Stenotrophomonas maltophilia is a Gram-negative bacterium found ubiquitously in the environment that has historically been regarded as nonpathogenic. S. maltophilia is increasingly observed in patient sputa in cystic fibrosis (CF), and while existing epidemiology indicates that patients with S. maltophilia have poorer diagnoses, its clinical significance remains unclear. Moreover, as multidrug resistance is common among S. maltophilia isolates, treatment options for these infections may be limited.
KEYWORDS: Stenotrophomonas maltophilia, Pseudomonas aeruginosa, polymicrobial infection, biofilm, inflammation, biofilms, cystic fibrosis, Pseudomonas, Stenotrophomonas
ABSTRACT
Stenotrophomonas maltophilia is a Gram-negative bacterium found ubiquitously in the environment that has historically been regarded as nonpathogenic. S. maltophilia is increasingly observed in patient sputa in cystic fibrosis (CF), and while existing epidemiology indicates that patients with S. maltophilia have poorer diagnoses, its clinical significance remains unclear. Moreover, as multidrug resistance is common among S. maltophilia isolates, treatment options for these infections may be limited. Here, we investigated the pathogenicity of S. maltophilia alone and during polymicrobial infection with Pseudomonas aeruginosa. Colonization, persistence, and virulence of S. maltophilia were assessed in experimental respiratory infections of mice. The results of this study indicate that S. maltophilia transiently colonizes the lung accompanied by significant weight loss and immune cell infiltration and the expression of early inflammatory markers, including interleukin 6 (IL-6), IL-1α, and tumor necrosis factor alpha (TNF-α). Importantly, polymicrobial infection with P. aeruginosa elicited significantly higher S. maltophilia counts in bronchoalveolar lavages and lung tissue homogenates. This increase in bacterial load was directly correlated with the density of the P. aeruginosa population and required viable P. aeruginosa bacteria. Microscopic analysis of biofilms formed in vitro revealed that S. maltophilia formed well-integrated biofilms with P. aeruginosa, and these organisms colocalize in the lung during dual-species infection. Based on these results, we conclude that active cellular processes by P. aeruginosa afford a significant benefit to S. maltophilia during polymicrobial infections. Furthermore, these results indicate that S. maltophilia may have clinical significance in respiratory infections.
INTRODUCTION
Cystic fibrosis (CF) is a genetic disease characterized by airway dehydration, hyperinflammation, and defects in mucociliary clearance in the lung, leading to chronic bacterial infections that are not efficiently resolved (1, 2). Even with recent advances in antibiotic regimens, pulmonary infections and their associated sequelae remain the primary drivers of morbidity and mortality in the CF patient population (3). It is well established that there are successional changes in the microbial population of the CF lung as patients age, with mucoid strains of Pseudomonas aeruginosa emerging as the primary pathogen for many patients (4). However, with the advent of culture-independent methods for microbial detection, a larger appreciation has developed for the complex microbiota present in CF airways (5), and many organisms once thought to be innocuous or only transiently present in the lung have emerged as possible drivers of disease progression (6–8). One organism recognized in recent years for its prevalence in this patient population is Stenotrophomonas maltophilia. Current data indicate that a significant proportion of the U.S. CF patient population is colonized with this organism, which has been increasing since its initial recognition in the mid-1990s (9).
S. maltophilia is a highly drug-resistant Gram-negative bacillus that is widely distributed in environmental sources and is known to cause nosocomial infections in immunocompromised hosts (10). However, the significance of S. maltophilia as an opportunistic pathogen in patients with CF remains uncertain (11, 12). While it is clear that S. maltophilia is frequently isolated from patients with lower lung function (as measured by predicted median forced expiratory volume in 1 s (FEV1)), at present it remains uncertain whether S. maltophilia merely colonizes a profoundly damaged lung environment or is the causative agent of the destruction (13–16).
Distinct from acute respiratory infections, thickened mucus and a decrease in mucociliary clearance in CF patients predispose them to bacterial colonization of the lung in the form of a chronic, complex polymicrobial biofilm (7). Epidemiologic data clearly show that S. maltophilia participates in polymicrobial infections in concert with many important pathogens in CF, including Staphylococcus aureus and P. aeruginosa (13, 15, 16). During late-stage disease, P. aeruginosa predominates within the CF lung environment, and many studies have shown that P. aeruginosa can interact with other microbes within the lung to alter infection dynamics (17–20). Several in vitro studies have confirmed that S. maltophilia and P. aeruginosa interact via quorum signaling pathways and that virulence factor expression differs during coculture (21–23); however, our current understanding of the impact of this cross-species interaction on disease progression is limited.
In this study, we address how S. maltophilia infects, colonizes, and persists in the lung alone and in concert with P. aeruginosa. We demonstrate that S. maltophilia can transiently colonize the lung, where it initiates an acute inflammatory response. Analysis of in vitro biofilms formed between S. maltophilia and P. aeruginosa indicate that they can form a polymicrobial biofilm with each organism in a distinct stratum. Notably, we found that the presence of P. aeruginosa facilitated S. maltophilia persistence in the lung during polymicrobial infections. This increased persistence required live P. aeruginosa, and S. maltophilia counts from the lung correlated directly with the amount of P. aeruginosa present. This study confirms the in vivo relevance of previous reports of the importance of S. maltophilia and P. aeruginosa dual-species interactions. From these data, we conclude that these organisms establish a bacterial interaction in which S. maltophilia benefits from a P. aeruginosa population in the lung.
RESULTS
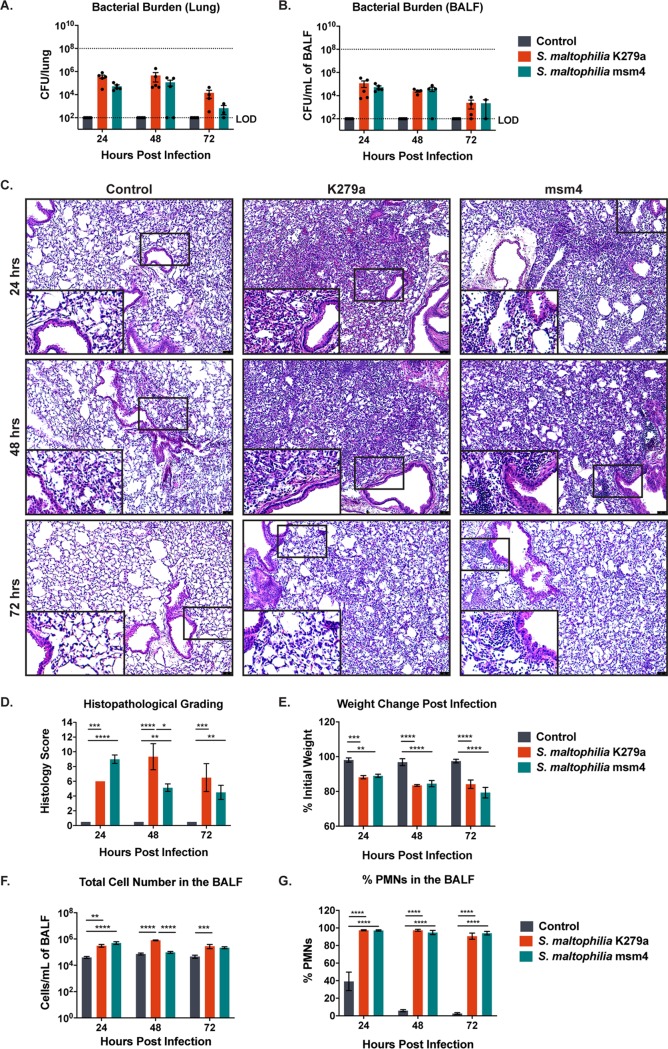
To test the virulence of S. maltophilia in respiratory infections, BALB/cJ mice were infected intratracheally (inoculum of ∼108 CFU/mouse) with S. maltophilia K279a or a clinical sputum isolate, S. maltophilia msm4. Both strains transiently colonized the lung, with detectable bacteria present in the lung homogenate (Fig. 1A) and bronchoalveolar lavage fluid (BALF) (Fig. 1B) for up to 72 hours postinfection. Histopathologic analysis (hematoxylin and eosin [H&E] staining) of lung sections revealed immune cell infiltration, peribronchial inflammation, and dense alveolar consolidation following infection with both strains of S. maltophilia that persisted to 72 hours postinfection (Fig. 1C). These results were scored in blind fashion for severity of inflammation, as quantified by the extent and severity of immune cell infiltration into the lung. Both groups of infected mice had significantly higher histology scores (indicating more severe immune cell infiltration) than uninfected animals at all time points (P < 0.0001). Results were similar between the two strains of S. maltophilia, with the exception of the 48-hour postinfection time point, at which S. maltophilia K279a evoked significantly greater levels of inflammation than the clinical isolate S. maltophilia msm4 (P = 0.013) (Fig. 1D). To look at a broader measure of virulence, we assessed weight changes of infected mice and found that both infected groups lost significantly more weight than their uninfected counterparts (P < 0.0001) and that weight loss became more severe as the infection progressed (P = 0.004) (Fig. 1E). The immune cell infiltrates seen in histopathological grading were defined via total and differential cell counts from the BALF following cytospin preparation and differential staining. As expected from the histopathology results, infected animals had significantly more immune cells in the BALF than uninfected controls (P < 0.0001) (Fig. 1F). Infection also significantly altered the composition of the infiltrate; infected groups had a greatly increased percentage of neutrophils in the BALF compared with control animals at all time points (P < 0.0001) (Fig. 1G).
FIG 1.
S. maltophilia persists in the lungs of BALB/cJ mice with pathological consequences. BALB/cJ mice were intratracheally infected with ∼108 CFU of S. maltophilia K279a or S. maltophilia msm4, and groups were euthanized at 24, 48, and 72 hours postinfection. Bacterial counts in the lung homogenate (A) or in the BALF (B) were enumerated via viable colony counting. (C) Representative images of H&E-stained lung sections from infected and control animals were taken. (D) Severity of infection as indicated by H&E-stained sections were examined and graded in blind fashion by a board-certified veterinary pathologist (T.S.) for a semiquantitative histology score. (E) Weight loss was monitored postinfection. Total immune cell number (G) and percentage of PMNs (F) and in the BALF were quantified by differential cell counts. Mean ± SEM, n = 3 to 6. Two-way ANOVA with Tukey’s post hoc comparisons, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Significant outliers were identified via ROUT method and removed. Groups with undetectable colony counts were represented at the limit of detection.
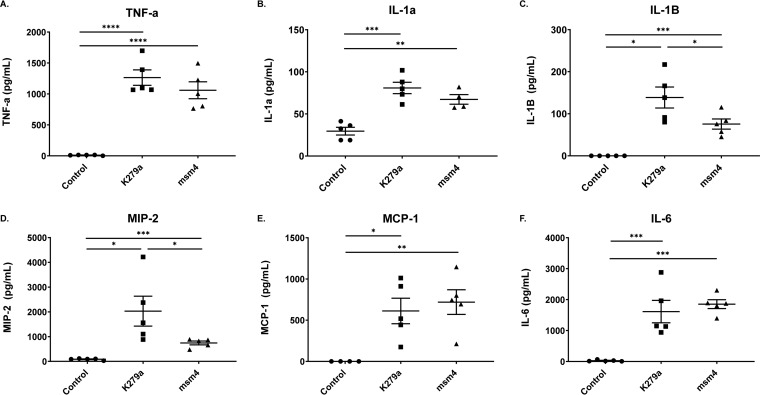
To better define the host inflammatory response in the lung following S. maltophilia infection, we performed Luminex multiplex analyses to measure cytokine and chemokine levels in the BALF. As reported in other models, tumor necrosis factor alpha (TNF-α) was upregulated in response to infection with both strains of S. maltophilia (P < 0.0001) (24) (Fig. 2A). Similarly, additional inflammatory markers, including interleukin 1α (IL-1α), IL-1β, and IL-6, were significantly higher in infected animals than in controls (Fig. 2B to F). Most of the cytokines measured indicate that S. maltophilia K279a elicits a similar inflammatory response to that of S. maltophilia msm4. However, there was one exception (IL-1β) in which S. maltophilia K279a appears to yield greater responses than S. maltophilia msm4 (P = 0.4111) (Fig. 2C and D).
FIG 2.
S. maltophilia infection induces a proinflammatory cytokine response. (A to F) Cytokine and chemokine analysis were performed on BALF supernatant from infected or mock-infected mice using a Luminex multiplex assay. Mean ± SEM, n = 5. One-way ANOVA with Tukey’s multiple-comparison test for post hoc analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Significant outliers were identified via ROUT method and removed.
Bacterial opportunists are thought to reside in the lungs of patients with CF in polymicrobial biofilm communities on the airway mucosal surface (25, 26). Consequently, we wanted to test whether these two species could be cultured in a polymicrobial biofilm. Static in vitro biofilms were inoculated with each species, or with both in equal proportions, and grown at 30°C up to 24 hours. S. maltophilia K279a survived up to 24 hours in coculture with a mucoid clinical isolate of P. aeruginosa (mPA0831) and a nonmucoid model strain (PAO1), with roughly equivalent amounts of each organism integrated into the biofilm, and no significant decrease in viable bacteria compared with single-species biofilms (see Fig. S1A and B in the supplemental material). These dual-species biofilms maintained an even composition of both organisms up to 12 hours for all strains, with a significant decrease in S. maltophilia viability at 24 hours postinoculation for the clinical strains S. maltophilia msm2 and S. maltophilia msm4 but not for S. maltophilia K279a (Fig. S1).
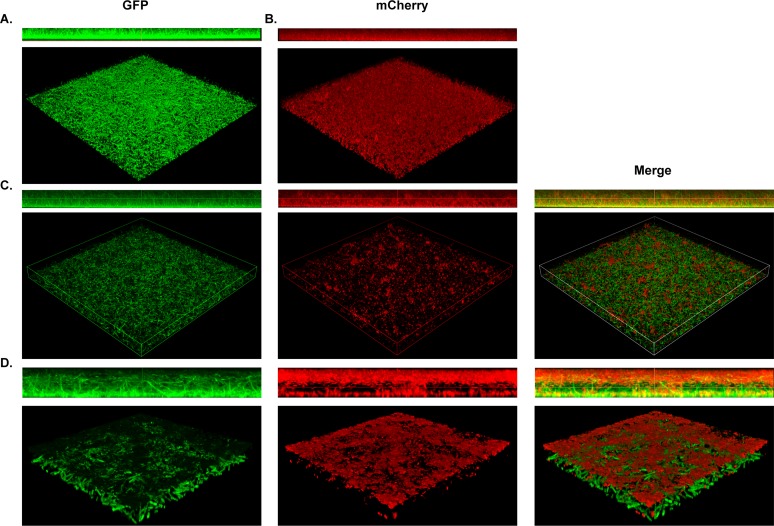
Next, we sought to define the spatial organization of these polymicrobial biofilms. Static single- and dual-species biofilms of S. maltophilia K279a (GFP+) and P. aeruginosa mPA0831 (mCherry+) were grown and imaged via confocal laser scanning microscopy (CLSM). In single-species biofilms, both organisms form a homogenous lawn structure. However, S. maltophilia shows a distinct filamentous morphology not seen in P. aeruginosa (Fig. 3A and B). In a dual-species context, we observed lawns of S. maltophilia with discrete foci of P. aeruginosa (Fig. 3C). The two organisms stratify into distinct layers, with S. maltophilia comprising the base of the biofilm and P. aeruginosa forming the upper layer. Furthermore, the filamentous structure of S. maltophilia is maintained in the presence of P. aeruginosa (Fig. 3D).
FIG 3.
S. maltophilia and P. aeruginosa stratify within polymicrobial biofilms in vitro. Structural composition of polymicrobial biofilms was assessed via confocal imaging of S. maltophilia K279a (GFP+), P. aeruginosa mPA0831 (mCherry+), or both grown at 30°C for 8 hours. Single-species biofilms of S. maltophilia K279a (GFP+) (A) and P. aeruginosa mPA0831 (mCherry+) (B) were imaged at ×40 magnification. Dual-species foci were imaged at ×40 magnification (C) and zoomed 4× for a total magnification of ×160 (D).
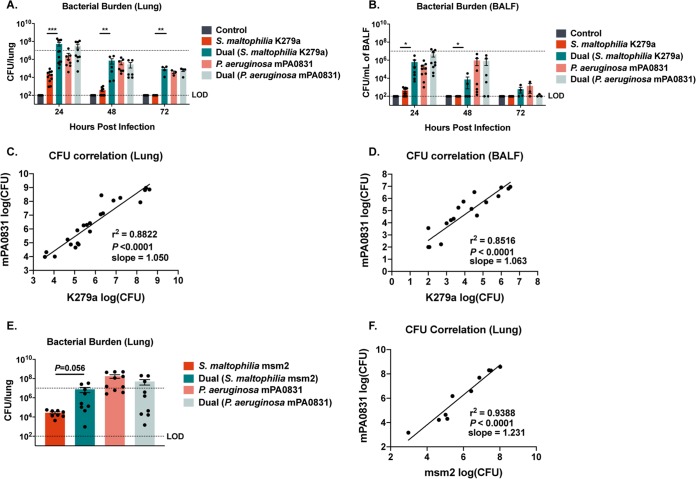
P. aeruginosa is responsible for a large degree of morbidity and mortality in patients with CF and can be cultured from nearly half of CF patients in the United States (3, 9). Notably, P. aeruginosa and S. maltophilia are frequently coisolated from patients (13, 15, 16), and thus, we hypothesized that these two species enhance each other’s virulence and survival during infection. To determine whether the presence of P. aeruginosa changes the infection kinetics of S. maltophilia, we infected mice concurrently with ∼107 CFU of S. maltophilia strain K279a and an equal dose of P. aeruginosa mPA0831, a mucoid clinical isolate from a chronically infected CF patient that was chosen to reflect the dominant infection type found in the CF lung. Notably, we found that while the presence of P. aeruginosa did not increase growth in dual-species biofilms in vitro (Fig. S1), during infection, the presence of P. aeruginosa conferred a 3-log increase in the S. maltophilia load in the lung at 24 hours (mean of 2.51 × 104 ± 8.91 × 103 compared with 5.326 × 107 ± 1.586 × 107, respectively; P = 0.0009) and 48 hours postinfection (3.87 × 102 ± 1.31 × 102 compared with 7.48 × 105 ± 4.47× 105, respectively; P = 0.0071) (Fig. 4A). Furthermore, in coinfected mice, we observed delayed clearance of S. maltophilia from the lung (Fig. 4A) and BALF (Fig. 4B), with detectable S. maltophilia present at 72 hours only in the dual-infected animals (mean, 9.28 × 104 ± 3.36 × 104; P = 0.0036). In contrast, the presence of S. maltophilia largely did not affect colonization, persistence, or clearance of P. aeruginosa. The bacterial loads of the two organisms in the lung (Fig. 4C) and in the BALF (Fig. 4D) were highly correlated, indicating that increasing the P. aeruginosa load in the lung results in a corresponding increase in S. maltophilia (P < 0.0001; in the lung and BALF). A similar increase in S. maltophilia counts was seen when a clinical isolate of S. maltophilia, msm2, was used (mean of 2.76 × 104 ± 6.60 × 103 compared with 7.63 × 106 ± 4.16 × 106, respectively; P = 0.0561) (Fig. 4E). In this case, the tight correlation in recoverable CFUs between the two organisms remained (Fig. 4F).
FIG 4.
Coinfection with mucoid P. aeruginosa confers an increase in S. maltophilia persistence. BALB/cJ mice were intratracheally infected with ∼107 CFU of S. maltophilia K279a and P. aeruginosa mPA0831 alone and in combination, and groups were euthanized at 24, 48, or 72 h postinfection. Bacterial load in lung homogenate (A) and in BALF (B) were enumerated via viable colony counting on differential medium. Mean ± SEM, n = 4 to 12. Two-way ANOVA with Tukey’s post hoc comparisons, *, P < 0.05; **, P < 0.01; ***, P < 0.001. Significant outliers were identified via ROUT method and removed. The correlation between bacterial burdens of S. maltophilia K279a and P. aeruginosa mPA0831 in the lung (C) and BALF (D) were calculated. n = 19 to 22. Linear regression with automatic outlier elimination and two-tailed Spearman correlation were performed on log-transformed bacterial counts. (E) Dual infections were also performed with S. maltophilia msm2 and P. aeruginosa mPA0831. Mean ± SEM, n = 8 to 10. Kruskal-Wallis test with Dunn’s post hoc comparisons. Significant outliers were identified via ROUT method and removed. (F) Calculations of the correlation between bacterial burdens of S. maltophilia msm2 and P. aeruginosa mPA0831 in the lung were also performed. n = 10. Linear regression with automatic outlier elimination and two-tailed Spearman correlation were performed on log-transformed bacterial counts. Groups with undetectable colony counts were represented at the limit of detection.
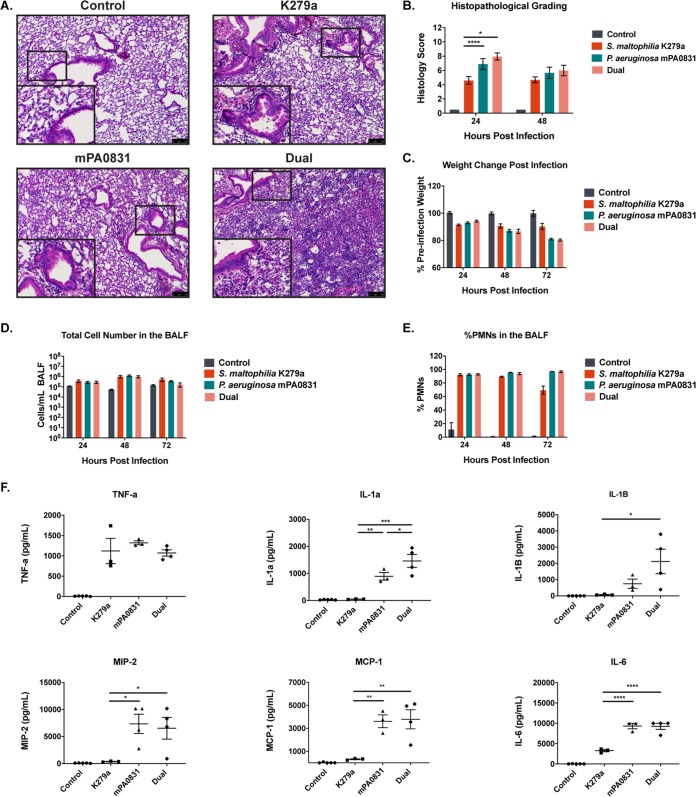
To determine whether this increase in bacterial burden was associated with a comparable increase in virulence, we assessed measures similar to those used in the mice infected with each species alone. H&E staining of lung sections again revealed significant pathological changes for all infection groups compared with control animals (Fig. 5A). When histology scores were quantified, coinfected mice had significantly higher scores than mice infected with S. maltophilia alone (indicating more severe immune cell infiltration) (P = 0.0009) but not significantly worse than mice infected with P. aeruginosa alone at 24 hours postinfection (P = 0.822) (Fig. 5B), a result that was supported by weight loss measurements (Fig. 5C). We again quantified immune cell infiltrates in the BALF via differential cell counts. All infection groups had an increase in total cells and in the percentage of neutrophils in the BALF, but no significant differences between groups were seen (Fig. 5D and E). To get a better overall picture of the immunological consequences of dual infection, we measured cytokine and chemokine levels from BALF supernatant using a Luminex multiplex assay. The majority of measured factors (IL-1β, IL-6, monocyte chemoattractant protein 1 [MCP-1], and MIP-2) showed levels in dual infection that were above S. maltophilia monoinfection but not significantly different from P. aeruginosa monoinfection. The two exceptions, TNF-α and IL-1α, showed opposing trends. Levels of TNF-α, an early proinflammatory cytokine published to be an important mediator of inflammation following S. maltophilia infection (24), was elevated in all groups compared with controls but was not significantly different between infection groups. IL-1α was the only cytokine elevated in dual-infected animals compared with both monoinfected groups (P = 0.036) (Fig. 5F). While most metrics of infection virulence indicated that polymicrobial infection was comparable to single-species infection with P. aeruginosa alone, total mortality was higher in this group (44.1% in polymicrobial infections compared to 18.4% with P. aeruginosa alone) (see Table S1 in the supplemental material).
FIG 5.
Inflammatory consequences of polymicrobial infection closely resemble those of P. aeruginosa single-species infection. (A) Representative images were taken of H&E-stained lung sections from mice infected with S. maltophilia K279a and P. aeruginosa mPA0831 alone and in combination. (B) Severity of infection as indicated by H&E staining was graded via a semiquantitative histology score for 24 and 48 hours postinfection. Mean ± SEM, n = 3 to 8. (C) Change in weight following infection was monitored. Mean ± SEM, n = 4 to 6. (D and E) Total immune cell number (D) and percentage of PMNs in the BALF (E) were quantified via differential cell counts. Mean ± SEM, n = 3 to 5. Two-way ANOVA with Tukey’s post hoc comparisons; *, P < 0.05; ****, P < 0.0001. (F) Cytokine/chemokine analysis performed on BALF supernatant using a Luminex multiplex assay. Mean ± SEM, n = 3 to 4. One-way ANOVA with Tukey’s multiple-comparison test for post hoc analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Significant outliers were identified via ROUT method and removed.
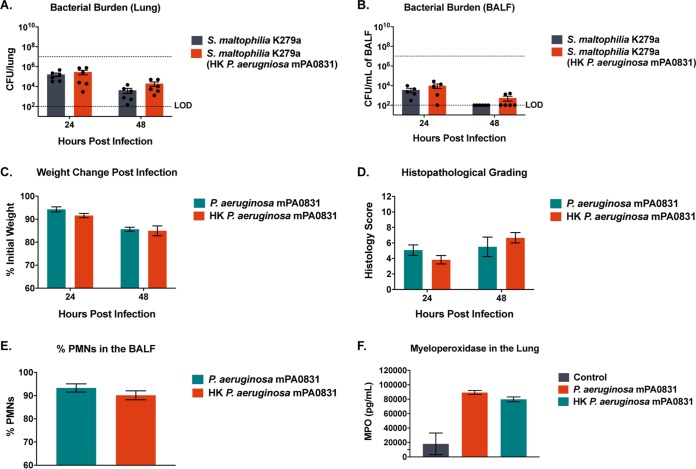
The polymicrobial infection data suggest that P. aeruginosa conferred a persistence benefit to S. maltophilia largely independent of immunological state. To determine if live P. aeruginosa is required for this persistence benefit, mice were challenged with S. maltophilia in the presence of an equivalent inoculum of heat-killed P. aeruginosa. We found that the heat-killed P. aeruginosa was unable to increase S. maltophilia persistence beyond what was seen in single infection in the lung or in BALF at 24 or 48 hours postinfection (Fig. 6A and B). Furthermore, we found that the early immune response to heat-killed and viable P. aeruginosa is not significantly different, as measured by weight change postinfection (Fig. 6C), histopathological grading of lung sections (Fig. 6D), percentage of polymorphonuclear leukocytes (PMNs) in the BALF (Fig. 6E), or neutrophil activation, as indicated by myeloperoxidase levels (Fig. 6F). These data indicate that activation of the innate immune response via bacterial pathogen-associated molecular patterns (PAMPs) is not responsible for the increase in S. maltophilia burden in the lung and that viable P. aeruginosa is required for our cooperative phenotype.
FIG 6.
The host inflammatory response to P. aeruginosa is not sufficient to confer benefit toward S. maltophilia. BALB/cJ mice were intratracheally infected with ∼107 CFU of S. maltophilia K279a alone or in the presence of heat-killed P. aeruginosa mPA0831 before being euthanized at 24 and 48 hours postinfection. Bacterial burden in the lung (A) and in the BALF (B) were enumerated via viable colony counting. Mean ± SEM, n = 6. Two-way ANOVA with Tukey’s post hoc comparisons. Significant outliers were identified via ROUT method and removed. To confirm that heat-killed P. aeruginosa elicits an acute inflammatory response similar to that of live P. aeruginosa, BALB/cJ mice were intratracheally infected with ∼107 CFU of heat-killed or live P. aeruginosa mPA0831 before being sacrificed at 24 and 48 hours postinfection. (C) Change in weight following infection was monitored. Mean ± SEM, n = 6. (D) Severity of infection as indicated by H&E staining was graded via a semiquantitative histology score. (E) Percentage of PMNs in the BALF were quantified via differential cell counts. (F) Myeloperoxidase concentration in the BALF was quantified via enzyme-linked immunosorbent assay (ELISA). Mean ± SEM, n = 3. Two-way ANOVA with Tukey’s post hoc comparisons. Significant outliers were identified via ROUT method and removed. Groups with undetectable colony counts were represented at the limit of detection.
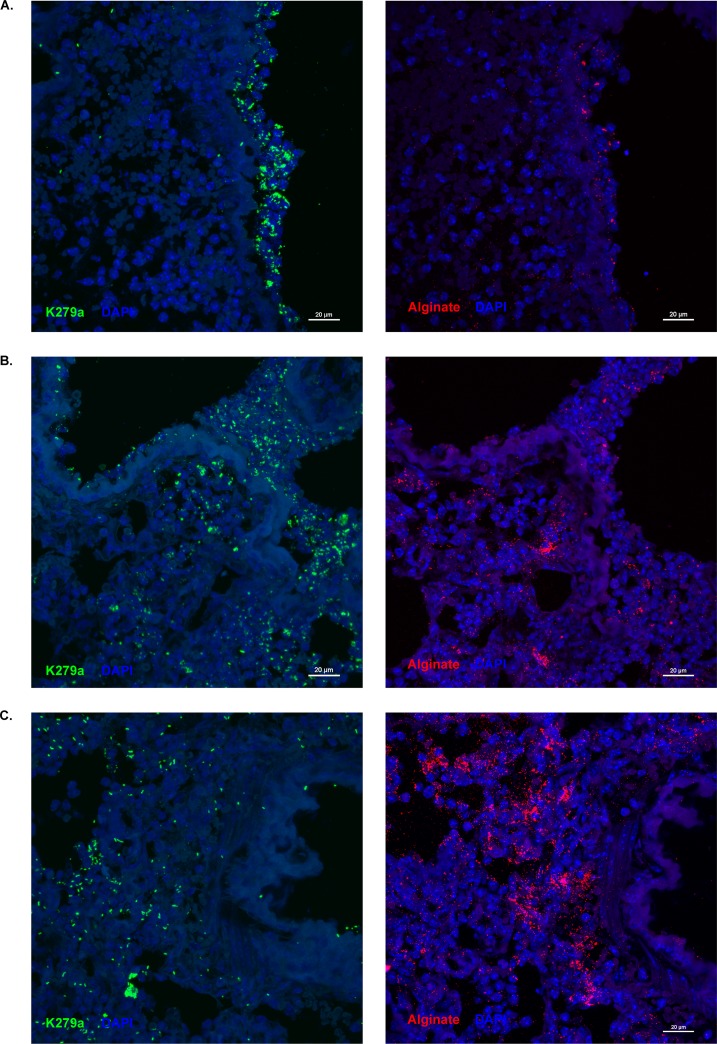
Although it has been reported that S. maltophilia and P. aeruginosa can be coisolated from CF patients, one hallmark of CF disease is the existence of microniches within the lung where bacteria are sequestered into distinct populations (27). In vitro biofilms of these two organisms indicated that they are able to live in integrated communities, but counts from lung homogenate alone do not determine whether the two populations detected are residing together in the lung during coinfection. We, therefore, investigated the localization of these two organisms in the lung. Due to problems with cross-reactivity between antibodies available for these organisms, we cut serial sections from dual-infected lungs and stained for either S. maltophilia or alginate, the predominant exopolysaccharide of P. aeruginosa during CF infections, and imaged corresponding lung structures from each. We found that foci of S. maltophilia localize in similar areas to alginate in the lung, including the airway epithelial surface (Fig. 7A and B), and the lung parenchyma (Fig. 7B and C).
FIG 7.
S. maltophilia colocalizes with alginate during polymicrobial infection in the lung. Representative images of fluorescently stained lung sections were taken via confocal laser scanning microscopy (CLSM). Ten-micrometer serial sections were cut from lungs of mice dually infected with ∼107 CFU/mouse of S. maltophilia K279a and P. aeruginosa mPA0831. Lung sections from dual species-infected mice were stained for S. maltophilia via antisera from rabbits immunized with heat-killed S. maltophilia (green) and for alginate via an anti-alginate polyclonal rabbit antibody (red). Lung structures were visualized with 4′,6-diamidino-2-phenylindole (DAPI; blue). Dual-species foci were imaged on the airway surface (A) and in the lung parenchyma (B and C) at ×40 magnification.
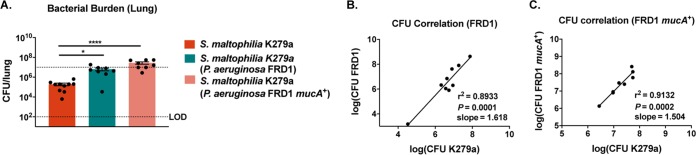
The production of alginate by P. aeruginosa has been shown to promote the persistence of other organisms within polymicrobial biofilms; most notably, this passive protection has been demonstrated for Staphylococcus aureus and commensal Streptococcus strains (28, 29). To determine whether alginate production by P. aeruginosa is responsible for the increase in S. maltophilia persistence, we intratracheally infected mice concurrently with ∼107 CFU of S. maltophilia strain K279a and an equal dose of an alginate overproducing strain, P. aeruginosa FRD1, or a nonmucoid isogenic mutant, P. aeruginosa FRD1/mucA+. Concurrent infection with both FRD1 and FRD1mucA+ resulted in significantly higher titers of S. maltophilia in lung homogenate than S. maltophilia alone (P = 0.0348 and < 0.0001, respectively) (Fig. 8A). Furthermore, the close correlation between S. maltophilia and P. aeruginosa counts was maintained, regardless of alginate production (Fig. 8B and C).
FIG 8.
The benefit conferred to S. maltophilia by P. aeruginosa is not alginate dependent. BALB/cJ mice were intratracheally infected with ∼107 CFU of S. maltophilia K279a and P. aeruginosa FRD1 or a nonmucoid isogenic mutant, P. aeruginosa FRD1/mucA+, at 24 hours postinfection. (A) Bacterial burden in the lung homogenate was enumerated via viable colony counting on differential medium. Mean ± SEM, n = 8 to 10. Two-way ANOVA with Tukey’s post hoc comparisons; *, P < 0.05; ****, P < 0.0001. Significant outliers were identified via ROUT method and removed. Correlation between bacterial burdens of S. maltophilia K279a and P. aeruginosa FRD1 (B) and P. aeruginosa FRD1/mucA+ (C) was calculated; n = 8 to 9. Linear regression with automatic outlier elimination and two-tailed Spearman correlation were performed on log-transformed bacterial counts.
DISCUSSION
With the recent advent and widespread use of culture-independent methodologies for profiling the microbial populations in the upper and lower airways of CF patients, it is now clear that the scope and breadth of bacterial phyla found within the CF lung is much broader than what was previously appreciated (6, 8, 19). Furthermore, it has become evident that shifts in the dominant species of this microbial population, as well as changes in the total amount of diversity present, can have a profound impact on the host inflammatory responses within the lung, and on overall pulmonary function (30–32). Currently, there is a need for a better understanding of how specific components of the lung microbiome impact health and disease in patients with CF.
Carriage of S. maltophilia is quite common in patients with CF and has been increasing in prevalence since the mid-1990s (9, 33). S. maltophilia is widely distributed in a range of environmental sites, including water and soil. However, an extensive body of research has made it clear that this organism can also act as an opportunist in human infections (10). Previous work has indicated that S. maltophilia can transiently colonize mice, but there remains a need for clarification of its full pathogenic potential in pulmonary infections (24, 34, 35). In our single-species infections, we found that the introduction of S. maltophilia resulted in transient lung colonization and a potent inflammatory response. This was characterized by weight loss, increased expression of proinflammatory cytokines and chemokines, and a subsequent recruitment of innate immune cells to the lung up to 72 hours postinfection (Fig. 1 and 2). While similar studies have varied in the route of infection, strains used, and mouse genotype, many common themes have emerged, including severe neutrophilia and a potent TNF-α response (24, 35, 36). Others have seen wide variability between clinical isolates, but we found infection dynamics of S. maltophilia K279a and the clinical strain S. maltophilia msm4 to be similar.
Reports have indicated that polymicrobial infection with S. maltophilia and P. aeruginosa might increase virulence in patients (37). While epidemiological data are often not explicit about polymicrobial infection rates, it is clear that a large proportion of S. maltophilia-infected patients are also P. aeruginosa positive, highlighting the importance of understanding how this dual-species interaction changes infection dynamics and patient outcomes (13, 15, 16, 37). Importantly, we found that infection with S. maltophilia in the presence of P. aeruginosa greatly increases S. maltophilia burden in lung homogenate and in BALF at every time point tested. Tight correlations between the lung burden of two organisms in all strain combinations indicate that as the amount of P. aeruginosa goes up in the lung, we get a subsequent increase in S. maltophilia counts. Looking at the inflammatory consequences, most metrics measured indicated that infection with P. aeruginosa was more inflammatory than infection with S. maltophilia but that dual infection was not significantly more damaging than P. aeruginosa alone. The exceptions to this were cytokine levels of TNF-α, which showed similar levels in all infection groups, and IL-1α, which was significantly upregulated in dual-infected animals compared with infection with either species alone (Fig. 5). We found that despite the lack of evidence for heightened pathological consequences in the dual-infected animals, they did have a higher rate of mortality than infection with P. aeruginosa alone.
Other groups have found mechanisms of cooperativity with P. aeruginosa that rely on a change in the inflammatory state of the lung due to the presence of bacterial PAMPs rather than on active processes by the bacterium (38). To determine if this was the case, we infected animals with S. maltophilia in the presence of heat-killed P. aeruginosa, a scenario that introduced PAMPs but did not recapitulate the active processes of secretion or biofilm formation. We found that in the absence of live P. aeruginosa, our persistence benefit was lost, indicating that active cellular processes by P. aeruginosa, rather than the host inflammatory response to bacterial PAMPs, are required for S. maltophilia to derive a persistence benefit.
The localization of P. aeruginosa on the top of dual-species biofilms, and the colocalization of S. maltophilia and alginate in the lung indicated that P. aeruginosa might be providing mechanical or chemical protection from the stressful lung environment via alginate production. This mechanism has been shown to mediate the interaction of P. aeruginosa with several other bacterial species (28, 29). However, the fact that S. maltophilia continued to derive benefit from P. aeruginosa presence even when alginate production was disrupted indicates that this is not the mechanism at play (Fig. 8).
One potential mechanism contributing to the increase in S. maltophilia persistence is the previously described interactions between the quorum signaling pathways of these two organisms. These systems include the diffusible signaling factor (DSF), an interkingdom signaling factor produced by most Xanthomonas species and sensed by P. aeruginosa, and smoR, a LuxR family regulatory protein in S. maltophilia that responds to acyl-homoserine lactone (AHL) signals, the canonical quorum signaling (QS) molecules of P. aeruginosa. Signaling through DSF is largely one-sided. S. maltophilia can produce the signal molecule cis-11-methyl-dodecenoic acid, which can act as an intraspecies autoinducer or can signal through PA1396, a sensor kinase of P. aeruginosa. The described phenotypes governed by this signaling in P. aeruginosa include increased expression of bacterial stress tolerance genes and antimicrobial resistance, both of which could ultimately be relevant to the cooperative behavior of these species in CF patients. In contrast, AHL signals in this system are produced exclusively by P. aeruginosa and can be sensed by both organisms. Signaling through smoR has been shown to increase S. maltophilia swarming motility (21), a virulence factor found in a wide variety of bacterial pathogens (39).
Another potential mechanism for the increase in S. maltophilia persistence is via localized host immune modulation due to the type III secretion system of P. aeruginosa. This is a complex secretion apparatus constructed to inject effector proteins into host cells, ultimately weakening epithelial barrier integrity and dampening local immune response by mediating phagocytic cell death or impairment (40). This mechanism would require active processes by P. aeruginosa, and close localization of these two organisms in the lung, both of which are supported in our model of infection. There are indications that P. aeruginosa isolates from chronic infection tend to have lower expression of or even mutations in genes involved in the type three secretion system (T3SS) (41, 42), and there have been indications that mucoid conversion of P. aeruginosa via a mucA mutation is accompanied by a downregulation of the T3SS (43, 44). However, it is not clear whether this is broadly applicable to the diverse array of P. aeruginosa strains found in patients, and the presence/regulation of a T3SS in our particular isolate has not been confirmed.
Ultimately, this study furthers our understanding of the consequences of dual-species interactions on the disease progression in CF. This work gives us a better idea of the factors that predispose patients to S. maltophilia acquisition, ultimately leading to better eradication strategies for an incredibly drug-resistant organism. Ongoing work in our laboratory is aimed at further characterizing the molecular mediators responsible for the synergism between these two bacterial species.
MATERIALS AND METHODS
Strains and growth conditions.
S. maltophilia K279a is a widely used model strain, and its genome has been fully annotated and sequenced (45); this strain and the S. maltophilia K279a-GFP derivative were provided by M. Herman (Kansas State University). S. maltophilia msm2 and S. maltophilia msm4 are sputum-derived isolates from patients with chronic CF and were provided by W. Benjamin (University of Alabama at Birmingham). P. aeruginosa PAO1 was provided by D. Wozniak (Ohio State University); P. aeruginosa mPA08-31 was obtained from S. Birket (University of Alabama at Birmingham); and P. aeruginosa FRD1 and an isogenic, nonmucoid derivative (P. aeruginosa FRD1mucA+) were obtained from J. Scoffield (University of Alabama at Birmingham) (29, 46). P. aeruginosa mPA08-31-mCherry+ was constructed by transforming parent strains with plasmid pUCP19+mCherry provided by D. Wozniak (Ohio State University). All strains were routinely cultured on Luria-Bertani (LB) agar (Difco) or in LB broth. S. maltophilia strains were streaked for colony isolation before inoculation into LB broth and shaking overnight at 30°C and 200 rpm. P. aeruginosa strains were streaked for colony isolation before inoculation into LB broth and shaking overnight at 37°C and 200 rpm.
Static biofilm assay.
In vitro biofilm assays were performed according to an established microtiter assay protocol (47), with some modifications. Biofilms were prepared from overnight broth cultures of S. maltophilia and P. aeruginosa diluted to an optical density at 600 nm (OD600) of 0.15 (∼108 CFU/ml) in LB broth and were inoculated into a 96-well microtiter dish (200 μl/well). Biofilms were incubated at 30°C or 37°C for the appropriate time before being washed, resuspended in phosphate-buffered saline (PBS), and plated on M9 minimal medium (48) to select for P. aeruginosa and LB agar with gentamicin (30 μg/ml) to select for S. maltophilia.
Confocal microscopy.
Confocal laser scanning microscopy (CLSM) was performed using a Nikon-A1R HD25 confocal laser microscope (Nikon, Tokyo, Japan). Images were acquired and processed using the NIS-elements 5.0 software. Single- and dual-species biofilms were grown by diluting overnight cultures of S. maltophilia K279a-GFP and P. aeruginosa mPA08-31 (mCherry+) to an OD600 of 0.15 (∼108 CFU/ml) in LB broth and were inoculated into a 35-mm glass-bottom confocal dish (MatTek) in 2-ml increments such that the final density in each well was ∼108 CFU of one or of both organisms. Dishes were incubated at 30°C for 8 hours before being washed with sterile PBS and fixed with 4% paraformaldehyde (Alfa Aesar, Tewksbury, MA).
Mouse respiratory infections.
BALB/cJ mice (8 to 10 weeks old) were obtained from Jackson Laboratories (Bar Harbor, ME). Mice were anesthetized with isoflurane and intratracheally infected with S. maltophilia (∼108 CFU in 100 μl PBS) for single-species infections. For polymicrobial infections, mice were infected intratracheally with either S. maltophilia, P. aeruginosa, or both (∼107 CFU each in 100 μl PBS). Bronchoalveolar lavage was performed by flushing lungs with 5 ml of cold PBS in 1-ml increments. Collected BALF was stored on ice until processing and was centrifuged (100 × g, 10 min) to separate the supernatant from immune cells. Cytokine analyses were performed on supernatants, and the cell pellet was used for total and differential cell counts. The left lung of each mouse was harvested and homogenized in 500 μl of sterile PBS for viable plate counting. Homogenate from single-species infections were serially diluted in PBS and plated onto LB agar to obtain viable CFU counts. Homogenate from polymicrobial infections were plated on M9 minimal medium (48) for P. aeruginosa and LB agar containing gentamicin (30 μg/ml) to enumerate S. maltophilia. All samples from polymicrobial infections were also plated on LB for total bacterial counts. The right lung of each animal was inflated with 10% buffered formalin and stored at 4°C for histological analysis. For heat-killed P. aeruginosa experiments, bacteria were washed with sterile PBS, resuspended at an OD600 equivalent to ∼108 CFU/ml in live bacteria, and incubated at 65°C for 1 h. The efficacy of bacterial killing was confirmed by a complete lack of growth on LB plates. All mouse infection protocols were approved by the University of Alabama at Birmingham (UAB) Institutional Animal Care and Use Committees.
Cytokine analysis and cell counts.
Cytokine analyses were performed on the supernatant from the first microliter of collected BALF via a custom Luminex panel (Millipore Sigma, Burlington, MA). For total and differential cell counts, cell pellets from BALF were resuspended in fresh PBS and mounted on a slide via cytospin at 500 rpm for 5 minutes. Cell spots were stained via a Kwik-Diff differential cell stain (ThermoFisher Scientific, Waltham, MA). Three representative fields from each spot were counted to determine the composition of immune cells.
Histological analysis.
Inflated right lungs from infected animals were stored in 10% neutral buffered formalin (Fisher Scientific, Waltham, MA) at 4°C until processing. Sections from each lobe of the right lung were trimmed and sent to the UAB Comparative Pathology Laboratory to be paraffin embedded and hematoxylin and eosin (H&E) stained. Images of stained lung sections were taken on a Leica LMD6 scope (Wetzlar, Germany) at ×10 and ×40 magnifications. Semiquantitative grading of all lung sections was performed by a board-certified veterinary pathologist in a blind fashion (T.S.). Semiquantitative histopathological scores were assigned using a scoring matrix based primarily on neutrophilic influx. Severity was rated on a scale of 0 to 4, where 0 represents no observable neutrophils and 1 represents ≤25%, 2 represents 25% to 50%, 3 represents 50% to 75%, or 4 represents 75% to 100% of the section affected of independently viewed fields of view. The density of PMNs in affected fields was scored from 1 to 3, to represent mild, moderate, and severe, respectively. The final histopathological score was calculated by multiplication of extent and severity scores.
Immunofluorescent staining.
For immunofluorescent staining of lung sections, lungs of infected animals were inflated with 10% neutral buffered formalin (Fisher Scientific) and stored at 4°C until processing. Sections from each lobe of the right lung were embedded in OCT medium (Sakura Finetek, Torrance, CA) before storage at –80°C. Tissue was cut via cryostat in 10-μm serial sections and mounted on Superfrost plus adhesion slides (Thermo Scientific, Waltham, CA) for staining. S. maltophilia K279a was probed via serum from rabbits immunized with heat-killed S. maltophilia (kindly provided by William Benjamin). Alginate was stained via polyclonal rabbit antibody (49). Stained sections were imaged on a Nikon-A1R HD25 confocal laser microscope.
Statistical analyses.
Unless otherwise noted, graphs represent sample means ± standard error of the mean (SEM). For nonparametric analyses, differences between groups were analyzed by Kruskal-Wallis test with the uncorrected Dunn’s test for multiple comparisons. For normally distributed data sets (as determined by Shapiro-Wilk normality test) a one-way analysis of variance (ANOVA) was used with Tukey’s multiple-comparison test. Those experiments with two factors (for example, CFU changes over time) were analyzed via two-way ANOVA. Outliers were detected via the ROUT method (Q, 1%) and excluded from the analysis. All statistical tests were performed using GraphPad Prism 8 (San Diego, CA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Cystic Fibrosis Foundation (CFFSWORDS1810) awarded to W.E.S. as well as NIH P30 center grant DK072482 and the Cystic Fibrosis Foundation Basic Research Center awarded to Steve Rowe. M.S.M. was supported by an NHBLI T32 UAB predoctoral training program in lung diseases (1T32HL134640-01; W.E.S. principal investigator [PI]) and as a CF trainee on the UAB CF center grant.
We thank Shawn Williams and Robert Grabski at the University of Alabama at Birmingham High Resolution Imaging Facility for their assistance with the Nikon A1 confocal microscope and imaging analysis. We also thank Bill Benjamin (University of Alabama at Birmingham), Daniel Wozniak (Ohio State University), Jessica Scoffield (The University of Alabama at Birmingham), and Susan Birket (The University of Alabama at Birmingham) for providing bacterial strains for this study.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bruscia EM, Bonfield TL. 2016. Innate and adaptive immunity in cystic fibrosis. Clin Chest Med 37:17–29. doi: 10.1016/j.ccm.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 2.Ratjen F, Doring G. 2003. Cystic fibrosis. Lancet 361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 3.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/cmr.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med 168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 5.Pattison SH, Rogers GB, Crockard M, Elborn JS, Tunney MM. 2013. Molecular detection of CF lung pathogens: current status and future potential. J Cyst Fibros 12:194–205. doi: 10.1016/j.jcf.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkins MD, Floto RA. 2015. Emerging bacterial pathogens and changing concepts of bacterial pathogenesis in cystic fibrosis. J Cyst Fibros 14:293–304. doi: 10.1016/j.jcf.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Surette MG. 2014. The cystic fibrosis lung microbiome. Ann Am Thorac Soc 11:S61–S65. doi: 10.1513/AnnalsATS.201306-159MG. [DOI] [PubMed] [Google Scholar]
- 8.Caverly LJ, Zhao J, LiPuma JJ. 2015. Cystic fibrosis lung microbiome: opportunities to reconsider management of airway infection. Pediatr Pulmonol 50:S31–S38. doi: 10.1002/ppul.23243. [DOI] [PubMed] [Google Scholar]
- 9.Cystic Fibrosis Foundation. 2018. Patient registry. 2017 annual data report. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 10.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg G, Martinez JL. 2015. Friends or foes: can we make a distinction between beneficial and harmful strains of the Stenotrophomonas maltophilia complex? Front Microbiol 6:241. doi: 10.3389/fmicb.2015.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen CR. 2012. Stenotrophomonas maltophilia: to be or not to be a cystic fibrosis pathogen. Curr Opin Pulm Med 18:628–631. doi: 10.1097/MCP.0b013e328358d4f8. [DOI] [PubMed] [Google Scholar]
- 13.Talmaciu I, Varlotta L, Mortensen J, Schidlow DV. 2000. Risk factors for emergence of Stenotrophomonas maltophilia in cystic fibrosis. Pediatr Pulmonol 30:10–15. doi:. [DOI] [PubMed] [Google Scholar]
- 14.Waters V, Yau Y, Prasad S, Lu A, Atenafu E, Crandall I, Tom S, Tullis E, Ratjen F. 2011. Stenotrophomonas maltophilia in cystic fibrosis: serologic response and effect on lung disease. Am J Respir Crit Care Med 183:635–640. doi: 10.1164/rccm.201009-1392OC. [DOI] [PubMed] [Google Scholar]
- 15.Marchac V, Equi A, Le Bihan-Benjamin C, Hodson M, Bush A. 2004. Case-control study of Stenotrophomonas maltophilia acquisition in cystic fibrosis patients. Eur Respir J 23:98–102. doi: 10.1183/09031936.03.00007203. [DOI] [PubMed] [Google Scholar]
- 16.Berdah L, Taytard J, Leyronnas S, Clement A, Boelle PY, Corvol H. 2018. Stenotrophomonas maltophilia: a marker of lung disease severity. Pediatr Pulmonol 53:426–430. doi: 10.1002/ppul.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Toole GA. 2018. Cystic fibrosis airway microbiome: overturning the old, opening the way for the new. J Bacteriol 200:e00561-17. doi: 10.1128/JB.00561-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. 2016. The biogeography of polymicrobial infection. Nat Rev Microbiol 14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang YJ, LiPuma JJ. 2016. The microbiome in cystic fibrosis. Clin Chest Med 37:59–67. doi: 10.1016/j.ccm.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. 2008. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog 4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez P, Huedo P, Martinez-Servat S, Planell R, Ferrer-Navarro M, Daura X, Yero D, Gibert I. 2015. Stenotrophomonas maltophilia responds to exogenous AHL signals through the LuxR solo SmoR (Smlt1839). Front Cell Infect Microbiol 5:41. doi: 10.3389/fcimb.2015.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM. 2008. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol 68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 23.Twomey KB, O'Connell OJ, McCarthy Y, Dow JM, O'Toole GA, Plant BJ, Ryan RP. 2012. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J 6:939–950. doi: 10.1038/ismej.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waters VJ, Gómez MI, Soong G, Amin S, Ernst RK, Prince A. 2007. Immunostimulatory properties of the emerging pathogen Stenotrophomonas maltophilia. Infect Immun 75:1698–1703. doi: 10.1128/IAI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudkjøbing VB, Thomsen TR, Alhede M, Kragh KN, Nielsen PH, Johansen UR, Givskov M, Høiby N, Bjarnsholt T. 2012. The microorganisms in chronically infected end-stage and non-end-stage cystic fibrosis patients. FEMS Immunol Med Microbiol 65:236–244. doi: 10.1111/j.1574-695X.2011.00925.x. [DOI] [PubMed] [Google Scholar]
- 26.Sibley CD, Rabin H, Surette MG. 2006. Cystic fibrosis: a polymicrobial infectious disease. Future Microbiol 1:53–61. doi: 10.2217/17460913.1.1.53. [DOI] [PubMed] [Google Scholar]
- 27.Markussen T, Marvig RL, Gómez-Lozano M, Aanæs K, Burleigh AE, Høiby N, Johansen HK, Molin S, Jelsbak L. 2014. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. mBio 5:e01592-14. doi: 10.1128/mBio.01592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limoli DH, Whitfield GB, Kitao T, Ivey ML, Davis MR Jr, Grahl N, Hogan DA, Rahme LG, Howell PL, O’Toole GA, Goldberg JB. 2017. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio 8:e00186-17. doi: 10.1128/mBio.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scoffield JA, Duan D, Zhu F, Wu H. 2017. A commensal streptococcus hijacks a Pseudomonas aeruginosa exopolysaccharide to promote biofilm formation. PLoS Pathog 13:e1006300. doi: 10.1371/journal.ppat.1006300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, Karaoz U, Andersen GL, Brown R, Fujimura KE, Wu B, Tran D, Koff J, Kleinhenz ME, Nielson D, Brodie EL, Lynch SV. 2010. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One 5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zemanick ET, Wagner BD, Robertson CE, Ahrens RC, Chmiel JF, Clancy JP, Gibson RL, Harris WT, Kurland G, Laguna TA, McColley SA, McCoy K, Retsch-Bogart G, Sobush KT, Zeitlin PL, Stevens MJ, Accurso FJ, Sagel SD, Harris JK, Zemanick ET, Wagner BD, Robertson CE, Ahrens RC, Chmiel JF, Clancy JP, Gibson RL, Harris WT, Kurland G, Laguna TA, McColley SA, McCoy K, Retsch-Bogart G, Sobush KT, Zeitlin PL, Stevens MJ, Accurso FJ, Sagel SD, Harris JK. 2017. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J 50:1700832. doi: 10.1183/13993003.00832-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatziagorou E, Orenti A, Drevinek P, Kashirskaya N, Mei-Zahav M, De Boeck K. 2019. Changing epidemiology of the respiratory bacteriology of patients with cystic fibrosis—data from the European cystic fibrosis society patient registry. J Cyst Fibros doi: 10.1016/j.jcf.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Rouf R, Karaba SM, Dao J, Cianciotto NP. 2011. Stenotrophomonas maltophilia strains replicate and persist in the murine lung, but to significantly different degrees. Microbiology 157:2133–2142. doi: 10.1099/mic.0.048157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Bonaventura G, Pompilio A, Zappacosta R, Petrucci F, Fiscarelli E, Rossi C, Piccolomini R. 2010. Role of excessive inflammatory response to Stenotrophomonas maltophilia lung infection in DBA/2 mice and implications for cystic fibrosis. Infect Immun 78:2466–2476. doi: 10.1128/IAI.01391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pompilio A, Ciavardelli D, Crocetta V, Consalvo A, Zappacosta R, Di Ilio C, Di Bonaventura G. 2014. Stenotrophomonas maltophilia virulence and specific variations in trace elements during acute lung infection: implications in cystic fibrosis. PLoS One 9:e88769. doi: 10.1371/journal.pone.0088769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin C, Yang W, Meng J, Lv Y, Wang J, Huang B. 2017. Co-infection of Pseudomonas aeruginosa and Stenotrophomonas maltophilia in hospitalised pneumonia patients has a synergic and significant impact on clinical outcomes. Eur J Clin Microbiol Infect Dis 36:2231–2235. doi: 10.1007/s10096-017-3050-4. [DOI] [PubMed] [Google Scholar]
- 38.Jones-Nelson O, Hilliard JJ, DiGiandomenico A, Warrener P, Alfaro A, Cheng L, Stover CK, Cohen TS, Sellman BR. 2018. The neutrophilic response to Pseudomonas damages the airway barrier, promoting infection by Klebsiella pneumoniae. Am J Respir Cell Mol Biol 59:745–756. doi: 10.1165/rcmb.2018-0107OC. [DOI] [PubMed] [Google Scholar]
- 39.Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain M, Ramirez D, Seshadri R, Cullina JF, Powers CA, Schulert GS, Bar-Meir M, Sullivan CL, McColley SA, Hauser AR. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol 42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu W, Badrane H, Arora S, Baker HV, Jin S. 2004. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J Bacteriol 186:7575–7585. doi: 10.1128/JB.186.22.7575-7585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones AK, Fulcher NB, Balzer GJ, Urbanowski ML, Pritchett CL, Schurr MJ, Yahr TL, Wolfgang MC. 2010. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J Bacteriol 192:5709–5717. doi: 10.1128/JB.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scoffield J, Silo-Suh L. 2016. Glycerol metabolism promotes biofilm formation by Pseudomonas aeruginosa. Can J Microbiol 62:704–710. doi: 10.1139/cjm-2016-0119. [DOI] [PubMed] [Google Scholar]
- 47.Merritt JH, Kadouri DE, O’Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 49.Theilacker C, Coleman FT, Mueschenborn S, Llosa N, Grout M, Pier GB. 2003. Construction and characterization of a Pseudomonas aeruginosa mucoid exopolysaccharide-alginate conjugate vaccine. Infect Immun 71:3875–3884. doi: 10.1128/iai.71.7.3875-3884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.