Antibiotic treatment of patients undergoing complex medical treatments can deplete commensal bacterial strains from the intestinal microbiota, thereby reducing colonization resistance against a wide range of antibiotic-resistant pathogens. Loss of colonization resistance can lead to marked expansion of vancomycin-resistant Enterococcus faecium (VRE), Klebsiella pneumoniae, and Escherichia coli in the intestinal lumen, predisposing patients to bloodstream invasion and sepsis.
KEYWORD: immunology
ABSTRACT
Antibiotic treatment of patients undergoing complex medical treatments can deplete commensal bacterial strains from the intestinal microbiota, thereby reducing colonization resistance against a wide range of antibiotic-resistant pathogens. Loss of colonization resistance can lead to marked expansion of vancomycin-resistant Enterococcus faecium (VRE), Klebsiella pneumoniae, and Escherichia coli in the intestinal lumen, predisposing patients to bloodstream invasion and sepsis. The impact of intestinal domination by these antibiotic-resistant pathogens on mucosal immune defenses and epithelial and mucin-mediated barrier integrity is unclear. We used a mouse model to study the impact of intestinal domination by antibiotic-resistant bacterial species and strains on the colonic mucosa. Intestinal colonization with K. pneumoniae, Proteus mirabilis, or Enterobacter cloacae promoted greater recruitment of neutrophils to the colonic mucosa. To test the hypothesis that the residual microbiota influences the severity of colitis caused by infection with Clostridioides difficile, we coinfected mice that were colonized with ampicillin-resistant bacteria with a virulent strain of C. difficile and monitored colonization and pathogenesis. Despite the compositional differences in the gut microbiota, the severity of C. difficile infection (CDI) and mortality did not differ significantly between mice colonized with different ampicillin-resistant bacterial species. Our results suggest that the virulence mechanisms enabling CDI and epithelial destruction outweigh the relatively minor impact of less-virulent antibiotic-resistant intestinal bacteria on the outcome of CDI.
INTRODUCTION
The human colonic microbiota consists of diverse, predominantly anaerobic bacterial strains belonging to the phyla Bacteroidetes and Firmicutes and represents an ecosystem within the host that is defined by multiple layers of cooperation and competition between its component microbes. The microbiota and its products impact host metabolism and immune development and protect against infection by activating a range of direct and indirect antimicrobial mechanisms that function in parallel and are referred to collectively as colonization resistance (1). Marked differences in baseline microbiota composition and colonization resistance between individuals likely account, at least in part, for the wide range of disease severities associated with infection by specific microbial pathogens. Although many factors influence the composition and diversity of the microbiota, including diet, environmental exposure, and host immune status, widespread use of antibiotics has had a particularly large impact on the microbiota of human and domesticated animal populations (2). While antibiotics dramatically reduce mortality resulting from bacterial infections, antibiotic-induced collateral damage to the microbiota impairs colonization resistance and, paradoxically, increases susceptibility to infection (3).
Loss of commensal bacteria is particularly common in patients undergoing cancer treatment and is associated with the expansion of antibiotic-resistant pathobionts, such as vancomycin-resistant Enterococcus faecium (VRE), Klebsiella pneumoniae, and Escherichia coli. In patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HSCT), the incidence of intestinal domination by VRE exceeds 50%, while domination by K. pneumoniae or E. coli occurs in as many as 20% of patients (4). Intestinal domination in allo-HSCT patients is associated with an increased incidence of bloodstream infection by the respective colon-dominating pathogen. However, the impact of intestinal domination by these antibiotic-resistant bacteria on the host’s mucosal immune cells and antimicrobial defenses is largely undefined.
Bone marrow-derived neutrophils and monocytes play essential roles in the mucosal immune defense against intestinal infections (5). Proinflammatory cytokines, such as interleukin 23 (IL-23), and inflammasome-mediated production of IL-1β by inflammatory monocyte-derived mononuclear phagocytes in the lamina propria (Lp) promote intestinal defense upon bacterial infection (6, 7). It is unclear whether the antibiotic-resistant pathobionts that commonly dominate the gut and cause systemic infections in hospitalized patients impact mucosal immune defenses and trigger inflammatory responses. Furthermore, whether intestinal colonization by common, antibiotic-resistant bacterial species impacts patient susceptibility to one of the leading causes of nosocomial infection, Clostridioides difficile, is unknown.
C. difficile is a common cause of antibiotic-associated diarrhea and is particularly prevalent in patients who have sustained damage to their intestinal microbiota. The interactions between the host, indigenous microbiota, and pathogens are complex, and each component contributes to the pathogenesis of C. difficile infection (CDI) (8). The severity of C. difficile infections differs, however; some patients have only mild disease, and others develop extensive necrotizing colitis. Host factors, such as age, underlying diseases, and immune status, and virulence differences between C. difficile strains contribute to disease severity but do not completely explain the wide range of disease manifestations (9, 10). Whether the residual microbiota following antibiotic treatment influences the severity of colitis caused by infection with C. difficile is unknown.
To address these issues, we used a mouse model to investigate intestinal colonization by a range of ampicillin-resistant bacterial strains isolated from patients undergoing cancer treatment. We found that intestinal colonization by different bacterial species resulted in differences in the recruitment of neutrophils to the intestinal lamina propria and in variable levels of inflammatory cytokine expression. Despite differences in the state and intensity of immune activation, we did not detect a significant impact of intestinal colonization by different antibiotic-resistant bacterial strains on the course and severity of infection by C. difficile. These findings indicate that the pathogenetic mechanisms engaged by C. difficile to cause colitis, and the host’s immune defenses leading to resolution, override the relatively minor impact of residual bacterial species on colonic immune defenses.
RESULTS
Intestinal domination by ampicillin-resistant bacterial strains.
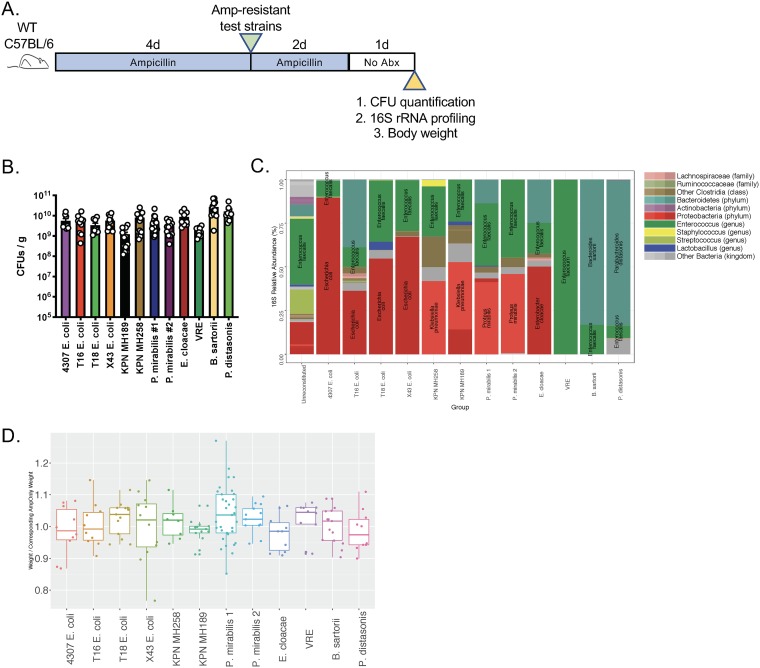
We obtained 10 ampicillin-resistant clinical isolates of K. pneumoniae, E. coli, Enterobacter cloacae, Proteus mirabilis, and vancomycin-resistant Enterococcus faecium (11, 12) from patients undergoing cancer treatment, as well as ampicillin-resistant strains of Bacteroides sartorii and Parabacteroides distasonis from a mouse colony that had been treated for >10 years with ampicillin (13). To establish colonization by these ampicillin-resistant strains, we depleted the indigenous microbiota of wild-type (WT) C57BL/6 mice with ampicillin for 4 days prior to inoculating them with 2 × 104 CFU of each strain by oral gavage. Infected mice were maintained on ampicillin for 2 additional days, and fecal pellets were obtained 1 day after antibiotic discontinuation in order to quantify CFU by selective plating (Fig. 1A). Although there were some differences in the densities of fecal colonization by the different ampicillin-resistant bacterial strains, all strains achieved densities exceeding 109 CFU per g of feces (Fig. 1B).
FIG 1.
The microbiota of antibiotic-treated mice are receptive to ampicillin-resistant bacterial species. (A) Wild-type C57BL/6 mice (n, ≥10 per group; results from 17 independent experiments) were treated with ampicillin in drinking water (0.5 g/liter) for 4 days and were then inoculated with 2 × 104 CFU of 1 of 12 ampicillin-resistant bacterial strains. Two days after inoculation, ampicillin was removed from the drinking water. Fecal pellets were collected 1 day later. Abx, antibiotics. (B) The abundance of inoculated bacteria in fecal pellets (expressed as CFU per gram) was determined. (C) DNA was extracted from fecal pellets and was subjected to 16S rRNA PCR amplification and sequencing of the V4–V5 region. The average relative abundances of bacterial taxa for all mice of the same group are plotted. (D) The weight of mice in each group was divided by the weight of control mice in order to assess weight loss associated with colonization by ampicillin-resistant bacterial strains.
To determine the microbiota composition of mice colonized with ampicillin-resistant bacterial strains, we performed 16S rRNA sequencing. Mice receiving ampicillin followed by inoculation with an ampicillin-resistant bacterial strain developed domination, defined here as exceeding 30% of relative abundance, as demonstrated in Fig. 1C. Of note, because ampicillin treatment had been discontinued for 24 h prior to fecal pellet collection, indigenous bacterial strains were detected in all mice. Bacterial populations following ampicillin selection were also detected in the unreconstituted group (Fig. 1C, left), albeit at lower densities, as determined by quantitative 16S rRNA gene PCR (data not shown). These strains likely represent residual bacteria that survived ampicillin treatment and underwent reexpansion following the discontinuation of ampicillin treatment. Additionally, mice did not lose weight or demonstrate evidence of pathology upon colonization with the resistant strains that were tested (Fig. 1D). These data show that we can substantially alter the microbiota of ampicillin-treated mice by administering a range of ampicillin-resistant bacterial isolates.
Ampicillin-resistant bacterial strains activate the mucosal immune system distinctly.
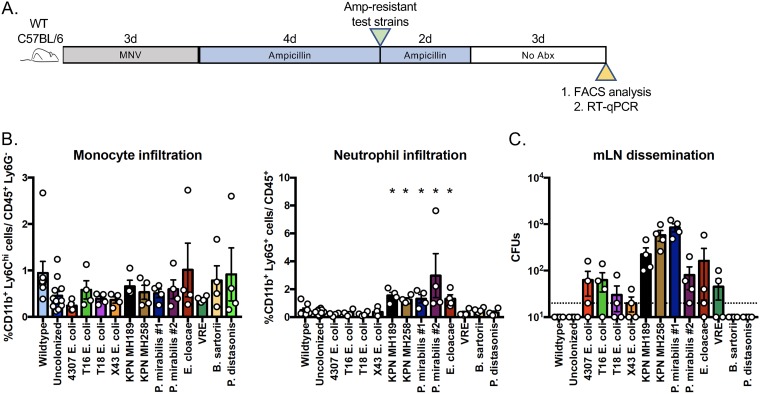
Although each member of our panel of ampicillin-resistant bacterial strains colonized mice treated with ampicillin, and none of the colonized mice lost weight or developed morbidity, it was unclear whether the different bacterial strains impacted the mucosal immune system. To address these questions, we treated wild-type C57BL/6 mice with metronidazole, neomycin, and vancomycin (MNV) for 3 days, followed by ampicillin for 4 days, with the goal of markedly reducing the indigenous bacterial populations. However, the additional MNV treatment did not completely eliminate the indigenous bacterial populations in mice colonized with ampicillin-resistant bacterial strains (see Fig. S2 in the supplemental material), suggesting a high tolerance of a subset of residual intestinal bacteria to antibiotic treatment. The mice were then individually administered 1 of the 12 ampicillin-resistant strains by oral gavage (Fig. 2A). Monocyte and neutrophil infiltration of the colonic lamina propria was determined 5 days following inoculation. Flow cytometric analyses demonstrated that monocyte infiltration of the colonic lamina propria was not influenced by intestinal colonization with different ampicillin-resistant bacterial species (Fig. 2B). In contrast, neutrophils were more abundant in the colonic laminae propriae of mice colonized with K. pneumoniae, P. mirabilis, or E. cloacae than in those of mice colonized with other species, and these bacterial strains had the highest levels of dissemination to mesenteric lymph nodes (mLNs) (Fig. 2B and C). Interestingly, B. sartorii and P. distasonis densely colonized the colon; however, these obligate anaerobes were not detected by culture in mLNs (Fig. 2C).
FIG 2.
Antibiotic-resistant bacterial strains exhibit distinct capacities to disseminate to mLNs and to recruit neutrophils to the colonic lamina propria. (A) Wild-type C57BL/6 mice (n, 4 to 5 per group) were treated with metronidazole, vancomycin, and neomycin (MNV) for 3 days, followed by ampicillin for 4 days. Then the mice were inoculated with 2 × 104 CFU of 1 of 12 antibiotic-resistant bacterial strains. Ampicillin was maintained in the drinking water for an additional 2 days. (B) Five days following inoculation, cells isolated from the colonic lamina propria were profiled by flow cytometry. (C) Bacterial dissemination to mLNs was determined by selective plating of whole mLNs.
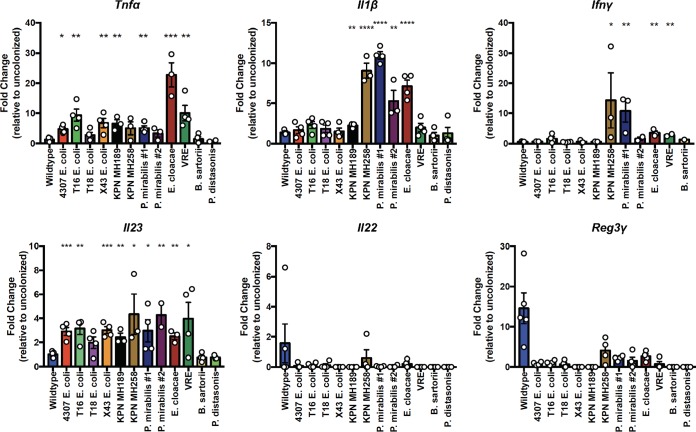
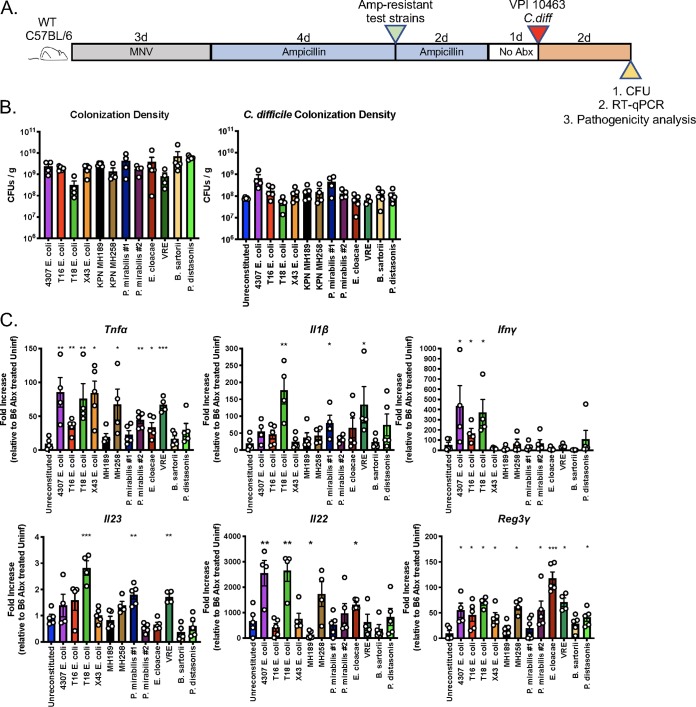
To determine whether members of the ampicillin-resistant bacterial panel triggered the transcription of genes encoding inflammatory cytokines, we performed reverse transcription (RT)-PCR for Tnfα, Il1β, Ifnγ, Il23, Il22, and Reg3γ (Fig. 3). These cytokines have been implicated in the mucosal defense against intestinal pathogens, including C. difficile (14–17). Tnfα and Il23 transcription was significantly increased in most mice that had been colonized with an ampicillin-resistant bacterial strain. Il1β expression generally correlated with increased neutrophil infiltration, with the exception of colonization by K. pneumoniae MH189, which was associated with increased neutrophil levels but baseline Il1β expression. Mice colonized with K. pneumoniae MH258, P. mirabilis #1, VRE, or E. cloacae had increased Ifnγ transcription. Previous studies have demonstrated that MyD88-mediated induction of IL-23 drives the expression of IL-22 by innate lymphocytes in the lamina propria, which induces Reg3γ expression in intestinal epithelial cells (18–20). Here, we have seen that while B. sartorii and P. distasonis did not induce Il23 transcription, the other ampicillin-resistant strains induced 2- to 4-fold-increased Il23 transcription, but this did not lead to increased transcription of either Il22 or Reg3γ. Our results suggest that the low level of IL-23 transcript induction by the ampicillin- resistant strains is insufficient to promote IL-22 and Reg3γ expression.
FIG 3.
Impact of antibiotic-resistant bacterial strains on the transcription of cytokines in the colon. The experimental procedure followed the schematic in Fig. 2A. Five days following inoculation, RT-PCR was performed on whole colonic tissue. The fold induction of the indicated cytokines relative to levels in antibiotic-treated but uncolonized mice is plotted. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Data shown are means ± SEM.
The composition of the microbiota does not significantly influence survival following C. difficile infection.
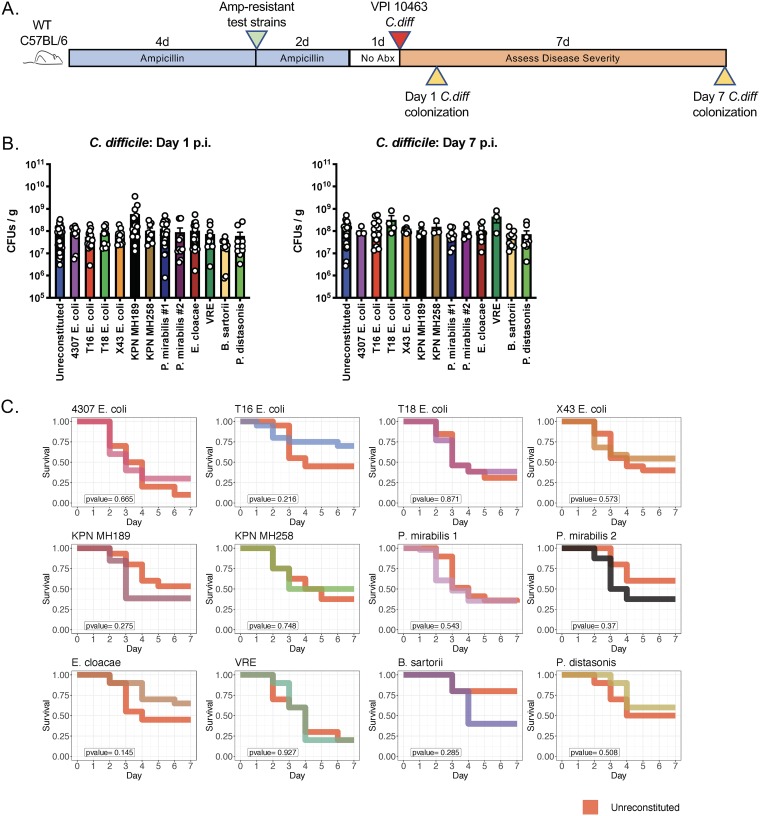
One question that remains unexplored is whether the residual microbiota composition at the time of a C. difficile infection contributes to the severity of colitis. The host mechanisms underlying recovery from acute CDI remain incompletely defined and might be influenced by microbiota composition. To address this question, wild-type C57BL/6 mice were first colonized with each of the ampicillin-resistant strains and then inoculated by oral gavage with approximately 200 spores of the virulent C. difficile strain VPI 10463 (Fig. 4A).
FIG 4.
The composition of the residual intestinal microbiota does not significantly influence survival following C. difficile infection. (A) Schematic of experimental design to determine the impact of residual microbiota composition on the outcome of C. difficile infection. Wild-type C57BL/6 mice (n, ≥10 per group; results from 17 independent experiments) were treated with ampicillin for 4 days and were then inoculated with 2 × 104 CFU of the antibiotic-resistant bacterial strains. Ampicillin was maintained in the drinking water for an additional 2 days. Mice were challenged with 200 C. difficile VPI 10463 spores via oral gavage 1 day after ampicillin withdrawal, and C. difficile colonization and mouse survival were monitored as indicated. (B) On day 1 and day 7 postinfection (p.i.), C. difficile CFU in fecal pellets were quantified using selective plating. (C) Survival was monitored for 1 week post-C. difficile infection.
As noted above, all 12 ampicillin-resistant strains achieved high density in the mouse intestine at the time of C. difficile infection (Fig. 1B and C). None of the strains provided colonization resistance or clearance of C. difficile for 7 days following inoculation (Fig. 4B; see also Fig. S1 in the supplemental material). The densities of C. difficile were similar in mice colonized with the different bacterial strains, and there were no statistically significant differences in survival following C. difficile infection (Fig. 4C). K. pneumoniae, P. mirabilis, and E. cloacae strains were the most immunomodulatory in terms of inducing neutrophil recruitment to the colon, induction of proinflammatory cytokines, and dissemination to mLNs. Despite these features, these strains did not impact survival following C. difficile infection.
The background microbiota influences the host immune response, the virulence of C. difficile, and bacterial dissemination in the setting of CDI.
To determine whether the presence of different antibiotic-resistant bacterial strains influences the mucosal immune response to C. difficile infection, wild-type C57BL/6 mice were first colonized with each ampicillin-resistant bacterial strain and then infected with C. difficile VPI 10463, and cecal contents and tissues were isolated 2 days following infection (Fig. 5A). All mice had comparable cecal content colonization levels with the ampicillin-resistant isolate they were administered and C. difficile (Fig. 5B). While C. difficile competes with the domination of the ampicillin-resistant strains in the intestinal contents, in contrast to the situation for uninfected mice, ampicillin-resistant strains persist in the intestinal niche upon C. difficile infection (Fig. S2 in the supplemental material). We found that the colonization of antibiotic-treated mice with antibiotic-resistant bacterial strains prior to C. difficile infection resulted in increased transcription of inflammatory cytokine genes (Fig. 5C). Most bacterial strains induced significant increases in Tnfα transcription over that for C. difficile-infected mice that had not received antibiotic-resistant bacterial strains. Colonization with E. coli expressing T18, P. mirabilis #1, or VRE increased the transcription of genes encoding IL-1β and IL-23, which have been shown to adversely impact CDI outcomes (21). Il22 transcription was dramatically increased in the setting of CDI and was associated with increased levels of Reg3γ transcription.
FIG 5.
Impact of the residual microbiota on host immune responses during CDI. (A) Schematic of the experimental design to determine the impact of residual microbiota composition on the outcome of C. difficile infection. Mice were inoculated with each antibiotic-resistant isolate followed by 200 spores of C. difficile VPI 10463 and were euthanized 2 days later. (B) The abundance (expressed in CFU per gram) of the antibiotic-resistant strain and C. difficile in cecal contents was determined by selective plating. (C) Fold induction of immune cytokines in colonic tissue as determined by quantitative RT-PCR. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) from results for control mice. Data shown are means ± SEM.
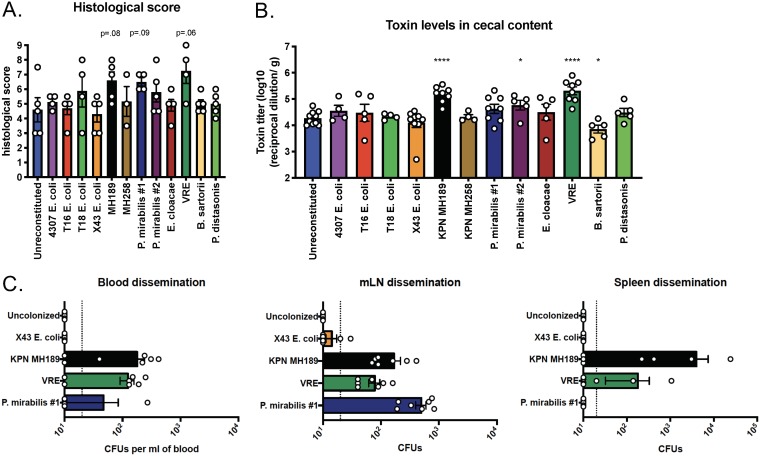
We also performed hematoxylin-and-eosin (H&E) staining on proximal colonic tissue sections (Fig. 6A; see also Fig. S3 in the supplemental material). Slides were assessed in a blinded fashion and were histologically scored, revealing that K. pneumoniae MH189 and VRE induced increased tissue pathology, in agreement with the finding that the cecal contents of mice colonized with these strains had higher C. difficile toxin levels. The toxin titer was influenced by the background microbiota composition (Fig. 6B); colonization with K. pneumoniae MH189, P. mirabilis #2, or VRE prior to C. difficile infection resulted in significantly higher toxin levels in cecal contents, and mice colonized with B. sartorii had reduced levels.
FIG 6.
Impact of the residual microbiota on host pathology, cecal toxin levels, and bacterial dissemination during CDI. The experimental design follows the schematic in Fig. 5A. (A) Pathology scores of histological tissue sections based on cellular infiltration, edema (host response), and epithelial layer damage. Data shown are means ± SEM. (B) C. difficile toxin titers in cecal contents were determined by an in vitro cytotoxicity assay. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (C) Mice (n, 7 from two independent experiments) were colonized with one of four strains. The mice were inoculated with 200 spores of C. difficile VPI 10463 and were euthanized on day 2 post-C. difficile infection. Blood, mesenteric lymph nodes (mLNs), and spleens were isolated and homogenized, and bacterial dissemination post-C. difficile infection was determined via selective plating.
To determine if epithelial damage caused by C. difficile enhances the dissemination of antibiotic-resistant bacterial strains, we quantified CFU in tissues on day 2 after C. difficile VPI 10463 inoculation of mice colonized with E. coli X43, K. pneumoniae MH189, VRE, or P. mirabilis #1 (Fig. 6C). K. pneumoniae MH189, VRE, and P. mirabilis #1 disseminated to the blood and mesenteric lymph nodes, and viable K. pneumoniae MH189 and VRE were detected in spleens. However, in spite of reaching comparable levels of intestinal colonization and mLN dissemination, no MH189 dissemination in the spleen was observed in the absence of C. difficile coinfection (see Fig. S4 in the supplemental material). E. coli X43 did not disseminate, except in low abundance to the mesenteric lymph nodes. Our results demonstrate that the residual microbiota influences the inflammatory response to C. difficile infection and that C. difficile infection is associated with greater dissemination of some bacterial strains that persist in the cecum and colon.
DISCUSSION
Antibiotic treatment can deplete commensal bacterial populations that provide colonization resistance against bacterial pathogens, including highly antibiotic-resistant bacterial strains that are major causes of infection in hospitalized patients. Loss of colonization resistance can result in the marked expansion of antibiotic-resistant strains of E. faecium, E. coli, and K. pneumoniae and many strains belonging to the Enterobacteriaceae family. Although intestinal domination by these strains is associated with a markedly increased risk of bacteremia and sepsis, particularly in the setting of cancer treatment with hematopoietic stem cell transplantation (4), the impact of domination on the mucosal immune response is incompletely defined. A previous study demonstrated that VRE and K. pneumoniae differ with respect to their abilities to penetrate the dense mucus layer and access MLNs (22), suggesting that immune activation by antibiotic-resistant bacterial strains likely differs. Our results confirm this notion by demonstrating that the induction of inflammatory cytokines can differ between bacterial strains. The mechanisms by which dominating bacterial strains traverse the intestinal epithelium and the pathways involved in immune activation remain to be defined.
The balance between inflammatory and regulatory immune responses influences the initiation, progression, and resolution of colitis. Inflammatory responses in the gut are generally provoked by intestinal microbes and their molecular products, which can differ dramatically in composition. We tested the hypothesis that the composition of the microbiota upon antibiotic selection in the intestine during the course of C. difficile infection would alter the severity of colitis and potentially alter recovery. We chose to introduce antibiotic-resistant bacterial species that commonly reside in the intestine, including K. pneumoniae, E. coli, E. cloacae, and P. mirabilis, as well as two commensal bacterial species belonging to the Bacteroidetes phylum. Surprisingly, we did not find that these bacterial strains significantly altered the course of C. difficile infection.
Previous studies have demonstrated that the gut microbiota can impact the severity of C. difficile infection (23, 24). For example, the presence of Lachnospiraceae in the gut has been associated with a lower risk of CDI (23, 25), and certain species of this commensal bacterial family, such as Clostridium scindens, mediate resistance against C. difficile infection by the production of secondary bile acids (26). Other commensal bacterial strains deplete nutrients that support the growth of C. difficile (27). Our study differs from previous work by focusing on the aberrant expansion of bacterial strains that are highly antibiotic resistant. The ampicillin-resistant bacterial strains that we introduced into antibiotic-treated mice do not convert primary to secondary bile acids. Furthermore, we did not detect any impact of these strains on colitis associated with C. difficile infection, suggesting that these ampicillin-resistant strains do not deplete nutrients that are essential for C. difficile virulence. It is possible that other antibiotic-resistant bacterial strains, perhaps those expressing potential enterotoxins, could enhance C. difficile colitis. Certain strains of Bacteroides fragilis, for examples, express an enterotoxin that has been associated with the development of colon cancer (28). Further investigation is required to determine whether such strains can enhance C. difficile colitis. Our results, however, suggest that the residual microbiota after antibiotic selection is unlikely to provide an explanation for the range of colitis severities seen in patients with C. difficile infection.
The determinants of C. difficile virulence have been of immense research interest for decades. C. difficile toxins (TcdA and TcdB) have been demonstrated to be critical factors causing colitis and pathogenesis in a hamster model (29). However, in our mouse model, the toxin level measured at 2 days post-C. difficile infection does not correlate with the survival rate of infected mice. Upon C. difficile infection of B. sartorii-inoculated mice, a lower toxin level was produced yet a worse survival rate was observed, suggesting that alternative factors likely contribute to pathogenicity in mice. E. cloacae triggers dramatic induction of Reg3γ levels upon C. difficile coinfection and is associated with a better (though not significantly so) survival rate of infected mice, suggesting that host immune responses also regulate C. difficile virulence.
In summary, we tested a group of ampicillin-resistant bacterial strains, all of which expanded to abnormally high densities in the guts of mice treated with ampicillin, and demonstrated their impact on the induction of inflammatory cytokines. Despite differences in cytokine induction, we failed to detect significant impacts of intestinal domination by these antibiotic-resistant bacterial strains on the severity of C. difficile-associated colitis. Our mouse model mimics clinical settings in which antibiotic-resistant bacterial strains dominate the guts of patients with C. difficile infection. Our data suggest that many, if not most, of these residual antibiotic-resistant bacterial strains have a minimal impact on the severity of C. difficile infection.
MATERIALS AND METHODS
Mice.
WT C57BL/6 mice were purchased from The Jackson Laboratory (Room MP14). All mice were bred and maintained under specific-pathogen-free conditions at the Memorial Sloan Kettering Research Animal Resource Center. Sex- and age-matched controls were used in all experiments according to institutional guidelines for animal care. All animal procedures were approved by the Institutional Animal Care and Use Committee of the Memorial Sloan Kettering Cancer Center.
Antibiotic pretreatment, ampicillin-resistant test strain administration, CDI, and mouse monitoring.
Wild-type C57BL/6 mice were cohoused for 2 weeks prior to antibiotic treatment. Mice were first treated with ampicillin in drinking water (0.5 g/liter) for 4 days and then inoculated with 2 × 104 CFU of 1 of 12 ampicillin-resistant bacterial strains grown in LB broth supplemented with ampicillin. Ampicillin in water was maintained for 2 additional days after inoculation. Twenty-four hours post-ampicillin cessation, mice received 200 C. difficile spores (strain VPI 10463 [ATCC 43255]) via oral gavage. Following infection, mice were monitored and were scored for disease severity by four parameters (14): weight loss (a score of 0 was assigned for >95% of initial weight, 1 for 95% to 90% of initial weight, 2 for 90% to 80% of initial weight, and 3 for <80% of initial weight), surface body temperature (a score of 0 was assigned for >32°C, 1 for 32°C to 30°C, 2 for 30°C to 28°C, and 3 for <28°C), diarrhea severity (a score of 0 was assigned for formed pellets, 1 for loose pellets, 2 for liquid discharge, and 3 for no pellets [discharge caked on fur]), and morbidity (1 point was assigned for each of the following symptoms, with a maximum score of 3: ruffled fur, hunched back, lethargy, ocular discharge).
Isolation of cells from the mLNs and intestine.
Cells were isolated on the basis of a previous description (30). Briefly, lymphocytes were isolated from the mLNs by mechanical disruption through 100-μm cell strainers. Single-cell suspensions were obtained from the ileum Lp by longitudinally cutting the ileum and then washing out the contents in phosphate-buffered saline (PBS). Intestinal tissues were incubated at 37°C under gentle agitation in stripping buffer (30) for 10 min to remove epithelial cells and then for another 20 min for the intraepithelial lymphocytes (IEL). Supernatants containing the IEL fraction were passed through 100-μm cell strainers and were resuspended in 40% Percoll. The remaining tissue was digested with collagenase IV (1.5 mg/ml; 500 U/ml) and DNase (20 μg/ml) in complete medium (Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin-streptomycin [10 μg/ml], gentamicin [50 μg/ml], 10 mM HEPES, 0.5 mM β-mercaptoethanol, and l-glutamine [20 μg/ml]) for 30 min at 37°C under gentle agitation. Supernatants containing the Lp fraction were passed through 100-μm cell strainers and were resuspended in 40% Percoll. Samples were then centrifuged for 20 min at 600 × g to obtain the IEL and Lp cell fractions.
FACS analysis.
Cell suspensions were surface stained in fluorescence-activated cell sorter (FACS) buffer (PBS, 2% bovine serum albumin, 0.2 mg of sodium azide, 2 mM EDTA) using a standard flow cytometric staining protocol with fluorescently conjugated antibodies specific to Ly6c (AL-21; BD Biosciences), major histocompatibility complex class II (MHCII) (M5/114.15.2; BD Biosciences), CD103 (2E7; BioLegend), F480 (BM8; Thermo Fisher), Ly6G (1A8; BD Biosciences), CD45.2 (104; BD Biosciences), CD11b (RM2817; Thermo Fisher), and CD11c (HL3; BD Biosciences). Cell viability was assessed with LIVE/DEAD Aqua stain (Thermo Fisher). Samples were collected by an LSR II flow cytometer (Becton, Dickinson). All flow cytometry data were analyzed by FlowJo, version 9.9 (Tree Star).
Histology sectional and pathology scoring.
Colon tissues were fixed with 4% paraformaldehyde and embedded in paraffin, and 5-μm sections were cut and stained with hematoxylin and eosin. H&E-stained colon tissue sections were blindly scored as described elsewhere (31) based on epithelial degeneration/cell death, edema, and cellular infiltration, with each parameter scored from 0 to 3.
Quantification of C. difficile burden and toxin.
Fecal pellets or cecal contents were resuspended in deoxygenated PBS, and 10-fold dilutions were plated onto brain heart infusion (BHI) agar supplemented with yeast extract, taurocholate, l-cysteine, cycloserine, and cefoxitin at 37°C in an anaerobic chamber (Coy Labs) overnight (32). The presence of C. difficile toxin B was determined using a cell-based cytotoxicity assay as described previously (33). Briefly, human embryonic lung fibroblast WI-38 cells (ATCC CCL-75) were incubated in a 96-well plate overnight at 37°C. Tenfold dilutions of supernatants from resuspended cecal contents were added to WI-38 cells, which were then incubated overnight at 37°C, and the presence of cell rounding and death was observed the next day. The presence of C. difficile toxin B was confirmed by neutralization by antitoxin antisera (Techlab, Blacksburg, VA). The data are expressed as the log10 reciprocal value of the last dilution where cell rounding was observed.
DNA extraction and 16S rRNA sequencing.
DNA was extracted from fecal pellets and intestinal contents as described previously (34). In brief, a frozen aliquot (∼100 mg) of each sample was suspended, while frozen, in a solution containing 500 μl of extraction buffer (200 mM Tris [pH 8.0], 200 mM NaCl, and 20 mM EDTA), 200 μl of 20% sodium dodecyl sulfate (SDS), 500 μl of phenol-chloroform–isoamyl alcohol (24:24:1), and 500 μl of 0.1-mm-diameter zirconia/silica beads (BioSpec Products). Microbial cells were lysed by mechanical disruption with a bead beater (BioSpec Products) for 2 min, after which two rounds of phenol-chloroform–isoamyl alcohol extraction were performed. DNA was precipitated with ethanol and was resuspended in 50 μl of Tris-EDTA (TE) buffer with 100 μg RNase ml−1. The isolated DNA was subjected to additional purification with QIAamp Mini Spin columns (Qiagen). For each sample, duplicate 50-μl PCRs were performed, with each reaction mixture containing 50 ng of purified DNA, 0.2 mM deoxynucleoside triphosphate s (dNTPs), 1.5 mM MgCl2, 2.5 U Platinum Taq DNA polymerase, 2.5 μl of 10× PCR buffer, and 0.5 μM each primer designed to amplify the V4–V5 region: 563F (5′-nnnnnnnn-NNNNNNNNNNNN-AYTGGGYDTAAAGNG-3′) and 926R (5′-nnnnnnnn-NNNNNNNNNNNN-CCGTCAATTYHTTTRAGT-3′). A unique 12-base Golay barcode (represented by N’s in the sequences) precedes each primer for sample identification (35), and 1 to 8 additional nucleotides (represented by n’s) were placed in front of the barcode to offset the sequencing of the primers. Cycling conditions were as follows: 94°C for 3 min, followed by 27 cycles of 94°C for 50 s, 51°C for 30 s, and 72°C for 1 min; 72°C for 5 min was used for the final elongation step. Replicate PCR products were pooled, and amplicons were purified using the QIAquick PCR purification kit (Qiagen). PCR products were quantified and pooled at equimolar amounts before Illumina barcodes and adaptors were ligated on using the Illumina TruSeq Sample Preparation protocol. The completed library was sequenced on an Illumina MiSeq platform by following the Illumina-recommended procedures with a paired-end 250 × 250-bp kit. 16S rRNA (V4–V5) paired-end reads were merged and demultiplexed. Merged reads of <300 bp or >360 bp were removed. Maximum expected error filtering (Emax = 1) (36), amplicon sequence variant (ASV) grouping, and chimeric and singleton sequence removal were performed using the Dada2 pipeline (37). Taxonomic classification of ASVs was performed by nucleotide BLAST of representative sequences from each ASV, with NCBI RefSeq as the reference training set. A minimum E value threshold value of 1e−10 was used for assignments.
Tissue RNA isolation, cDNA preparation, and RT-PCR.
RNA was isolated from colon tissue using mechanical homogenization and TRIzol isolation (Invitrogen) according to the manufacturer’s instructions. cDNA was generated using QuantiTect reverse transcriptase (Qiagen). RT-PCR was performed on cDNA using TaqMan primers and probes in combination with TaqMan PCR master mix (ABI), and reactions were run on an RT-PCR system (StepOnePlus; Applied Biosystems). Gene expression is displayed in the figures as the fold increase over expression in uninfected C57BL/6 mice and is normalized to hypoxanthine phosphoribosyltransferase (HPRT) expression. Quantitative PCR (qPCR) was run under the standard conditions recommended by StepOne software, version 2.3, as follows: holding phase, 50°C for 2 min and 95°C for 10 min; cycling phase, 95°C for 15 s and 60°C for 1 min (40 cycles). TaqMan probes used in this study were as follows: Reg3γ (Mm00441127_m1), IL-22 (Mm00444241_m1), IL-23 (Mm00518984_m1), IL1β (Mm01336189_m1), TNFα (Mm00443258_m1), IFNγ (Mm01168134_m1), and Hprt (Mm00446968_m1).
Statistical analysis.
The results presented here represent means ± standard errors of the means (SEM). Statistical significance was determined by the unpaired t test, by the Mann-Whitney test for sample size n ≤ 5, by two-way analysis of variance (ANOVA) for time course experiments, and by the log rank test for survival curves. Statistical analyses were performed using GraphPad Prism software, version 6.0. Significance is indicated by asterisks in figures (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Data availability.
Raw 16S rRNA sequencing reads have been deposited in BioProject under accession number PRJNA576440.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grants R01 AI095706 and R01 AI042135 from the National Institutes of Health.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sorbara MT, Pamer EG. 2019. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol 12:1–9. doi: 10.1038/s41385-018-0053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S, Covington A, Pamer EG. 2017. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev 279:90–105. doi: 10.1111/imr.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales M-A, Jenq RR, van den Brink MRM, Pamer EG. 2012. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caballero S, Pamer EG. 2015. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol 33:227–256. doi: 10.1146/annurev-immunol-032713-120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franchi L, Muñoz-Planillo R, Núñez G. 2012. Sensing and reacting to microbes through the inflammasomes. Nat Immunol 13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 8.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BB, Carter RA, Ling L, Leiner I, Taur Y, Kamboj M, Dubberke ER, Xavier J, Pamer EG. 2017. Pathogenicity locus, core genome, and accessory gene contributions to Clostridium difficile virulence. mBio 8:e00885-17. doi: 10.1128/mBio.00885-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loo VG, Bourgault A-M, Poirier L, Lamothe F, Michaud S, Turgeon N, Toye B, Beaudoin A, Frost EH, Gilca R, Brassard P, Dendukuri N, Béliveau C, Oughton M, Brukner I, Dascal A. 2011. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med 365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 11.Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, Seok R, Leiner IM, Taur Y, Peled JU, van den Brink MRM, Litvak Y, Bäumler AJ, Chaubard J-L, Pickard AJ, Cross JR, Pamer EG. 2019. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med 216:84–98. doi: 10.1084/jem.20181639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong H, Carter RA, Leiner IM, Tang Y-W, Chen L, Kreiswirth BN, Pamer EG. 2015. Distinct contributions of neutrophils and CCR2+ monocytes to pulmonary clearance of different Klebsiella pneumoniae strains. Infect Immun 83:3418–3427. doi: 10.1128/IAI.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, Miller L, Kim GJ, Ling L, Pamer EG. 2017. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 21:592–602.e4. doi: 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Sušac B, Ling L, Leiner I, Pamer EG. 2015. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowardin CA, Kuehne SA, Buonomo EL, Marie CS, Minton NP, Petri WA. 2015. Inflammasome activation contributes to interleukin-23 production in response to Clostridium difficile. mBio 6:e02386-14. doi: 10.1128/mBio.02386-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasegawa M, Kamada N, Jiao Y, Liu MZ, Núñez G, Inohara N. 2012. Protective role of commensals against Clostridium difficile infection via an IL-1β-mediated positive-feedback loop. J Immunol 189:3085–3091. doi: 10.4049/jimmunol.1200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa M, Yada S, Liu MZ, Kamada N, Muñoz-Planillo R, Do N, Núñez G, Inohara N. 2014. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity 41:620–632. doi: 10.1016/j.immuni.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. 2007. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J Exp Med 204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A 105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buonomo EL, Madan R, Pramoonjago P, Li L, Okusa MD, Petri WA. 2013. Role of interleukin 23 signaling in Clostridium difficile colitis. J Infect Dis 208:917–920. doi: 10.1093/infdis/jit277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caballero S, Carter R, Ke X, Sušac B, Leiner IM, Kim GJ, Miller L, Ling L, Manova K, Pamer EG. 2015. Distinct but spatially overlapping intestinal niches for vancomycin-resistant Enterococcus faecium and carbapenem-resistant Klebsiella pneumoniae. PLoS Pathog 11:e1005132. doi: 10.1371/journal.ppat.1005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YJ, Arguello ES, Jenq RR, Littmann E, Kim GJ, Miller LC, Ling L, Figueroa C, Robilotti E, Perales M-A, Barker JN, Giralt S, van den Brink MRM, Pamer EG, Taur Y. 2017. Protective factors in the intestinal microbiome against Clostridium difficile infection in recipients of allogeneic hematopoietic stem cell transplantation. J Infect Dis 215:1117–1123. doi: 10.1093/infdis/jix011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seekatz AM, Rao K, Santhosh K, Young VB. 2016. Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Med 8:47. doi: 10.1186/s13073-016-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert AM, Sinani H, Schloss PD. 2015. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. mBio 6:e00974-15. doi: 10.1128/mBio.00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 30.Abt MC, Buffie CG, Sušac B, Becattini S, Carter RA, Leiner I, Keith JW, Artis D, Osborne LC, Pamer EG. 2016. TLR-7 activation enhances IL-22–mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med 8:327ra25. doi: 10.1126/scitranslmed.aad6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarchum I, Liu M, Shi C, Equinda M, Pamer EG. 2012. Critical role for MyD88-mediated neutrophil recruitment during Clostridium difficile colitis. Infect Immun 80:2989–2996. doi: 10.1128/IAI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorg JA, Dineen SS. 2009. Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol Chapter 9:Unit 9A.1. doi: 10.1002/9780471729259.mc09a01s12. [DOI] [PubMed] [Google Scholar]
- 33.Jarchum I, Liu M, Lipuma L, Pamer EG. 2011. Toll-like receptor 5 stimulation protects mice from acute Clostridium difficile colitis. Infect Immun 79:1498–1503. doi: 10.1128/IAI.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, Pamer EG. 2012. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med 209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC, Flyvbjerg H. 2015. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 37.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S rRNA sequencing reads have been deposited in BioProject under accession number PRJNA576440.