Invasive aspergillosis (IA) is a life-threatening infection that affects an increasing number of patients undergoing chemotherapy or allo-transplantation, and recent studies have shown that genetic factors contribute to disease susceptibility. In this two-stage, population-based, case-control study, we evaluated whether 7 potentially functional single nucleotide polymorphisms (SNPs) within the ARNT2 and CX3CR1 genes influence the risk of IA in high-risk hematological patients.
KEYWORDS: ARNT2, CX3CR1, host immunity, invasive aspergillosis, genetic susceptibility
ABSTRACT
Invasive aspergillosis (IA) is a life-threatening infection that affects an increasing number of patients undergoing chemotherapy or allo-transplantation, and recent studies have shown that genetic factors contribute to disease susceptibility. In this two-stage, population-based, case-control study, we evaluated whether 7 potentially functional single nucleotide polymorphisms (SNPs) within the ARNT2 and CX3CR1 genes influence the risk of IA in high-risk hematological patients. We genotyped selected SNPs in a cohort of 500 hematological patients (103 of those had been diagnosed with proven or probable IA), and we evaluated their association with the risk of developing IA. The association of the most interesting markers of IA risk was then validated in a replication population, including 474 subjects (94 IA and 380 non-IA patients). Functional experiments were also performed to confirm the biological relevance of the most interesting markers. The meta-analysis of both populations showed that carriers of the ARNT2rs1374213G, CX3CR1rs7631529A, and CX3CR1rs9823718G alleles (where the RefSeq identifier appears as a subscript) had a significantly increased risk of developing IA according to a log-additive model (P value from the meta-analysis [PMeta] = 9.8 · 10−5, PMeta = 1.5 · 10−4, and PMeta =7.9 · 10−5, respectively). Haplotype analysis also confirmed the association of the CX3CR1 haplotype with AGCGG with an increased risk of IA (P = 4.0 · 10−4). Mechanistically, we observed that monocyte-derived macrophages (MDM) from subjects carrying the ARNTR2rs1374213G allele or the GG genotype showed a significantly impaired fungicidal activity but that MDM from carriers of the ARNT2rs1374213G and CX3CR1rs9823718G or CX3CR1rs7631529A alleles had deregulated immune responses to Aspergillus conidia. These results, together with those from expression quantitative trait locus (eQTL) data browsers showing a strong correlation of the CX3CR1rs9823718G allele with lower levels of CX3CR1 mRNA in whole peripheral blood (P = 2.46 · 10−7) and primary monocytes (P = 4.31 · 10−7), highlight the role of the ARNT2 and CX3CR1 loci in modulating and predicting IA risk and provide new insights into the host immune mechanisms involved in IA development.
INTRODUCTION
Invasive aspergillosis (IA) is a life-threatening infection in which Aspergillus spp. colonize lung or sinus tissues and spread through the bloodstream to other sites in the body (1). Recent studies have consistently reported that IA is increasing in incidence among immunocompromised (1), postoperative (2), critically ill (3), and solid-organ transplantation (1, 4) patients. Following a specific exposure, the risk of developing IA depends on factors such as a weakened immune system (1, 5), graft versus host disease (6), hematological malignancy (7), long-term corticosteroid and/or immunomodulatory therapy (8, 9), lower respiratory tract and cytomegalovirus infections (10), AIDS (11), and lung disorders (12).
There is robust evidence that the combination of some of these clinical risk factors and a specific host genetic background may render individuals more vulnerable to IA (13, 14) and increase the risk of infection-related hospitalizations and deaths (15, 16). In particular, it is widely known that the presence of single nucleotide polymorphisms (SNPs) within macrophage-related genes has an impact in modulating the risk of developing invasive fungal infections. It has been reported, for instance, that polymorphisms within tumor necrosis factor receptors (TNFRs; TNFR1 and TNFR2) (17, 18), Toll-like receptors (TLRs; TLR2, TLR3, TLR4, TLR5, TLR6, and TLR9) (19–23), dectins (Dectin-1 and Dectin-2) (24–26), cytokines (interleukin 1 [IL-1], IL-10) (27–29), chemokines (CXCL10) (30), and dectin, cytokine, and chemokine receptors substantially influence the risk of developing IA in high-risk patients.
Recent studies have also pointed to the important role played by macrophage-related genes other than those mentioned above, such as CX3CR1 and ARTN2. CX3CR1 is a G-coupled transmembrane chemokine receptor that, after the interaction with its ligand fractalkine, is able to induce downstream signaling leading to NF-κB activation and thereby the production of fractalkine itself and some other proinflammatory cytokines (31). Furthermore, it has been demonstrated that CX3CR1 is implicated in the differentiation of phagocytes (32) and in modulating the interaction of immune phagocytes with fungal pathogens (33). On the other hand, it has also been found that the AHR/ARNT2 complex plays a role in regulating the activity and differentiation of phagocytic cells (including macrophages) and lymphocytes but also in modulating the transcription of multiple immune-related genes, including cytokines (TNF alpha [TNF-α], IL-1β, IL-2, transforming growth factor α [TGF-α], and TGF-β) (34, 35) and the shift of the Th1/Th2 balance toward Th1 (36).
Taking this under consideration, the aim of this study was to conduct a two-stage case-control association study to evaluate whether 7 potentially functional SNPs within the ARNT2 and CX3CR1 genes were associated with the risk of developing IA in 2 independent cohorts of hematological patients at high risk of infection. We also conducted functional assays to determine the effect of these markers on the immune response to Aspergillus fumigatus.
(Parts of this study were reported at the 6th Advances against Aspergillosis Conference, 27 February to 1 March 2014, in Madrid, Spain, and at the 44th Annual Meeting of the European Society for Blood and Marrow Transplantation, 18 to 21 March 2018, in Lisbon, Portugal.)
RESULTS
Characteristics of study subjects.
The study population included 197 cases with proven or probable IA and 777 disease-matched patients without signs of infection. Demographic and clinical characteristics of the patients included are summarized in Table 1. Briefly, IA and non-IA groups had similar ages and underlying disease distributions in both the discovery and replication cohorts. Furthermore, the two populations showed similar proportions of patients who underwent allogeneic hematopoietic stem cell transplantation (allo-SCT) (Table 1). Overall, IA was more frequently found in men than in women (P = 0.001) and among those patients diagnosed with acute lymphoblastic leukemia (ALL) (P = 0.028). As expected, IA tended to be less frequent among patients who completed antifungal prophylaxis (P = 0.003).
TABLE 1.
Baseline and clinical characteristic of patients with or without IAa
| Population | Variableb | Overall (n = 500) |
IA patients (n = 103) |
Non-IA patients (n = 397) |
P value |
|---|---|---|---|---|---|
| Discovery | Demographics | ||||
| Age (avg no. of yrs ± SD) | 51.96 ± 15.20 | 51.94 ± 12.80 | 51.97 ± 15.78 | 0.986 | |
| Sex ratio (no. of males/no. of females) | 1.16 (269/231) | 2.03 (69/34) | 1.02 (200/197) | 0.003 | |
| No. (%) with hematological disease: | |||||
| AML | 354 (70.80) | 66 (64.08) | 288 (72.54) | 0.092 | |
| ALL | 65 (13.00) | 17 (16.50) | 48 (12.09) | 0.235 | |
| Other | 81 (16.20) | 20 (19.42) | 61 (15.37) | 0.320 | |
| Allo-SCT | 202 (40.40) | 46 (44.66) | 156 (39.29) | 0.323 | |
| No. (%) who ever received prophylaxis | 268 (66.83) | 36 (49.31) | 232 (70.94) | 0.0008 | |
| (n = 474) | (n = 94) | (n = 380) | |||
| Replication | Demographics | ||||
| Age (avg no. of yrs ± SD) | 52.48 ± 16.57 | 51.38 ± 17.32 | 52.75 ± 16.43 | 0.512 | |
| Sex ratio (no. of males/no. of females) | 1.53 (287/187) | 2.24 (65/29) | 1.41 (222/158) | 0.056 | |
| No. (%) with hematological disease: | |||||
| AML | 252 (53.28) | 50 (53.19) | 203 (53.42) | 0.912 | |
| ALL | 75 (15.86) | 21 (22.58) | 54 (14.21) | 0.076 | |
| Other | 146 (30.87) | 23 (24.73) | 123 (32.37) | 0.174 | |
| Allo-SCT | 157 (33.19) | 31 (33.33) | 127 (33.42) | 0.968 | |
| No. (%) who ever received prophylaxis | 182 (63.85) | 38 (61.29) | 144 (64.57) | 0.744 | |
| (n = 974) | (n = 197) | (n = 777) | |||
| Overall | Demographics | ||||
| Age (avg no. of yrs ± SD) | 52.23 ± 15.89 | 51.74 ± 15.10 | 52.36 ± 16.10 | 0.612 | |
| Sex ratio (no. of males/no. of females) | 1.33 (556/418) | 2.12 (134/63) | 1.19 (422/355) | 0.001 | |
| No. (%) with hematological disease: | |||||
| AML | 606 (62.22) | 116 (58.88) | 491 (63.19) | 0.265 | |
| ALL | 140 (14.37) | 38 (19.29) | 102 (13.13) | 0.028 | |
| Other | 227 (23.31) | 43 (21.83) | 184 (23.68) | 0.583 | |
| Allo-SCT | 359 (36.86) | 77 (39.09) | 283 (36.42) | 0.489 | |
| No. (%) who ever received prophylaxis | 450 (65.59) | 74 (54.81) | 376 (68.36) | 0.003 |
Abbreviations: Allo-SCT, allogeneic stem cell transplantation; AML, acute myeloid leukemia; ALL, acute lymphoid leukemia. P values of ≤0.05 were considered significant and are shown in bold.
Prophylaxis status was available for 400 subjects from the discovery cohort (73 IA and 327 non-IA patients) and 285 subjects from the replication cohort (62 IA and 223 non-IA patients).
Association analysis.
All SNPs were in Hardy-Weinberg equilibrium (HWE) in the control group (P > 0.05). The logistic-regression analysis, adjusted for age, gender, allo-SCT, and country of origin, revealed that 3 polymorphisms within the ARTN2 and CX3CR1 genes showed a statistically significant association with the risk of IA when log-additive and dominant models of inheritance were assumed (Table 2). Carriers of the CX3CR1rs7631529A, CX3CR1rs9823718G, and ARNT2rs1374213G alleles (where the RefSeq identifier [rsID] appears as a subscript) had an increased risk of developing IA (per-allele odds ratio [OR] = 2.64; 95% confidence intervals [95%CI], 1.52 to 4.60; P = 0.00080; per-allele OR = 2.38; 95%CI, 1.51 to 3.76; P = 0.00030; and dominant-model OR [ORDom] = 2.88; 95%CI, 1.49 to 5.55; P = 0.0006). Importantly, the association of the CX3CR1rs7631529, CX3CR1rs9823718, and ARNT2rs1374213 SNPs survived after correction for multiple testing (study-wide threshold = 0.0024) (Table 2). In addition, although information about prophylaxis status was available for about 66% of the patients, the association of the CX3CR1rs7631529 and CX3CR1rs9823718 SNPs with IA risk remained statistically significant when the use of antifungal prophylaxis was added as a covariate for adjustment (per-allele OR = 3.08; 95%CI, 1.62 to 5.87; P = 0.0008, and per-allele OR = 2.54; 95%CI, 1.47 to 4.30; P = 0.0009, respectively). After correcting for antifungal prophylaxis status, the association of ARNT2rs1374213 with IA remained close to the multiple testing significance threshold when a dominant model was assumed (ORDom = 2.41; 95%CI, 1.18 to 4.92; PDom = 0.0098). In line with these findings, we also found a significant effect of the haplotype CX3CR1AGCGG (CX3CR1 with AGCGG, containing the risk alleles) in determining the risk of developing the infection (PHap = 0.0004), whereas only a modest effect on IA risk was found for the ARNT2GT haplotype (PHap = 0.011) (Table 3).
TABLE 2.
Summary results for the ARNT2 and CX3CR1 SNPs associated with IA risk in the discovery populationa
| Gene | SNP rsID | bp | Chr. | Risk allele | MAF of IA cases | MAF of non-IA cases | MAF of healthy controlsb | HWE for non-IA group | OR (range) (additive) | P value | OR (range) (dominant) | P value | OR (range) (recessive) | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARNT2 | rs1374213 | 80732053 | 15 | G | 0.560 | 0.430 | 0.464 | 0.30 | 1.58 (1.11–2.23) | 0.0093 | 2.88 (1.49–5.55) | 0.00060 | 1.29 (0.74–2.25) | 0.37 |
| ARNT2 | rs12439281 | 80980052 | 15 | T | 0.180 | 0.200 | 0.166 | 1.00 | 0.86 (0.56–1.31) | 0.48 | 0.90 (0.55–1.47) | 0.67 | 0.47 (0.10–2.15) | 0.29 |
| CX3CR1 | rs7631529 | 39286825 | 3 | A | 0.130 | 0.060 | 0.074 | 0.39 | 2.64 (1.52–4.60) | 0.00080 | 2.35 (1.30–4.28) | 0.0062 | NA (NA) | NA |
| CX3CR1 | rs9823718 | 39293757 | 3 | G | 0.190 | 0.100 | 0.094 | 0.38 | 2.38 (1.51–3.76) | 0.00030 | 2.44 (1.43–4.15) | 0.0012 | 6.22 (1.67–23.2) | 0.0091 |
| CX3CR1 | rs9862876 | 39311583 | 3 | G | 0.210 | 0.230 | 0.209 | 0.89 | 0.86 (0.58–1.29) | 0.46 | 0.81 (0.50–1.31) | 0.39 | 0.98 (0.34–2.78) | 0.96 |
| CX3CR1 | rs9868689 | 39312941 | 3 | T | 0.180 | 0.190 | 0.172 | 0.32 | 0.96 (0.63–1.48) | 0.86 | 0.89 (0.54–1.45) | 0.63 | 1.59 (0.48–5.29) | 0.46 |
| CX3CR1 | rs12107527 | 39369682 | 3 | T | 0.280 | 0.330 | 0.329 | 0.87 | 0.76 (0.52–1.11) | 0.15 | 0.84 (0.53–1.35) | 0.48 | 0.36 (0.12–1.04) | 0.071 |
Abbreviations: SNP, single nucleotide polymorphism; rsID, RefSeq identifier; bp, base pair position; Chr., chromosome; MAF, minor allele frequency; OR, odds ratio; NA, not applicable. The major allele for each SNP was considered the reference allele, and the minor allele was considered the effect allele. Boldface indicates a significant difference.
Adjusted for age, sex, allo-SCT status, and country of origin. MAFs for healthy controls were estimated using immuno-ChIP data from a European population of 756 aged-matched subjects selected from the general population. Immuno-ChIP data were available in our laboratory at GENYO (Granada, Spain).
TABLE 3.
Haplotype association analysis for ARNT2 and CX3CR1 SNPsa
| Gene | Nucleotide in: |
Freq | OR (95%CI) of discovery population (n = 424) | P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs1374213 | rs12439281 | rs7931529 | rs9823718 | rs9862876 | rs9868689 | rs12107527 | ||||
| ARNT2b | A | G | 0.424 | 1.00 | ||||||
| G | G | 0.387 | 1.13 (0.73–1.76) | 0.58 | ||||||
| A | T | 0.120 | 0.32 (0.08–1.28) | 0.11 | ||||||
| G | T | 0.070 | 2.62 (1.25–5.47) | 0.011 | ||||||
| CX3CR1c | G | C | C | G | G | 0.4645 | 1.00 | |||
| G | C | C | G | A | 0.1874 | 0.55 (0.31–1.00) | 0.050 | |||
| G | C | G | A | A | 0.1056 | 1.46 (0.79–2.71) | 0.23 | |||
| G | C | G | A | G | 0.0602 | 0.37 (0.13–1.09) | 0.073 | |||
| A | G | C | G | G | 0.0563 | 3.56 (1.78–7.13) | 0.0004 | |||
| G | C | G | G | G | 0.0497 | 0.49 (0.17–1.41) | 0.19 | |||
| G | G | C | G | G | 0.0325 | 1.88 (0.77–4.60) | 0.17 | |||
| A | G | C | G | A | 0.0176 | 0.29 (0.01–13.5) | 0.52 | |||
Nucleotides that are bold and underlined represent those alleles that were significantly associated with IA risk in the single SNP analysis. Association estimates were adjusted for age, sex, and country of origin. The minimum haplotype frequency (Freq) was set at 0.01. P values that were ≤0.05 are in bold.
The global haplotype association P value was 0.014.
The global haplotype association P value was 0.00019.
Interestingly, when we attempted to replicate these relevant associations in other populations of European ancestry, we could confirm that carriers of the ARNT2rs1374213G allele had a significantly increased risk of IA (per-allele OR = 1.63; 95%CI, 1.18 to 2.27; P = 0.003), whereas carriers of the CX3CR1rs9823718G and CX3CR1rs7631529A alleles also tended to have an increased risk of developing the infection (OR = 1.58; 95%CI, 0.97 to 2.57; P = 0.073; and OR = 1.77; 95%CI, 0.98 to 3.21; P = 0.059, respectively). The direction of the effect for these associations was similar to the one observed in the discovery population, and the combined analysis confirmed that each copy of the minor allele for the ARNT2 and CX3CR1 SNPs was strongly associated with a 1.61- to 2.19-fold-increased risk of IA when a log-additive model was assumed (PMeta= 0.000098, 0.00015, 0.000079) (Table 4). Although slightly weaker, the association of the ARNT2rs1374213, CX3CR1rs7631529, and CX3CR1rs9823718 SNPs with IA risk was also statistically significant when dominant or even recessive models of inheritance were assumed (Table 4). In addition, the distribution of the ARNT2rs1374213G, CX3CR1rs7631529A, and CX3CR1rs9823718G alleles was consistent across different countries, which undoubtedly pointed to a relevant role for these genes in modulating the risk of IA (Table 4 and see Table S1 in the supplemental material).
TABLE 4.
Meta-analyses of ARNT2 and CX3CR1 polymorphismsa
| Gene | SNP rsID | Chr. | Risk allele | Discovery population |
Replication population |
Meta-analysis |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | P value | OR | P value | OR | PMeta | PHet | I2 (%) | ||||
| ARNT2 (additive) | rs1374213 | 15 | G | 1.58 (1.11–2.23) | 0.0093 | 1.63 (1.18–2.27) | 0.003 | 1.61 (1.27–2.04) | 0.000098 | 0.890 | 0.00 |
| ARNT2 (dominant) | rs1374213 | 15 | G | 2.88 (1.49–5.55) | 0.00060 | 1.84 (1.05–3.22) | 0.032 | 2.22 (1.45–3.40) | 0.00024 | 0.309 | 3.2 |
| ARNT2 (recessive) | rs1374213 | 15 | G | 1.29 (0.74–2.25) | 0.37 | 2.09 (1.24–3.53) | 0.006 | 1.67 (1.14–2.44) | 0.0086 | 0.215 | 34.8 |
| CX3CR1 (additive) | rs9823718 | 3 | G | 2.38 (1.51–3.76) | 0.00030 | 1.58 (0.97–2.57) | 0.073 | 1.97 (1.41–2.74) | 0.000079 | 0.230 | 30.9 |
| CX3CR1 (dominant) | rs9823718 | 3 | G | 2.44 (1.43–4.15) | 0.0012 | 1.62 (0.90–2.91) | 0.10 | 2.03 (1.37–3.01) | 0.00044 | 0.310 | 2.5 |
| CX3CR1 (recessive) | rs9823718 | 3 | G | 6.22 (1.67–23.2) | 0.0091 | 2.61 (0.60–11.38) | 0.20 | 4.23 (1.59–11.28) | 0.0040 | 0.389 | 0.0 |
| CX3CR1 (additive) | rs7631529 | 3 | A | 2.64 (1.52–4.60) | 0.00080 | 1.77 (0.98–3.21) | 0.059 | 2.19 (1.46–3.29) | 0.00015 | 0.334 | 0.0 |
| CX3CR1 (dominant) | rs7631529 | 3 | A | 2.35 (1.30–4.28) | 0.0062 | 1.71 (0.88–3.31) | 0.11 | 2.04 (1.31–3.18) | 0.0016 | 0.480 | 0.0 |
| CX3CR1 (recessive) | rs7631529 | 3 | A | NA (NA) | NA | 7.34 (0.64–84.0) | 0.11 | NA (NA) | NA | NA | NA |
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; PHet, pheterogeneity; I2, amount of dispersion in the meta-analysis; NA, not applicable. Values in boldface indicate a significant difference at nominal level (P < 0.05). The major allele for each SNP was considered the reference allele, and the minor allele was considered the effect allele. ORs for the discovery and replication populations were adjusted for age, sex, allo-SCT, and country of origin. Meta-analyses were conducted by assuming a fixed-effect model.
Functional evidence of the impact of ARNT2 and CX3CR1 SNPs on the immune response.
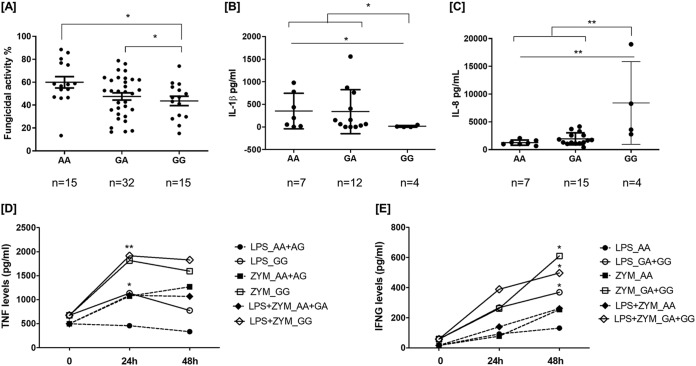
Next, we evaluated whether the ARNTR2rs1374213, CX3CR1rs7631529, and CX3CR1rs9823718 variants influenced the strength of immune responses against A. fumigatus. Interestingly, we observed that MDM from subjects carrying the ARNTR2rs1374213GG genotype showed an impaired capacity to kill A. fumigatus conidia compared with MDM from carriers of the AA genotype (Fig. 1A). We also observed that bronchoalveolar lavage (BAL) fluid samples from IA patients carrying the ARNTR2rs1374213GG genotype had a significantly decreased release of IL-1β and an exacerbated production of IL-8 (Fig. 1B and 2C). Furthermore, we observed that, after stimulation with zymosan for 24 or 48 h, peripheral blood mononuclear cells (PBMCs) from carriers of the ARNTR2rs1374213GG genotype and the ARNTR2rs1374213G allele showed a significantly increased production of TNF-α (Fig. 1D) and gamma interferon (IFN-γ) (Fig. 1E).
FIG 1.
(A) Fungicidal activities of monocyte-derived macrophages (A); (B to E) cytokine levels on BAL samples (B and C) and produced by PBMCs (D and E) according to the ARNT2rs1374213 genotype. MDM were stimulated with Aspergillus conidia (1:10), and PBMCs were stimulated with zymosan (5 μg/ml) alone or in combination with lipopolysaccharide (LPS; 100 ng/ml). Supernatants were harvested for cytokine analysis at 24 and/or 48 h. (A) MDM from subjects carrying the ARNT2rs1374213GG genotype or the ARNT2rs1374213G allele showed an impaired capacity to kill A. fumigatus conidia compared with that of carriers of the AA genotype (AA genotype, 59.92% of conidia killed, versus GG genotype, 43.61% [P = 0.0176], or AA genotype, 59.92%, versus GA + GG genotype, 46.27% [P = 0.0122], respectively). BAL samples from IA patients carrying the ARNT2rs1374213GG genotype had a significantly decreased release of IL-1β (B) and a significantly exacerbated production of IL-8 (C) in comparison with those of patients carrying the AA genotype or the A allele [PIL-1β (AA versus GG) = 0.042 and PIL-1β (AA + AG versus GG) = 0.026; PIL8(AA versus GG) = 0.0061 and PIL8 (AA + AG versus GG) = 0.0024, respectively]. (D) After stimulation with zymosan (ZYM) for 24 h, PBMCs from carriers of the ARNT2rs1374213GG genotype (n = 3) showed a significantly increased production of TNF-α compared with that of subjects carrying the A allele [n = 18; PTNF-LPS (24 h) = 0.017, PTNF-ZYM (24 h) = 0.068 PTNF-LPS + ZYM (24 h) = 0.001] (Fig. 2D). (E) Similarly, after stimulation with zymosan for 48 h, PBMCs from carriers of the ARNT2rs1374213G allele (n = 6) showed a significantly increased production of IFN-γ [n = 14, PIFN-γ–LPS (48 h) = 0.042, PIFN-γ–ZYM (48 h) = 0.045, and PIFN-γ–LPS + ZYM (48 h) = 0.040].
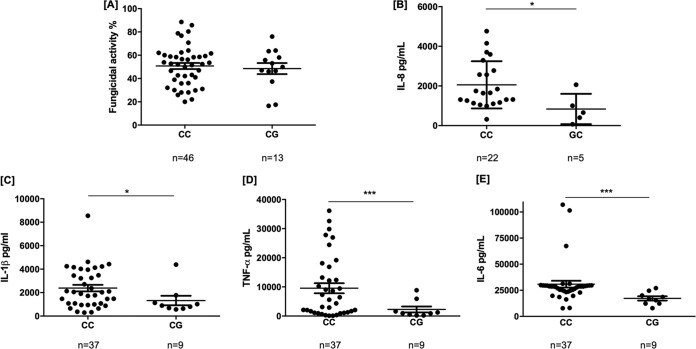
FIG 2.
Fungicidal activities of monocyte-derived macrophages (A) and cytokine levels on BAL samples from IA patients (B) or from stimulated MDM (C to E) according to the CX3CR1rs9823718 genotype. MDM were stimulated with Aspergillus conidia (1:10). Supernatants were harvested for cytokine analysis at 24 and/or 48 h. (A) No differences in fungicidal activity were observed among carriers of the different CX3CR1rs9823718 genotypes. (B) BAL samples from patients carrying the CX3CR1rs9823718G allele, which strongly correlates with lower levels of CX3CR1 mRNA in whole peripheral blood (Z-score = –5.16, P = 2.46 · 10−7, and Z-score = –5.05, P = 4.41 · 10−7) and primary monocytes (P = 4.31 · 10−7), had a significantly decreased release of IL-8 (P = 0.019). (C to E) Likewise, MDM from healthy individuals carrying the CX3CR1rs9823718G allele showed a significantly decreased production of IL-1β (PIL-1 = 0.0418) (C), TNF-α (PTNF = 0.0007) (D), and IL-6 (PIL-6 = 0.0003) (E) after coincubation for 24 h with A. fumigatus conidia. Given the relatively high linkage disequilibrium between CX3CR1 markers, results obtained for the CX3CR1rs7631529 SNP were almost identical.
On the other hand, although we could not detect differences in capacities to kill A. fumigatus conidia among carriers of the different CX3CR1rs9823718 genotypes (Fig. 2A), we observed that BAL samples from IA patients carrying the CX3CR1rs9823718G allele had a significantly decreased release of IL-8 (Fig. 2B) and that MDM from healthy individuals carrying this allele also showed a significantly decreased production of IL-1β, TNF-α, and IL-6 after coincubation for 24 h with A. fumigatus conidia (Fig. 2C to E). These results, together with those from the expression quantitative trait locus (eQTL) data browsers, which report a strong correlation of the CX3CR1rs9823718G allele with lower levels of CX3CR1 mRNA in whole peripheral blood (P = 2.46 · 10−7) and primary monocytes (P = 4.31 · 10−7) (37), suggested that the CX3CR1 locus might be implicated in modulating the immune response against A. fumigatus. Given the moderate linkage disequilibrium between the CX3CR1rs7631529 and CX3CR1rs9823718 variants (r2 = 0.62) (Fig. S1), the results for carriers of the CX3CR1rs7631529A allele were similar to those observed for carriers of the CX3CR1rs9823718G allele (data not shown). No significant differences in cytokine production according to the CX3CR1rs9823718 and CX3CR1rs7631529 genotypes were observed in PBMCs from healthy subjects after stimulation with zymosan for 24 or 48 h (data not shown).
DISCUSSION
To the best of our knowledge, this is the first association study evaluating the role of polymorphisms within ARNT2 and CX3CR1 genes and the risk of IA. Despite the relatively modest sample size of the populations analyzed, our results showed a consistent association with IA risk for 3 SNPs within the ARNT2 and CX3CR1 genes. In the combined analysis of the discovery and replication populations, the strongest effect was observed for the ARNT2rs1374213 and CX3CR1rs9823718 SNPs.
The ARNT2 gene (15q25.1) encodes a member of the basic helix-loop-Per-Arnt-Sim (bHLH-PAS) superfamily of transcription factors, which form heterodimers with the aryl hydrocarbon receptor (AHR) (38), a key regulator of the innate and adaptive immunity implicated in the transcriptional regulation of a wide range of immunity genes, including cytokines (34). The AHR/ARNT2 complex influences the activity and differentiation of phagocytic cells and lymphocytes and the shift in the Th1/Th2 balance toward Th1 (36). Early studies have also described that this protein complex modulates the cell cycle (39) and the response to chemical compounds (40). Although the specific role of ARNT2 in the modulation of the antifungal effector function of innate cells remains elusive, a recent study has demonstrated that ARNT2 is a late-phase TNF response gene that is induced by IL-10 (41) and that, therefore, might play a role in the modulation of the disease tolerance defense pathway and host fitness (42). Although genetic studies have suggested that ARNT2 polymorphisms are involved in neurological diseases (43), little is known about their relationship with infectious diseases (44). In support of a role for the ARNT2 SNPs in the control of host innate immunity against Aspergillus, we observed that the MDM from carriers of the ARNT2rs1374213GG genotype showed an impaired ability to kill Aspergillus conidia that might be due, at least in part, to an impaired macrophage-mediated production of IL-1β and the consequent depletion of phagocyte-dependent antifungal activities. We also observed that MDM from carriers of the ARNT2rs1374213GG genotype showed an exacerbated release of IL-8, which might promote infection by inducing the recruitment of neutrophils to the site of infection and, consequently, the release of TNF-α and IFN-γ, which might cause severe inflammation of the lung tissue and defective fungal clearance. In line with these results, we observed that PBMCs from subjects carrying the ARNT2rs1374213G allele showed a sustained and significantly increased production of TNF-α and IFN-γ after stimulation with fungal antigens. These findings, along with those from previous studies suggesting that IL-8 may also induce the synthesis of metalloproteinase-9 (45), suggest that ARNT2 might increase an individual’s susceptibility to IA by depleting the antifungal activity of innate immune cells and by promoting aggressive adaptive proinflammatory responses mediated by IL-8 that might lead to degradation of the extracellular matrix, lung tissue damage, and a defective fungal clearance.
On the other hand, it is well established that CX3CR1 is involved in the pathogenesis of fungal infections (33). The CX3CR1 gene (3p22.2) encodes the fractalkine receptor, a gastrointestinal tract-coupled transmembrane protein and chemokine receptor that contributes to controlling host innate and adaptive immunity at multiple levels. CX3CL1, its unique ligand, along with CCL26, is markedly upregulated under inflammatory conditions, suggesting that the CX3CL1/CX3CR1 axis might play an essential role during infection. CX3CR1 is expressed mainly in airway and alveolar epithelial cells of the lungs (46) but also in a wide range of immune cells, such as cytotoxic effector lymphocytes, low-expression CCR2 (CCR2low) LY6Clow and CX3CR1hi monocytes, macrophages, and subsets of dendritic cells (47) that interact with fungal pathogens in a CX3CR1-dependent manner (33). CX3CR1 participates in the differentiation of myeloid progenitors to the monocyte, macrophage, and dendritic cell lineage (32), and its depletion impairs the survival of these cells and their capacities to promote human leukocyte adhesion, migration, and extravasation into inflamed tissues. In line with these findings, this study showed that the presence of the CX3CR1rs7631529A and CX3CR1rs9823718G alleles, which strongly correlate with lower expression of the CX3CR1 protein in cytotoxic effector lymphocytes and phagocytes (37, 48), rendered patients more likely to develop IA. In addition, our data showed that the CX3CR1AGGCC haplotype containing risk alleles was strongly associated with an increased risk of developing IA. Given the relatively high linkage disequilibrium (LD) value of the CX3CR1 risk alleles, we found that the effect of this haplotype was of a magnitude similar to those observed for single SNPs. These findings together with those from publicly available eQTL data browsers suggest that the effect attributable to the CX3CR1 locus on IA risk might depend on eQTL alleles. However, additional research is now warranted to clarify whether the effect of CX3CR1 SNPs on CX3CR1 mRNA expression is led by a single SNP or a larger haplotype. In addition, in support of the idea suggesting a functional role of CX3CR1 SNPs in biological processes, such as cell adhesion and migration (47), we also found a reduced production of IL-8 in the lungs of infected patients carrying the CX3CR1rs9823718G allele, whereas MDM from carriers of this allele showed a significantly reduced release of IL-1β and TNF-α after coincubation with Aspergillus conidia. These results are consistent with those of previous studies suggesting that CX3CR1 is essential for an effective immune response against Aspergillus (49) but also with those of previous studies suggesting a functional impact of CX3CR1 SNPs in modulating susceptibility to other lethal infections (50) and disease progression (51, 52). In line with our results, it has been observed that defects in CX3CR1 expression increases susceptibility to fungal pathogens by reducing macrophage accumulation in tissues and survival (33) and that this receptor is essential for the clearance of fungal pathogens from the mucosa and lower gastrointestinal tract (53). Therefore, although the information provided by our and other studies is relevant, an in-depth analysis of the biological role of CX3CR1 in IA, including mechanistic insights, is still needed.
Finally, it is necessary to mention that even though the influence of ARNT2 and CX3CR1 variants on the risk of the infection was expected to be modest, the allelic distribution of these SNPs was consistent across different countries, and overall, our study was sufficiently powered to detect such small effects. Based on the genotype frequencies observed in the discovery cohort, we had 80% of the power (log-additive model) to detect an odds ratio of 1.62 at an alpha of 0.0024 (multiple-testing threshold) for a polymorphism with a minor allele frequency (MAF) of 0.25 and an OR of 1.95 for an SNP with a MAF of 0.10. Importantly, our results were confirmed in a replication cohort, supporting the notion of a role of the ARNT2 and CX3CR1 loci in IA susceptibility.
In conclusion, this association study identifies for the first time ARNT2 and CX3CR1 as new susceptibility loci for IA and provides new insights about the possible role of these loci in modulating innate and adaptive immune responses to A. fumigatus.
MATERIALS AND METHODS
Study population.
The discovery population consisted of 500 European hematological patients at high risk of invasive fungal infection, 103 hematological patients diagnosed with proven or probable IA, and 397 noninfected and disease-matched patients. Hematological patients were allo-transplanted or diagnosed with acute leukemia receiving intensive remission-induction chemotherapy and were recruited through the AspBIOmics consortium (www.aspbiomics.eu) and from two Spanish medical institutions (the University Hospital of Salamanca and the Clinic University Hospital of Valencia). A Spanish multicenter clinical trial was registered with ClinicalTrials.gov (NCT01742026) and EudraCT (2010-019406-17). In accordance with the Declaration of Helsinki, all participants provided their written informed consent to participate in the study, and the ethical committees of the following participating centers and hospitals approved the study: Virgen de las Nieves University Hospital (0702/12; Granada, Spain), University Hospital of Salamanca (NCT01742026; Salamanca, Spain), Clinic University Hospital of Valencia (NCT01742026; Valencia, Spain), Centro Nacional de Microbiología (NCT01742026; Madrid, Spain), University of Würzburg (173/11; Würzburg, Germany), and Medical University of Innsbruck (UN4529 and 04/2014; Innsbruck, Austria). Ethical approval was also provided by the ethical review boards of the Università Cattolica del S. Cuore (0029458/16 and 0003932/17; Rome, Italy) and the University of Modena and Reggio Emilia (2629/16; Modena, Italy). Approval for the functional genomics studies was obtained from the ethics subcommittees for Virgen de las Nieves University Hospital (Spain; 02276/17), Life and Health Sciences of the University of Minho (Portugal; SECVS 014/2015), and the National Commission for the Protection of Data (Portugal; 1950/015). Proven and probable IA cases were diagnosed according to the revised EORTC/MSG criteria (54). Proven IA cases were diagnosed after microscopic analysis or culture of sterile material (biopsy specimens or needle aspirations) and subsequent identification of the Aspergillus mold by experienced pathologists. All centers used the presence of galactomannan as a microbiological criterion for the diagnosis of probable IA. Patients with no sign of infection and lacking pulmonary infiltrates for a period of at least 12 months were classified as non-IA cases.
DNA extraction, sample selection, genotyping, quality control, and filtering.
Genomic DNA from saliva or blood samples from hematological patients at high risk of IA and healthy controls was isolated using the Oragene DNA self-collection kit (Oragene) or the Maxwell 16 blood DNA purification kit (Promega) according to the manufacturers’ instructions. DNA samples from donors were considered when IA infection occurred after transplantation. In order to avoid cellular chimerism, donor DNA was extracted directly from the peripheral blood of each allo-SCT donor.
SNPs were selected based on their potential functionality according to HaploReg (www.broadinstitute.org/mammals/haploreg/haploreg.php), RegulomeDB (www.regulomedb.org/), the Blood eQTL browser (https://genenetwork.nl/bloodeqtlbrowser/), and the Genotype-Tissue Expression Portal (GTEx Portal) (www.gtexportal.org/home/) and their linkage disequilibria (defined as r2 values) and because of the plausible implication of the ARNT2 and CX3CR1 loci in the modulation of immune responses against fungal pathogens (Table 5) (33). Genotyping of selected SNPs in the discovery population was performed using KASP probes according to the manufacturer’s instructions (LGC Genomics, Hoddesdon, UK). For quality control purposes, 5% of samples were randomly included as duplicates, and the concordance between duplicate samples was ≥99.5.
TABLE 5.
List of selected SNPs according to their potential functionality in the GTEx portal, HaploReg, Encode, etc.a
| Gene | SNP rsID | bp | Location | Chr. | Risk allele | Remarks about known function |
|---|---|---|---|---|---|---|
| ARNT2 | rs1374213 | 80732053 | Intron | 15 | G | Regulome score = 5 (TF binding or DNase peak); histone modifications; regulation of changes in the promoter H3K9ac in primary monocytes (H3K9ac_Pro) |
| ARNT2 | rs12439281 | 80980052 | Near gene (downstream) | 15 | T | eQTL for ABHD17C in whole blood (P = 2.40 · 10–5) |
| CX3CR1 | rs7631529 | 39286825 | Intron | 3 | A | eQTL for CX3CR1 in whole blood (P = 2.25 · 10–6) and most of the primary immune cell types (primary monocytes, primary B cells, neutrophils and natural killer from peripheral blood); Regulome score = 5 (TF binding or DNase peak); bound protein (HFH1); regulation of histone marks in primary monocytes (H3K4me1_Enh and H3K27ac_Enh) |
| CX3CR1 | rs9823718 | 39293757 | Intron | 3 | G | eQTL for CX3CR1 in whole blood (P = 2.46 · 10–7 and 4.41 · 10–7); Regulome score = 4 (TF binding + DNase peak); regulation of histone marks in primary monocytes (13_EnhA1, H3K4me1_Enh, H3K4me3_Pro, and H3K27ac_Enh), primary B cells, neutrophils, and primary mononuclear cells from peripheral blood |
| CX3CR1 | rs9862876 | 39311583 | Intron | 3 | G | Histone modifications; the gene regulates multiple histone marks in primary mononuclear cells, T cells (CD8+), neutrophils, and natural killer cells (H3K4me1, H3K4me3, and H3K27ac) |
| CX3CR1 | rs9868689 | 39312941 | Intron | 3 | T | Regulome score = 5 (TF binding or DNase peak); regulation of multiple histone marks in primary mononuclear cells, primary monocytes, B and T cells (CD8+), neutrophils, and natural killer cells (H3K4me1, H3K4me3, and H3K27ac) |
| CX3CR1 | rs12107527 | 39369682 | Near gene (upstream) | 3 | T | eQTL of CCR8 in EBV-transformed lymphocytes (P = 5.3 · 10–7); Regulome score = 4 (TF binding + DNase peak); bound protein (NF-κB1) in multiple immune-related cell lines; regulation of enhancer histone marks in primary T regulatory cells and primary T helper memory cells from peripheral blood. |
Abbreviations: SNP, single nucleotide polymorphism; Chr., chromosome; TF, transcription factor.
Association analysis.
The Hardy-Weinberg equilibrium (HWE) test was performed on the control group (non-IA patients) by means of a standard observed-expected χ2 test. Logistic regression analyses adjusted for age, sex, allo-SCT, prophylactic status, and country of origin were used to assess the main effects of the selected SNPs on IA risk. Associations for each marker with IA risk were tested according to log-additive, dominant, and recessive models of inheritance. Statistical power was calculated using Quanto v.12.4 (http://biostats.usc.edu/software), assuming a log-additive model.
LD and haplotype analysis.
We performed haplotype frequency estimation and haplotype association analysis adjusted for age, sex, allo-SCT, and country of origin using the haplo.stats package. Haplotype frequencies were determined using the expectation-maximization (EM) algorithm, haplotypes were reconstructed using SNPtool and Haploview, and block structures were determined according to the method of Gabriel et al. (55) (see Fig. S1 in the supplemental material).
Replication population.
Four hundred seventy-four hematological patients (94 with proven or probable IA) were recruited from a Spanish institution (Virgen de las Nieves University Hospital) and two Italian medical institutions (Università Cattolica del S. Cuore, Rome, Italy, and the University of Modena and Reggio Emilia, AOU Policlinico, Modena, Italy) as described elsewhere (56). Ethical approval was provided by the ethical review boards of these institutions (approvals 0029458/16 and 0003932/17 [Rome, Italy] and approval 2629/16 [Modena, Italy]). Genotyping of this replication cohort was performed using the same genotyping technology, and again 5% of samples were randomly included as duplicates. Genotyping concordance was ≥99%.
Correction for multiple testing was performed using the Bonferroni method but also with consideration of the number of inheritance models tested (log additive, dominant, and recessive). Therefore, the study-wide significance threshold used for the risk analysis was 0.0024 ([0.05]/7 [independent markers]/3 inheritance models).
Meta-analysis.
The meta-analysis of the discovery data with the data from the replication cohort was performed using Stata (v.12) by following an additive model. The I2 statistic, a statistic used to measure dispersion in the meta-analysis, was used to assess heterogeneity across populations, and the pooled odds ratio (OR) was computed using the fixed-effect model.
Cell isolation and differentiation.
After we obtained informed consent (PI12/02688 and SECVS_014/2015 protocols), peripheral blood mononuclear cells (PBMCs) and monocyte-derived macrophages (MDM) were obtained from healthy blood donors according to standard procedures (56). Briefly, PBMCs were isolated by gradient centrifugation using Ficoll-Paque Plus (GE Healthcare Bio-Sciences). They were then washed twice in phosphate-buffered saline (PBS) and resuspended in 2 ml of RPMI 1640 culture medium with l-glutamine and phenol red but without sodium pyruvate (Gibco/Life Technologies) and supplemented with 10% sterile heat-inactivated fetal bovine serum (FBS) and an antibiotic mixture containing penicillin, streptomycin, and neomycin (Gibco/Life Technologies) at 37°C in 5% CO2. Monocytes were isolated by immunomagnetic selection of CD14+ cells (Miltenyi Biotec), and the purity of the obtained CD14+ population was assessed by fluorescence-activated cell sorting analysis. Monocytes were then plated at a density of 5 × 105 cells/ml in 24-well plates (Corning) and cultivated for 7 days in complete RPMI 1640 medium supplemented with human serum and 20 ng/ml of granulocyte-macrophage colony-stimulating factor (GM-CSF; Miltenyi Biotec) to allow differentiation into macrophages. The culture medium was replaced every 3 days, and acquisition of macrophage morphology was confirmed by phase-contrast microscopy (Olympus DX51; Olympus). PBMCs and MDM were grouped according to ARNT2 and CX3CR1 genotypes.
Assessment of fungicidal activity and in vitro stimulation assays.
The AF293 strain (ATCC) was grown on Sabouraud dextrose agar (Difco) for 7 days at 28°C. The conidia were then separated from the mycelium with PBS (Sigma) with 0.1% Tween 20 (Sigma), followed by gentle agitation and subsequent filtration (40 μM). The concentration of conidia per milliliter was determined using a Neubauer chamber. Human MDM from 64 healthy subjects were then infected with live A. fumigatus conidia at an effector-to-target ratio of 1:10 for 1 h, after which the noningested conidia were removed and the MDM were allowed to kill internalized conidia for 2 h. To measure the fungicidal ability, MDM were lysed by quickly freezing them at –80°C and thawing them at 37°C (releasing ingested conidia), cell lysates were mixed thoroughly, and serial dilutions were made in PBS and plated on Sabouraud dextrose agar. Following a 2-day incubation, the number of CFU was determined, and the percentage of CFU inhibition was calculated. CFU counts of conidia treated under the same experimental conditions but in the absence of macrophages were used as controls. Stimulation assays were performed with MDM or PBMCs from 48 healthy donors selected according to their ARNT2rs1374213 and CX3CR1rs9823718 genotypes. PBMCs (1 × 106) were incubated with zymosan (5 μg/ml; Sigma-Aldrich) alone or in combination with lipopolysaccharide (LPS; 100 ng/ml; Sigma-Aldrich) for 24 and 48 h, whereas MDM were left untreated as a negative control or infected with live conidia of A. fumigatus at a 1:10 effector-to-target ratio for 24 h. After the incubation period, supernatants were collected and stored at –80°C until cytokines were measured (a period usually less than 3 months). Cytokine levels were determined in triplicate using the ProcartaPlex multiplex immunoassay (eBioscience) or enzyme-linked immunosorbent assay (ELISA) MAX deluxe set (BioLegend).
BAL specimen collection.
BAL samples from 28 IA patients were collected at the National Reference Center for Medical Mycology in Leuven (Belgium) when a patient fulfilled the European Organisation for Research and Treatment of Cancer (EORTC) diagnostic criteria for probable IA. BAL specimens were obtained by instillation of two samples of 20 ml of 0.9% sterile saline solution to the most peripheral bronchus of the most radiologically involved lobe. BAL samples with comparable recovery rates and from patients that were not long-term smokers, who did not have any other relevant lung-associated diseases, and who were undergoing similar drug regimens were used, according to the standardization rules of the European Respiratory Society, to measure acellular components. BAL specimens were centrifuged at 3,000 rpm for 5 min at 4°C to remove cell debris. All samples were stored at –80°C until use. Cytokine levels were quantified using the human premixed multianalyte kit (R&D Systems).
Statistical analysis for functional analysis.
Cytokine levels in BAL samples or cell culture supernatants were compared between specific genotype groups. Statistical significance was evaluated using an unpaired t test with or without Welch’s correction or the Mann-Whitney U test. A P of ≤ 0.05 was considered significant (Prism v6.0).
Supplementary Material
ACKNOWLEDGMENTS
We thank Antonio Fernández-Montoya (Centro Regional de Transfuciones Sanguíneas Granada-Almería, Spain) and António Marques (Hospital de Braga, Braga, Portugal), who provided the buffy coat cells. We also thank Astellas Pharma Inc. and Consuelo González Moreno (AML survivor) for supporting this work.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We declare no conflict of interest.
This study was supported by grants PI12/02688 and PI17/02276 from the Fondo de Investigaciones Sanitarias (Madrid, Spain), the program PIM2010EPA-00756 from ERA-NET PathoGenoMics (grant 0315900A), the Collaborative Research Center/Transregio 124 FungiNet, the Northern Portugal Regional Operational Program (grant NORTE 2020) under the Portugal 2020 Partnership Agreement through the European Regional Development Fund (FEDER) (grant NORTE-01-0145-FEDER-000013), and the Fundação para a Ciência e Tecnologia (FCT) (grants IF/00735/2014 to A.C. and SFRH/BPD/96176/2013 to C.C.).This study was also supported by Astellas Pharma Inc. and by a donation of Consuelo González Moreno, an acute myeloid leukemia survivor.
M.J. and J. Sainz conceived the study and participated in its design and coordination. C.B.L. and J.M.S.-M. performed the genetic analyses. C.B.L., C.C., S.M.G., A.C., and J. Sainz performed in vitro functional analyses. J. Springer, M. Lackner, P.G.-S., J.B., J.S.-C., J.M.S.-M., R.R.-T., L.A.-F., M.A.L.-N., L.F., J.M.A., L. Pagano, L. Potenza, S.M.G., M. Luppi, C.S., M.C.-E., K.L., J.A.M., C.L.-F., H.E., L.V., J.L., A.C., M.J., J. Sainz, and the PCRAGA Study Group coordinated patient’s recruitment and provided the clinical data. M.A.-R. provided immuno-chromatin immunoprecipitation (immuno-ChIP) data from healthy controls. J. Sainz and M.M.-B. analyzed the data. M.J. and J. Sainz drafted the manuscript. All authors read and approved the final version of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Steinbach WJ, Marr KA, Anaissie EJ, Azie N, Quan SP, Meier-Kriesche HU, Apewokin S, Horn DL. 2012. Clinical epidemiology of 960 patients with invasive aspergillosis from the PATH Alliance registry. J Infect 65:453–464. doi: 10.1016/j.jinf.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Baddley JW. 2011. Clinical risk factors for invasive aspergillosis. Med Mycol 49(Suppl 1):S7–S12. doi: 10.3109/13693786.2010.505204. [DOI] [PubMed] [Google Scholar]
- 3.Taccone FS, Van den Abeele AM, Bulpa P, Misset B, Meersseman W, Cardoso T, Paiva JA, Blasco-Navalpotro M, De Laere E, Dimopoulos G, Rello J, Vogelaers D, Blot SI, Asp I, on behalf of the AspICU Study Investigators . 2015. Epidemiology of invasive aspergillosis in critically ill patients: clinical presentation, underlying conditions, and outcomes. Crit Care 19:7. doi: 10.1186/s13054-014-0722-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavalda J, Len O, San Juan R, Aguado JM, Fortun J, Lumbreras C, Moreno A, Munoz P, Blanes M, Ramos A, Rufi G, Gurgui M, Torre-Cisneros J, Montejo M, Cuenca-Estrella M, Rodriguez-Tudela JL, Pahissa A, Resitra . 2005. Risk factors for invasive aspergillosis in solid-organ transplant recipients: a case-control study. Clin Infect Dis 41:52–59. doi: 10.1086/430602. [DOI] [PubMed] [Google Scholar]
- 5.Marr KA, Carter RA, Boeckh M, Martin P, Corey L. 2002. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood 100:4358–4366. doi: 10.1182/blood-2002-05-1496. [DOI] [PubMed] [Google Scholar]
- 6.Labbe AC, Su SH, Laverdiere M, Pepin J, Patino C, Cohen S, Kiss T, Lachance S, Sauvageau G, Busque L, Roy DC, Roy J. 2007. High incidence of invasive aspergillosis associated with intestinal graft-versus-host disease following nonmyeloablative transplantation. Biol Blood Marrow Transplant 13:1192–1200. doi: 10.1016/j.bbmt.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Chamilos G, Luna M, Lewis RE, Bodey GP, Chemaly R, Tarrand JJ, Safdar A, Raad II, Kontoyiannis DP. 2006. Invasive fungal infections in patients with hematologic malignancies in a tertiary care cancer center: an autopsy study over a 15-year period (1989–2003). Haematologica 91:986–989. [PubMed] [Google Scholar]
- 8.Palmer LB, Greenberg HE, Schiff MJ. 1991. Corticosteroid treatment as a risk factor for invasive aspergillosis in patients with lung disease. Thorax 46:15–20. doi: 10.1136/thx.46.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warris A, Bjorneklett A, Gaustad P. 2001. Invasive pulmonary aspergillosis associated with infliximab therapy. N Engl J Med 344:1099–1100. [PubMed] [Google Scholar]
- 10.Husni RN, Gordon SM, Longworth DL, Arroliga A, Stillwell PC, Avery RK, Maurer JR, Mehta A, Kirby T. 1998. Cytomegalovirus infection is a risk factor for invasive aspergillosis in lung transplant recipients. Clin Infect Dis 26:753–755. doi: 10.1086/514599. [DOI] [PubMed] [Google Scholar]
- 11.Denning DW, Follansbee SE, Scolaro M, Norris S, Edelstein H, Stevens DA. 1991. Pulmonary aspergillosis in the acquired immunodeficiency syndrome. N Engl J Med 324:654–662. doi: 10.1056/NEJM199103073241003. [DOI] [PubMed] [Google Scholar]
- 12.Tutar N, Metan G, Koc AN, Yilmaz I, Bozkurt I, Simsek ZO, Buyukoglan H, Kanbay A, Oymak FS, Gulmez I, Demir R. 2013. Invasive pulmonary aspergillosis in patients with chronic obstructive pulmonary disease. Multidiscip Respir Med 8:59. doi: 10.1186/2049-6958-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunha C, Aversa F, Romani L, Carvalho A. 2013. Human genetic susceptibility to invasive aspergillosis. PLoS Pathog 9:e1003434. doi: 10.1371/journal.ppat.1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maskarinec SA, Johnson MD, Perfect JR. 2016. Genetic susceptibility to fungal infections: what is in the genes? Curr Clin Microbiol Rep 3:81–91. doi: 10.1007/s40588-016-0037-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menzin J, Meyers JL, Friedman M, Korn JR, Perfect JR, Langston AA, Danna RP, Papadopoulos G. 2011. The economic costs to United States hospitals of invasive fungal infections in transplant patients. Am J Infect Control 39:e15–e20. doi: 10.1016/j.ajic.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Baddley JW, Andes DR, Marr KA, Kontoyiannis DP, Alexander BD, Kauffman CA, Oster RA, Anaissie EJ, Walsh TJ, Schuster MG, Wingard JR, Patterson TF, Ito JI, Williams OD, Chiller T, Pappas PG. 2010. Factors associated with mortality in transplant patients with invasive aspergillosis. Clin Infect Dis 50:1559–1567. doi: 10.1086/652768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sainz J, Perez E, Hassan L, Moratalla A, Romero A, Collado MD, Jurado M. 2007. Variable number of tandem repeats of TNF receptor type 2 promoter as genetic biomarker of susceptibility to develop invasive pulmonary aspergillosis. Hum Immunol 68:41–50. doi: 10.1016/j.humimm.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Sainz J, Salas-Alvarado I, López-Fernández E, Olmedo C, Comino A, García F, Blanco A, Gómez-Lopera S, Oyonarte S, Bueno P, Jurado M. 2010. TNFR1 mRNA expression level and TNFR1 gene polymorphisms are predictive markers for susceptibility to develop invasive pulmonary aspergillosis. Int J Immunopathol Pharmacol 23:423–436. doi: 10.1177/039463201002300205. [DOI] [PubMed] [Google Scholar]
- 19.Bochud PY, Chien JW, Marr KA, Leisenring WM, Upton A, Janer M, Rodrigues SD, Li S, Hansen JA, Zhao LP, Aderem A, Boeckh M. 2008. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med 359:1766–1777. doi: 10.1056/NEJMoa0802629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho A, Cunha C, Carotti A, Aloisi T, Guarrera O, Di Ianni M, Falzetti F, Bistoni F, Aversa F, Pitzurra L, Rodrigues F, Romani L. 2009. Polymorphisms in Toll-like receptor genes and susceptibility to infections in allogeneic stem cell transplantation. Exp Hematol 37:1022–1029. doi: 10.1016/j.exphem.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Grube M, Loeffler J, Mezger M, Kruger B, Echtenacher B, Hoffmann P, Edinger M, Einsele H, Andreesen R, Holler E. 2013. TLR5 stop codon polymorphism is associated with invasive aspergillosis after allogeneic stem cell transplantation. Med Mycol 51:818–825. doi: 10.3109/13693786.2013.809630. [DOI] [PubMed] [Google Scholar]
- 22.Kesh S, Mensah NY, Peterlongo P, Jaffe D, Hsu K, van den Brink M, O'reilly R, Pamer E, Satagopan J, Papanicolaou GA. 2005. TLR1 and TLR6 polymorphisms are associated with susceptibility to invasive aspergillosis after allogeneic stem cell transplantation. Ann N Y Acad Sci 1062:95–103. doi: 10.1196/annals.1358.012. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho A, De Luca A, Bozza S, Cunha C, D'Angelo C, Moretti S, Perruccio K, Iannitti RG, Fallarino F, Pierini A, Latgé J-P, Velardi A, Aversa F, Romani L. 2012. TLR3 essentially promotes protective class I-restricted memory CD8(+) T-cell responses to Aspergillus fumigatus in hematopoietic transplanted patients. Blood 119:967–977. doi: 10.1182/blood-2011-06-362582. [DOI] [PubMed] [Google Scholar]
- 24.Sainz J, Lupiáñez CB, Segura-Catena J, Vazquez L, Ríos R, Oyonarte S, Hemminki K, Försti A, Jurado M. 2012. Dectin-1 and DC-SIGN polymorphisms associated with invasive pulmonary aspergillosis infection. PLoS One 7:e32273. doi: 10.1371/journal.pone.0032273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunha C, Di Ianni M, Bozza S, Giovannini G, Zagarella S, Zelante T, D'Angelo C, Pierini A, Pitzurra L, Falzetti F, Carotti A, Perruccio K, Latgé J-P, Rodrigues F, Velardi A, Aversa F, Romani L, Carvalho A. 2010. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood 116:5394–5402. doi: 10.1182/blood-2010-04-279307. [DOI] [PubMed] [Google Scholar]
- 26.Chai LY, de Boer MG, van der Velden WJ, Plantinga TS, van Spriel AB, Jacobs C, Halkes CJ, Vonk AG, Blijlevens NM, van Dissel JT, Donnelly PJ, Kullberg BJ, Maertens J, Netea MG. 2011. The Y238X stop codon polymorphism in the human beta-glucan receptor dectin-1 and susceptibility to invasive aspergillosis. J Infect Dis 203:736–743. doi: 10.1093/infdis/jiq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunha C, Goncalves SM, Duarte-Oliveira C, Leite L, Lagrou K, Marques A, Lupianez CB, Mesquita I, Gaifem J, Barbosa AM, Pinho Vaz C, Branca R, Campilho F, Freitas F, Ligeiro D, Lass-Florl C, Loffler J, Jurado M, Saraiva M, Kurzai O, Rodrigues F, Castro AG, Silvestre R, Sainz J, Maertens JA, Torrado E, Jacobsen ID, Lacerda JF, Campos A Jr, Carvalho A. 2017. IL-10 overexpression predisposes to invasive aspergillosis by suppressing antifungal immunity. J Allergy Clin Immunol 140:867–870.e9. doi: 10.1016/j.jaci.2017.02.034. [DOI] [PubMed] [Google Scholar]
- 28.Sainz J, Hassan L, Perez E, Romero A, Moratalla A, Lopez-Fernandez E, Oyonarte S, Jurado M. 2007. Interleukin-10 promoter polymorphism as risk factor to develop invasive pulmonary aspergillosis. Immunol Lett 109:76–82. doi: 10.1016/j.imlet.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Sainz J, Perez E, Gomez-Lopera S, Jurado M. 2008. IL1 gene cluster polymorphisms and its haplotypes may predict the risk to develop invasive pulmonary aspergillosis and modulate C-reactive protein level. J Clin Immunol 28:473–485. doi: 10.1007/s10875-008-9197-0. [DOI] [PubMed] [Google Scholar]
- 30.Mezger M, Steffens M, Beyer M, Manger C, Eberle J, Toliat MR, Wienker TF, Ljungman P, Hebart H, Dornbusch HJ, Einsele H, Loeffler J. 2008. Polymorphisms in the chemokine (C-X-C motif) ligand 10 are associated with invasive aspergillosis after allogeneic stem-cell transplantation and influence CXCL10 expression in monocyte-derived dendritic cells. Blood 111:534–536. doi: 10.1182/blood-2007-05-090928. [DOI] [PubMed] [Google Scholar]
- 31.Chandrasekar B, Mummidi S, Perla RP, Bysani S, Dulin NO, Liu F, Melby PC. 2003. Fractalkine (CX3CL1) stimulated by nuclear factor kappaB (NF-kappaB)-dependent inflammatory signals induces aortic smooth muscle cell proliferation through an autocrine pathway. Biochem J 373:547–558. doi: 10.1042/BJ20030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F. 2006. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 33.Lionakis MS, Swamydas M, Fischer BG, Plantinga TS, Johnson MD, Jaeger M, Green NM, Masedunskas A, Weigert R, Mikelis C, Wan W, Lee CC, Lim JK, Rivollier A, Yang JC, Laird GM, Wheeler RT, Alexander BD, Perfect JR, Gao JL, Kullberg BJ, Netea MG, Murphy PM. 2013. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest 123:5035–5051. doi: 10.1172/JCI71307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai ZW, Pineau T, Esser C. 1996. Identification of dioxin-responsive elements (DREs) in the 5′ regions of putative dioxin-inducible genes. Chem Biol Interact 100:97–112. doi: 10.1016/0009-2797(96)03691-5. [DOI] [PubMed] [Google Scholar]
- 35.Jeon MS, Esser C. 2000. The murine IL-2 promoter contains distal regulatory elements responsive to the Ah receptor, a member of the evolutionarily conserved bHLH-PAS transcription factor family. J Immunol 165:6975–6983. doi: 10.4049/jimmunol.165.12.6975. [DOI] [PubMed] [Google Scholar]
- 36.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. 2014. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol 32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 37.Westra H-J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, Van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, Hoen PAC, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Völker U, Perola M, Salomaa V, Brody J, Suchy-Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JBJ, Franke L. 2013. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dougherty EJ, Pollenz RS. 2008. Analysis of Ah receptor-ARNT and Ah receptor-ARNT2 complexes in vitro and in cell culture. Toxicol Sci 103:191–206. doi: 10.1093/toxsci/kfm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolluri SK, Weiss C, Koff A, Gottlicher M. 1999. p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev 13:1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu YZ, Hogenesch JB, Bradfield CA. 2000. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol 40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 41.Antoniv TT, Ivashkiv LB. 2011. Interleukin-10-induced gene expression and suppressive function are selectively modulated by the PI3K-Akt-GSK3 pathway. Immunology 132:567–577. doi: 10.1111/j.1365-2567.2010.03402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, Bicciato S, Mazza EM, Macchiarulo A, Vacca C, Iannitti R, Tissi L, Volpi C, Belladonna ML, Orabona C, Bianchi R, Lanz TV, Platten M, Della Fazia MA, Piobbico D, Zelante T, Funakoshi H, Nakamura T, Gilot D, Denison MS, Guillemin GJ, DuHadaway JB, Prendergast GC, Metz R, Geffard M, Boon L, Pirro M, Iorio A, Veyret B, Romani L, Grohmann U, Fallarino F, Puccetti P. 2014. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hovey D, Zettergren A, Jonsson L, Melke J, Anckarsater H, Lichtenstein P, Westberg L. 2014. Associations between oxytocin-related genes and autistic-like traits. Soc Neurosci 9:378–386. doi: 10.1080/17470919.2014.897995. [DOI] [PubMed] [Google Scholar]
- 44.Wagage S, John B, Krock BL, Hall AO, Randall LM, Karp CL, Simon MC, Hunter CA. 2014. The aryl hydrocarbon receptor promotes IL-10 production by NK cells. J Immunol 192:1661–1670. doi: 10.4049/jimmunol.1300497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson PG, Wark PA, Simpson JL, Meldrum C, Meldrum S, Saltos N, Boyle M. 2003. Induced sputum IL-8 gene expression, neutrophil influx and MMP-9 in allergic bronchopulmonary aspergillosis. Eur Respir J 21:582–588. doi: 10.1183/09031936.03.00001803. [DOI] [PubMed] [Google Scholar]
- 46.Jamieson WL, Shimizu S, D'Ambrosio JA, Meucci O, Fatatis A. 2008. CX3CR1 is expressed by prostate epithelial cells and androgens regulate the levels of CX3CL1/fractalkine in the bone marrow: potential role in prostate cancer bone tropism. Cancer Res 68:1715–1722. doi: 10.1158/0008-5472.CAN-07-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imai T, Hieshima K, Haskell C, Baba M, Nagira M, Nishimura M, Kakizaki M, Takagi S, Nomiyama H, Schall TJ, Yoshie O. 1997. Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell 91:521–530. doi: 10.1016/s0092-8674(00)80438-9. [DOI] [PubMed] [Google Scholar]
- 48.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, Eleftheriadis M, Sinning CR, Schnabel RB, Lubos E, Mennerich D, Rust W, Perret C, Proust C, Nicaud V, Loscalzo J, Hubner N, Tregouet D, Munzel T, Ziegler A, Tiret L, Blankenberg S, Cambien F. 2010. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One 5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Julia V, Staumont-Salle D, Dombrowicz D. 2016. Role of fractalkine/CX3CL1 and its receptor CX3CR1 in allergic diseases. Med Sci (Paris) 32:260–266. (In French.) doi: 10.1051/medsci/20163203010. [DOI] [PubMed] [Google Scholar]
- 50.Vidal F, Chemokines LTNP Study Group, Vilades C, Domingo P, Broch M, Pedrol E, Dalmau D, Knobel H, Peraire J, Gutierrez C, Sambeat MA, Fontanet A, Deig E, Cairo M, Montero M, Richart C, Mallal S, Chemokines L. 2005. Spanish HIV-1-infected long-term nonprogressors of more than 15 years have an increased frequency of the CX3CR1 249I variant allele. J Acquir Immune Defic Syndr 40:527–531. doi: 10.1097/01.qai.0000186362.50457.e0. [DOI] [PubMed] [Google Scholar]
- 51.Faure S, Meyer L, Costagliola D, Vaneensberghe C, Genin E, Autran B, Delfraissy JF, McDermott DH, Murphy PM, Debre P, Theodorou I, Combadiere C. 2000. Rapid progression to AIDS in HIV+ individuals with a structural variant of the chemokine receptor CX3CR1. Science 287:2274–2277. doi: 10.1126/science.287.5461.2274. [DOI] [PubMed] [Google Scholar]
- 52.Singh KK, Hughes MD, Chen J, Spector SA. 2005. Genetic polymorphisms in CX3CR1 predict HIV-1 disease progression in children independently of CD4+ lymphocyte count and HIV-1 RNA load. J Infect Dis 191:1971–1980. doi: 10.1086/430091. [DOI] [PubMed] [Google Scholar]
- 53.Break TJ, Jaeger M, Solis NV, Filler SG, Rodriguez CA, Lim JK, Lee CC, Sobel JD, Netea MG, Lionakis MS. 2015. CX3CR1 is dispensable for control of mucosal Candida albicans infections in mice and humans. Infect Immun 83:958–965. doi: 10.1128/IAI.02604-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group . 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. 2002. The structure of haplotype blocks in the human genome. Science 296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 56.Lupianez CB, Canet LM, Carvalho A, Alcazar-Fuoli L, Springer J, Lackner M, Segura-Catena J, Comino A, Olmedo C, Rios R, Fernandez-Montoya A, Cuenca-Estrella M, Solano C, Lopez-Nevot MA, Cunha C, Oliveira-Coelho A, Villaescusa T, Fianchi L, Aguado JM, Pagano L, Lopez-Fernandez E, Potenza L, Luppi M, Lass-Florl C, Loeffler J, Einsele H, Vazquez L, PCRAGA Study Group, Jurado M, Sainz J. 2015. Polymorphisms in host immunity-modulating genes and risk of invasive aspergillosis: results from the AspBIOmics Consortium. Infect Immun 84:643–657. doi: 10.1128/IAI.01359-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.