been implicated in persistence and general stress response (30). We analyzed the abundance of groEL_1 transcripts in both C. trachomatis and C. pneumoniae under each treatment (Fig. 7B). In agreement with previous findings, groEL_1 transcripts remained unchanged (<1.5× difference) in C. trachomatis between 10 and 24 hpi in IFN-γ-treated samples, whereas groEL_1 transcripts decreased in abundance during the same time frame in the untreated samples (18). Interestingly, indolmycin-treated samples closely mirrored the effects of IFN-γ, while AN3365 treatment resulted in a 4- to 5-fold increase of groEL_1 transcripts at the 24-h time point (P = 0.053). Conversely, for C. pneumoniae, all treatment conditions followed a similar transcript pattern as in untreated samples for groEL_1. This is also consistent with previous observations for IFN-γ-induced persistence in this organism (18). The reason for the groEL_1 differences between the chlamydial species is not known at this time and requires further investigation.
FIG 7.
Transcriptional changes in Chlamydia consistent with persistence can be detected during indolmycin and AN3365 treatment. (A) Transcripts of euo were elevated in C. trachomatis (Ctr) or C. pneumoniae (Cpn) following treatments with 120 μM indolmycin, 1 μg·ml−1 AN3365, or IFN-γ. (B) Under standard conditions, groEL_1 transcripts in C. trachomatis decreased between 10 and 24 hpi. However, treatment with indolmycin resulted in unchanged transcript levels, similar to what is seen in IFN-γ-treated samples, while AN3365 treatment resulted in slightly higher transcript levels (P = 0.053). In C. pneumoniae, no significant change was seen between 24 and 48 h under any treatment, in agreement with previous reports investigating IFN-γ exposure. (C) trpB transcripts accumulated at 14 hpi, as expected, in indolmycin- and IFN-γ-treated samples but not in those treated with AN3365. At 24 hpi, AN3365-treated samples exhibit a slight (3- to 4-fold) increase in trpB (P = 0.076). Student's t test was used to compare each 24 hpi value to that at 10 hpi untreated in C. trachomatis and each 48 hpi value to that at 24 hpi untreated in C. pneumoniae following Log10 transformation. *, P < 0.05.
Additionally, indolmycin, but not AN3365, treatment resulted in a rapid (4 h) and large increase in trpB transcripts (C. pneumoniae does not carry trpB), a gene that is repressed under Trp-replete conditions (Fig. 7C) (31). This suggests C. trachomatis detects and quickly responds to a lack of Trp. Moreover, there was a trend toward increased trpB transcripts at 24 hpi following AN3365 treatment (P = 0.076). Considering the lack of change observed at 14 hpi, the possible increase of trpB transcripts at 24 hpi may be an indirect effect caused by a cascade of responses rather than an immediate reaction to Leu limitation. It is important to note that while C. trachomatis detects Trp limitations when cultured in Trp-deplete media (no indolmycin), the abundance of euo transcripts does not change (see Fig. S5). Therefore, removing Trp from the medium cannot be used as a reliable IFN-γ-free model for persistence, particularly for faster growing species and strains of Chlamydia.
Transcript levels of the 3′ end of the ytg operon are reduced in Chlamydia during indolmycin or AN3365 treatment.
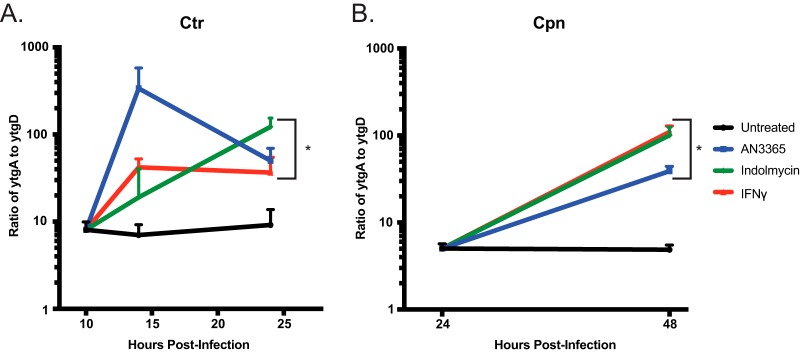
To determine whether indolmycin and AN3365 could mimic a more nuanced characteristic of IFN-γ-induced persistence, we looked to the ytgABCD operon (26). As previously described by Ouellette et al. (32), IFN-γ treatment results in Rho-dependent polarization of the ytg operon. This results in a skewed ratio of ytgA to ytgD transcripts. During the normal developmental cycle, this ratio is approximately 5 to 10 but increases to >30-fold or higher during IFN-γ-mediated Trp starvation. We hypothesized that this skew was caused by ribosome stalling at Trp codon-rich regions along the transcript due to the lack of charged tRNAs, which allows Rho to bind internal rut sites to terminate transcription prematurely (32). Of note, within the ytgC gene are three tandem W codons, and the gene encodes additional W residues. L residues are highly abundant in the operon, with 98 total residues in ytgB and ytgC and a total of 11 LL motifs in C. trachomatis. We reasoned that both indolmycin and AN3365 should produce the same phenotype as a result of the stalling of ribosomes on Trp codons, in the case of indolmycin treatment, and Leu codons, in the case of AN3365 treatment. We quantified ytgA and ytgD transcripts at different times after addition of the tRNA synthetase inhibitors (at 10 hpi for C. trachomatis, 24 hpi for C. pneumoniae) and compared the ratios to those from IFN-γ-treated cultures. As anticipated, both indolmycin- and AN3365-treated samples resembled the IFN-γ-induced persistent state with regard to the polarity of the ytg operon, shown in Fig. 8 as a disproportionate level of ytgA to ytgD transcripts. Interestingly, differences in the C. trachomatis ytgA:ytgD ratios were observed within 4 h of treatment (14 hpi) and were more pronounced for L limitation (AN3365) than for W limitation (indolmycin or IFN-γ) (Fig. 8A). This is consistent with the larger number of L versus W residues in the operon. However, by 24 hpi, the ytgA:ytgD ratios for all treatments were similar, possibly reflecting a recovery in read-through potential in AN3365-treated cultures. For C. pneumoniae, all treatments resulted in the expected increase in ytgA transcripts in proportion to ytgD transcripts when measured at 48 hpi (after treatment at 24 hpi). Overall, these data demonstrate that the tRNA synthetase inhibitors recapitulate key characteristics of amino acid starvation in Chlamydia.
FIG 8.
Transcriptional analysis shows a decrease in read-through efficiency of the ytgABCD operon during 120 μM indolmycin, 1 μg·ml−1 AN3365, or IFN-γ treatment in C. trachomatis (Ctr) (A) or C. pneumoniae (Cpn) (B). RT-qPCR was performed to determine nanograms of cDNA of both ytgA and ytgD. Each was normalized to genomic DNA (gDNA) collected from replicate wells and expressed as a ratio of nanograms cDNA per nanograms gDNA of ytgA over ytgD. Student's t test was used to compare each 24 hpi value to that at 10 hpi untreated in C. trachomatis and each 48 hpi value to that at 24 hpi untreated in C. pneumoniae following Log10 transformation. *, P < 0.05.
DISCUSSION
Persistence is an important but poorly understood aspect of chlamydial diseases. The immunological basis for inducing persistence in cell culture was first described by Beatty et al. in 1993, who described the effects of IFN-γ, and its reversibility, on chlamydial growth and morphology in human cell lines (12, 16, 25). These effects were connected with the ability of human IFN-γ to induce a tryptophan-limiting environment in the cell by activating IDO expression (12). Broadly speaking, these effects are likely mediated by the inability to efficiently translate key proteins enriched in Trp residues (Fig. 1) (18, 33, 34). More recent studies have characterized transcriptional and translational changes associated with IFN-γ-mediated persistence (18, 30). More importantly, these “persistence” characteristics as defined in cell culture models have recently been observed in patient samples (13). This underscores the need to have a better mechanistic understanding of how amino acid limitation results in a persistent phenotype.
In 2011, the Clarke group published the first study demonstrating stable transformation of Chlamydia trachomatis serovar L2 (35). This advance, common for decades in other bacterial systems, has allowed fluorescent tagging of target proteins and reverse genetic tools to be applied to Chlamydia (36–40). However, serovar L2 is among the fastest growing strains of C. trachomatis (41), and slower growing strains and species of Chlamydia have proven more difficult to transform. As it relates to genetic studies of IFN-γ-induced persistence, this creates a hurdle. When IFN-γ is added to cell cultures, removal of Trp from the cytosol by IDO is gradual and takes approximately 24 h. This is a time during the L2 developmental cycle when RBs are differentiating to EBs and EBs are rapidly accumulating (e.g., see Fig. 2A). For IFN-γ to be effective, pretreatment of host cells with IFN-γ prior to infection is required, yet this strategy is inconsistent. To induce a persistent state in serovar L2, we recently published a protocol that described the pretreatment of cells with IFN-γ prior to infection followed by the addition of IFN-γ-conditioned medium at 10 hpi (32). This allows IDO to be sufficiently expressed and Trp to be catabolized, resulting in small inclusions that contain relatively few aberrant organisms approximately 24 h postinfection (hpi). While this protocol elicited reproducibly persistent forms for us, differences between labs, the cell types used, and batches of IFN-γ (which require careful titration for effective dose) may not make it easily transferable to other systems. This can lead to discrepancies in findings, since too much IFN-γ exposure prevents primary differentiation of chlamydial EBs to RBs, while too little results in mixed populations of persistent and normal organisms within a culture (20). In sum, the field would benefit greatly from a tool that allows reproducible induction of persistence and that minimizes confounding variables while maximizing control and flexibility of experimental design parameters.
Here, we sought to evaluate the effects of characterized bacterial tRNA synthetase inhibitors (28, 29, 42) for their ability to induce persistence in Chlamydia as a first step in developing systems that would allow us to mechanistically address this alternative developmental state. Such an approach would offer immediate advantages over IFN-γ-mediated tryptophan limitation in that a translation block (i.e., starvation-mimicking condition) could be induced immediately upon addition of the inhibitors. By using Escherichia coli tRNA turnover rates as a guide, we hypothesize that the pool of charged Trp-tRNA or Leu-tRNA would be depleted within seconds after treatment (43). This would allow more direct comparisons between research groups, with less variability in experimental systems. In particular, the use of inhibitors circumvents the host cell’s ability to regenerate amino acid pools through autophagy, which would occur under conditions where an amino acid is omitted from the culture medium. Indeed, we observed that, under such conditions, although the absence of Trp was sensed, as demonstrated by increased trpB transcripts, euo transcripts, a marker of persistence, did not increase (see Fig. S5 in the supplemental material). This suggests that, for serovar L2, simply omitting amino acids from the culture medium is not sufficient to induce a bona fide persistence response.
These data indicate the ability of indolmycin and AN3365 to induce persistence by limiting Chlamydia’s use of a single specific amino acid by blocking the charging of its cognate tRNA. Considering the parallels in morphology, transcriptional response, and mode of stress caused by these compounds in comparison to those for IFN-γ within each species, we conclude that the use of indolmycin and AN3365 in place of IFN-γ is a viable method to study amino acid starvation stress responses. Interestingly, indolmycin treatment replicates the key transcriptional and morphological phenotypes associated with IFN-γ-induced persistence, further supporting that tryptophan limitation is the main antichlamydial inhibitory mechanism of IFN-γ in human cells (11, 19). The transcriptional changes of C. pneumoniae in response to the inhibitors more closely mirrored IFN-γ-mediated persistence than those in C. trachomatis. Given the lower growth rate of C. pneumoniae, this is not surprising. The greater heterogeneity in transcription responses between indolmycin and IFN-γ in C. trachomatis may be due to the organism’s quicker developmental cycle and asynchronous nature. That being said, the difference between the tryptophan starvation condition in C. pneumoniae versus that in C. trachomatis was not more than 4-fold and showed the same trends overall.
AN3365-induced Leu starvation displayed noteworthy differences from the Trp starvation conditions. First, groEL_1 transcripts generally increased during the analysis in C. trachomatis. This is consistent with what we previously characterized as codon-dependent transcriptional changes during amino acid starvation, as Hsp60_1 contains 45 L residues (26). Second, trpB transcripts were not increased 4 h after treatment, as expected, since Leu starvation should not activate expression of the trpRBA operon. However, 14 h posttreatment, there was an approximately 3-fold increase in trpB levels that approached statistical significance, suggesting some derepression of the operon. TrpR contains 11 Leu residues, including one LL motif. Therefore, the inability to efficiently translate the repressor over time may allow for gradual transcriptional activation of the operon. Alternatively, the recently described role for the iron-sensitive repressor YtgR in blocking transcription of trpBA may also be important (44). The YtgCR protein contains approximately 60 Leu residues with multiple LL motifs that likely prevent efficient translation of this sequence. This is under investigation, but we also observed transcriptional changes in the ytg operon (see below).
We recently demonstrated differences in transcript levels between the 5′ and 3′ ends of large monocistronic and polycistronic transcripts (26). We further connected this to Rho-dependent polarity prematurely terminating transcription in Trp codon-rich transcripts during IFN-γ-mediated Trp starvation (32). To determine whether the tRNA synthetase inhibitors could replicate the destabilization of the 3′ end of a large transcript, we analyzed the abundance of the ytgA and ytgD transcripts. Consistent with what was previously observed for IFN-γ-mediated Trp starvation, both indolmycin and AN3365 caused a disparity in transcript abundance between the 5′ and 3′ ends of the ytg operon. Interestingly, AN3365 caused a quick destabilization of ytgD transcripts in C. trachomatis before recovering to levels observed under Trp starvation conditions. The reasons for this are not clear but are under investigation.
The tools described here to mimic specific amino acid starvation states, by blocking tRNA charging, in the absence of chemokines or other significant alterations to culture conditions will facilitate broad comparisons of chlamydial persistence between species. For example, Chlamydia caviae resists IFN-γ-mediated persistence by recycling the product of Trp degradation, N-formylkynurenine, through a Trp scavenging pathway (45). Indolmycin treatment will facilitate studies of Trp starvation responses in this species. Likewise, these treatments can be used in mouse cells, where IDO is not the primary antichlamydial effector (10, 46). Also of interest are other intracellular pathogens such as Coxiella or Rickettsia, as studying their response mechanisms to amino acid starvation could lead to a greater understanding of evolutionary strategies employed by obligate intracellular pathogens, which typically lack functional stringent responses, to adapt to this stress.
Following the validation of these compounds as tools to study persistence, we aim to further investigate the role of amino acid limitation in regulating the persistent state. With the ability to limit an amino acid other than Trp, we can more rigorously test the hypothesis that Trp limitation increases transcription of Trp codon-containing genes to determine if this is a broad response to amino acid limitation or perhaps something specific to Trp (26). We can now also apply genetic tools to study amino acid starvation responses in C. trachomatis L2.
MATERIALS AND METHODS
Organisms and cell culture.
The human epithelial cell line HEp-2 was routinely cultivated at 37°C with 5% CO2 in Dulbecco’s modified Eagle medium (DMEM; Gibco, Dun Laoghaire, Ireland) supplemented with 10% fetal bovine serum (FBS). The HEp-2 cells were a kind gift of H. Caldwell (NIH/NIAID). C. trachomatis serovar L2 and C. pneumoniae AR39 EBs were harvested from infected HEp-2 cell cultures at 37°C and 35°C, respectively, with 5% CO2 and density gradient purified. The titers of purified EBs were determined for infectivity by determining inclusion-forming units (IFUs) on fresh cell monolayers. All bacterial and eukaryotic cell stocks were confirmed to be Mycoplasma negative using the LookOut Mycoplasma PCR Detection kit (Sigma, St. Louis, MO).
Indolmycin was purchased from Cayman Chemical (Ann Arbor, MI) and resuspended to 120 mM in dimethyl sulfoxide (DMSO; Sigma). Aliquots were kept at −80°C and used only once to avoid freeze-thawing. Indolmycin was used at 120 μM and added at 10 hpi (C. trachomatis) or 24 hpi (C. pneumoniae) in all experiments except where indicated. Immediately prior to treatment, cell medium was replaced with DMEM lacking Trp (made in-house) to enhance the inhibitory effects of indolmycin. DMEM lacking Trp was made using 10% fetal bovine serum that had been dialyzed to remove any additional amino acids. All custom medium components were purchased from Sigma.
AN3365 was purchased from Cayman Chemical and resuspended to 5 mg·ml−1 in DMSO. Aliquots were kept at −20°C and allowed one additional freeze-thaw. AN3365 concentration was titrated to induce persistence without completely stalling development and was used at 1 μg·ml−1 with treatment at 10 hpi (C. trachomatis) or 24 hpi (C. pneumoniae). No modifications to DMEM were necessary. In some experiments with C. trachomatis, AN3365 was added at different concentrations or at different times postinfection as indicated in the text, with inclusion-forming unit samples (see below) collected at 24 hpi.
Recombinant human gamma interferon (IFN-γ) was purchased from Cell Sciences (Canton, MA) and resuspended to 100 μg·ml−1 in 0.1% bovine serum albumin (BSA; Sigma) diluted in water. Aliquots were frozen at −80°C and used only once to avoid freeze-thawing. IFN-γ was titrated for its effect to induce persistence without killing the bacteria, and in our experiments, 0.5 ng·ml−1 was added to cells approximately 11 h prior to infection. Medium was replaced at 10 hpi with IFN-γ-conditioned medium (ICM) to induce persistence in C. trachomatis as described previously (32). ICM was prepared by adding 2 ng·ml−1 IFN-γ to uninfected HEp-2 cells for approximately 54 h prior to collection and filtration of the medium. For C. pneumoniae experiments, infected cells were treated with 2 ng·ml−1 at the time of infection.
Inclusion-forming unit assays.
Infectious progeny (i.e., EBs) were determined from a primary infection based on inclusions formed in a secondary infection (see reference 18). Primary infection samples were harvested by scraping cells in 2 sucrose-phosphate (2SP) solution. Samples were lysed via a single freeze-thaw cycle, serially diluted, and used to infect a fresh cell monolayer. The secondary infection was allowed to progress for 24 h prior to fixation and labeling for immunofluorescence microscopy (see below) using antibodies directed against the outer membranes of the organisms. Titers were enumerated by calculating the total number of inclusions per field based on counts from 15 fields of view.
Immunofluorescence microscopy.
Cells were cultured on glass coverslips in 24-well tissue culture plates and infected with C. trachomatis at a multiplicity of infection (MOI) of 1 or C. pneumoniae at an MOI of 2. All cells were fixed in 100% methanol. Organisms were stained using a primary goat or mouse antibody specific to either C. trachomatis or C. pneumoniae major outer membrane protein (MOMP), respectively, and a donkey anti-goat or anti-mouse secondary antibody conjugated to Alexa Fluor 488 (Jackson Laboratory, Bar Harbor, ME). Where applicable, a primary mouse (C. trachomatis) or rabbit (C. pneumoniae) antibody specific to chlamydial Hsp60 was also used in conjunction with a secondary donkey anti-mouse or anti-rabbit antibody conjugated to Alexa Fluor 594 (Jackson Laboratory). DAPI (4′,6-diamidino-2-phenylindole) was added to visualize DNA.
Nucleic acid extraction and RT-qPCR.
RNA extraction was performed on infected cell monolayers using TRIzol (Invitrogen/Thermo Fisher). Samples were treated with Turbo DNAfree (Ambion/Thermo Fisher) according to the manufacturer’s instructions to remove DNA contamination. cDNA was synthesized from DNA-free RNA using random nonamers (New England BioLabs, Ipswich, MA) and SuperScript III reverse transcriptase (RT; Invitrogen/Thermo Fisher) per the manufacturers’ instructions. Reaction end products were diluted 10-fold with molecular biology-grade water, aliquoted for later use, and stored at −80°C. Equal volumes of each reaction mixture were used in 25-μl quantitative PCR (qPCR) mixtures with SYBR green master mix (Applied Biosystems) and quantified on a QuantStudio 3 (Applied Biosystems/Thermo Fisher) using the standard amplification cycle with a melting curve analysis. Results were compared to a standard curve generated against purified C. trachomatis L2 or C. pneumoniae genomic DNA as appropriate. DNA samples were collected from replicate wells during the same experiments using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). Equal total DNA quantities were used in qPCR with a groEL_1 primer set to quantify chlamydial genomes. Genome values were used to normalize respective transcript data. All qPCR primer sequences can be found in Table S1 in the supplemental material. RT-qPCR results were normalized for efficiency, with typical results demonstrating an r2 of >0.995 and efficiencies greater than 90%. Student's t tests were used to compare each 24 hpi value to that at 10 hpi untreated in C. trachomatis and each 48 hpi value to that at 24 hpi untreated in C. pneumoniae following Log10 transformation.
Reactivation.
To determine the possibility of recovery from persistence, samples were treated or not with AN3365 or indolmycin as described above. After 24 hpi in C. trachomatis or 48 hpi in C. pneumoniae, all samples were washed three times with DPBS and given fresh medium. Samples were allowed to recover for an additional 24 or 48 h before collection for the IFU assay or fixation for immunofluorescence microscopy.
Electron microscopy.
HEp-2 cells were cultured in a 6-well plate and infected with C. trachomatis serovar L2 at an MOI of 2.5. Samples were treated or not with indolmycin or AN3365 at 10 hpi. Cells were trypsinized at 24 hpi and pelleted at 500 × g for 5 min. Pellets were washed 3× using DPBS. Following the final wash, pellets were resuspended in 1 ml of fixative containing 2% glutaraldehyde, 2% paraformaldehyde, and 0.1 M Sorenson’s phosphate buffer (pH 7.2). Postfixation was carried out using 1% osmium tetroxide followed by dehydration in a series of increasing EtOH concentrations. Ninety-nanometer sections were cut using a Leica UC6 Ultramicrotome with a Diatome diamond knife. Sections were stained in 2% uranyl acetate and Reynold’s lead citrate. Images were collected on an FEI Tecnai G2 transmission electron microscope (TEM) operated at 80 kV.
Cell activity assay.
HEp-2 cells (10,000 per well) were cultured in a 96-well plate for 24 h in the presence of 1 μg·ml−1 AN3365, 120 μM indolmycin in DMEM lacking Trp, DMEM lacking Trp, 0.5 ng·ml−1 IFN-γ, 2 μg·ml−1 cycloheximide, or 2 μg·ml−1 cycloheximide in DMEM lacking Trp. Additional wells containing only DMEM or DMEM lacking Trp without HEp-2 cells were included to correct for background absorbance. After 24 h of treatment, PrestoBlue (Invitrogen/Thermo Fisher) was added per the manufacturer’s instructions. Absorbance was measured at 570 nm and corrected using the appropriate background wells. Lower absorbance values correlate to decreased cellular activity. Student's t tests were used to compare background-corrected 570 nm absorbance values of each treated sample to that from an untreated control.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by start-up funds from the University of Nebraska Medical Center as well as a CAREER award (1810599) from the National Science Foundation and an NIH/NIAID award (1R01AI132406-01A1) to S.P.O.
We thank H. Caldwell (NIAID/NIH) for eukaryotic cell stocks and the antibody to C. pneumoniae MOMP, R. Morrison (UAMS) for the antibody to chlamydial Hsp60_1, E. Rucks (UNMC) for the antibody against Chlamydia and for critical review of the manuscript, and R. Carabeo (UNMC) for critical review of the manuscript. We also thank Tom Bargar and Nicholas Conoan of the Electron Microscopy Core Facility (EMCF) at the University of Nebraska Medical Center for technical assistance. The EMCF is supported by state funds from the Nebraska Research Initiative (NRI) and the University of Nebraska Foundation, and institutionally by the Office of the Vice Chancellor for Research.
This publication’s contents and interpretations are the sole responsibility of the authors.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.CDC. 2018. Sexually transmitted disease surveillance 2017. U.S. Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 2.Satterwhite CL, Torrone E, Meites E, Dunne EF, Mahajan R, Ocfemia MC, Su J, Xu F, Weinstock H. 2013. Sexually transmitted infections among US women and men: prevalence and incidence estimates, 2008. Sex Transm Dis 40:187–193. doi: 10.1097/OLQ.0b013e318286bb53. [DOI] [PubMed] [Google Scholar]
- 3.Brunham RC, Maclean IW, Binns B, Peeling RW. 1985. Chlamydia trachomatis: its role in tubal infertility. J Infect Dis 152:1275–1282. doi: 10.1093/infdis/152.6.1275. [DOI] [PubMed] [Google Scholar]
- 4.Kuo CC, Jackson LA, Campbell LA, Grayston JT. 1995. Chlamydia pneumoniae (TWAR). Clin Microbiol Rev 8:451–461. doi: 10.1128/CMR.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saikku P, Leinonen M, Mattila K, Ekman MR, Nieminen MS, Makela PH, Huttunen JK, Valtonen V. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet 2:983–986. doi: 10.1016/s0140-6736(88)90741-6. [DOI] [PubMed] [Google Scholar]
- 6.Hahn DL. 1995. Treatment of Chlamydia pneumoniae infection in adult asthma: a before-after trial. J Fam Pract 41:345–351. [PubMed] [Google Scholar]
- 7.AbdelRahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Moore ER, Ouellette SP. 2014. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front Cell Infect Microbiol 4:157. doi: 10.3389/fcimb.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdelrahman Y, Ouellette SP, Belland RJ, Cox JV. 2016. Polarized cell division of Chlamydia trachomatis. PLoS Pathog 12:e1005822. doi: 10.1371/journal.ppat.1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry LL, Feilzer K, Caldwell HD. 1997. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J Immunol 158:3344–3352. [PubMed] [Google Scholar]
- 11.Byrne GI, Lehmann LK, Landry GJ. 1986. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect Immun 53:347–351. doi: 10.1128/IAI.53.2.347-351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beatty WL, Morrison RP, Byrne GI. 1994. Immunoelectron-microscopic quantitation of differential levels of chlamydial proteins in a cell culture model of persistent Chlamydia trachomatis infection. Infect Immun 62:4059–4062. doi: 10.1128/IAI.62.9.4059-4062.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis ME, Belland RJ, AbdelRahman YM, Beatty WL, Aiyar AA, Zea AH, Greene SJ, Marrero L, Buckner LR, Tate DJ, McGowin CL, Kozlowski PA, O’Brien M, Lillis RA, Martin DH, Quayle AJ. 2014. Morphologic and molecular evaluation of Chlamydia trachomatis growth in human endocervix reveals distinct growth patterns. Front Cell Infect Microbiol 4:71. doi: 10.3389/fcimb.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfefferkorn ER. 1984. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc Natl Acad Sci U S A 81:908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehm U, Klamp T, Groot M, Howard JC. 1997. Cellular responses to interferon-gamma. Annu Rev Immunol 15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 16.Beatty WL, Byrne GI, Morrison RP. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci U S A 90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane CD, Vena RM, Ouellette SP, Byrne GI. 1999. Intracellular tryptophan pool sizes may account for differences in gamma interferon-mediated inhibition and persistence of chlamydial growth in polarized and nonpolarized cells. Infect Immun 67:1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouellette SP, Hatch TP, AbdelRahman YM, Rose LA, Belland RJ, Byrne GI. 2006. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol Microbiol 62:1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x. [DOI] [PubMed] [Google Scholar]
- 19.Ibana JA, Belland RJ, Zea AH, Schust DJ, Nagamatsu T, AbdelRahman YM, Tate DJ, Beatty WL, Aiyar AA, Quayle AJ. 2011. Inhibition of indoleamine 2,3-dioxygenase activity by levo-1-methyl tryptophan blocks gamma interferon-induced Chlamydia trachomatis persistence in human epithelial cells. Infect Immun 79:4425–4437. doi: 10.1128/IAI.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonhardt RM, Lee S-J, Kavathas PB, Cresswell P. 2007. Severe tryptophan starvation blocks onset of conventional persistence and reduces reactivation of Chlamydia trachomatis. Infect Immun 75:5105–5117. doi: 10.1128/IAI.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas SM, Garrity LF, Brandt CR, Schobert CS, Feng GS, Taylor MW, Carlin JM, Byrne GI. 1993. IFN-gamma-mediated antimicrobial response. Indoleamine 2,3-dioxygenase-deficient mutant host cells no longer inhibit intracellular Chlamydia spp. or Toxoplasma growth. J Immunol 150:5529–5534. [PubMed] [Google Scholar]
- 22.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 23.Zomorodipour A, Andersson S. 1999. Obligate intracellular parasites: Rickettsia prowazekii and Chlamydia trachomatis. FEBS Lett 452:11–15. doi: 10.1016/S0014-5793(99)00563-3. [DOI] [PubMed] [Google Scholar]
- 24.Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect Immun 72:1843–1855. doi: 10.1128/iai.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beatty WL, Morrison RP, Byrne GI. 1995. Reactivation of persistent Chlamydia trachomatis infection in cell culture. Infect Immun 63:199–205. doi: 10.1128/IAI.63.1.199-205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouellette SP, Rueden KJ, Rucks EA. 2016. Tryptophan codon-dependent transcription in Chlamydia pneumoniae during gamma interferon-mediated tryptophan limitation. Infect Immun 84:2703–2713. doi: 10.1128/IAI.00377-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouellette SP, Dorsey FC, Moshiach S, Cleveland JL, Carabeo RA. 2011. Chlamydia species-dependent differences in the growth requirement for lysosomes. PLoS One 6:e16783. doi: 10.1371/journal.pone.0016783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner RG, Thorpe LF, Reuter W, Nierhaus KH. 1976. Indolmycin inhibits prokaryotic tryptophanyl-tRNA ligase. Eur J Biochem 68:1–3. doi: 10.1111/j.1432-1033.1976.tb10758.x. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez V, Crépin T, Palencia A, Cusack S, Akama T, Baker SJ, Bu W, Feng L, Freund YR, Liu L, Meewan M, Mohan M, Mao W, Rock FL, Sexton H, Sheoran A, Zhang Y, Zhang Y-K, Zhou Y, Nieman JA, Anugula MR, Keramane EM, Savariraj K, Reddy DS, Sharma R, Subedi R, Singh R, O'Leary A, Simon NL, De Marsh PL, Mushtaq S, Warner M, Livermore DM, Alley MRK, Plattner JJ. 2013. Discovery of a novel class of boron-based antibacterials with activity against gram-negative bacteria. Antimicrob Agents Chemother 57:1394–1403. doi: 10.1128/AAC.02058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belland RJ, Nelson DE, Virok D, Crane DD, Hogan D, Sturdevant D, Beatty WL, Caldwell HD. 2003. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci U S A 100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akers JC, Tan M. 2006. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J Bacteriol 188:4236–4243. doi: 10.1128/JB.01660-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouellette SP, Messerli PR, Wood NA, Hajovsky H. 2018. Characterization of chlamydial Rho and the role of rho-mediated transcriptional polarity during interferon gamma-mediated tryptophan limitation. Infect Immun 86:e00240-18. doi: 10.1128/IAI.00240-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Østergaard O, Follmann F, Olsen AW, Heegaard NH, Andersen P, Rosenkrands I. 2016. Quantitative protein profiling of Chlamydia trachomatis growth forms reveals defense strategies against tryptophan starvation. Mol Cell Proteomics 15:3540–3550. doi: 10.1074/mcp.M116.061986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo C-C, Xie G, Bonner CA, Jensen RA. 2012. The alternative translational profile that underlies the immune-evasive state of persistence in Chlamydiaceae exploits differential tryptophan contents of the protein repertoire. Microbiol Mol Biol Rev 76:405–443. doi: 10.1128/MMBR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson CM, Fisher DJ. 2013. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One 8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouellette SP. 2018. Feasibility of a conditional knockout system for Chlamydia based on CRISPR interference. Front Cell Infect Microbiol 8:59. doi: 10.3389/fcimb.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller KE, Wolf K, Fields KA. 2016. Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. mBio 7:e01817-15. doi: 10.1128/mBio.01817-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agaisse H, Derré I. 2013. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One 8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyairi I, Mahdi OS, Ouellette SP, Belland RJ, Byrne GI. 2006. Different growth rates of Chlamydia trachomatis biovars reflect pathotype. J Infect Dis 194:350–357. doi: 10.1086/505432. [DOI] [PubMed] [Google Scholar]
- 42.Mendes RE, Alley MRK, Sader HS, Biedenbach DJ, Jones RN. 2013. Potency and spectrum of activity of AN3365, a novel boron-containing protein synthesis inhibitor, tested against clinical isolates of Enterobacteriaceae and nonfermentative Gram-negative bacilli. Antimicrob Agents Chemother 57:2849–2857. doi: 10.1128/AAC.00160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jakubowski H, Goldman E. 1984. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J Bacteriol 158:769–776. doi: 10.1128/JB.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pokorzynski ND, Brinkworth AJ, Carabeo R. 2019. A bipartite iron-dependent transcriptional regulation of the tryptophan salvage pathway in Chlamydia trachomatis. Elife 8:e42295. doi: 10.7554/eLife.42295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Read TD, Myers GSA, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia RC, McClarty G, Rank RG, Bavoil PM, Fraser CM. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche‐specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res 31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsey KH, Miranpuri GS, Sigar IM, Ouellette S, Byrne GI. 2001. Chlamydia trachomatis persistence in the female mouse genital tract: inducible nitric oxide synthase and infection outcome. Infect Immun 69:5131–5137. doi: 10.1128/IAI.69.8.5131-5137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.