Abstract
Chromatin remodelling complexes of the SWI/SNF family function in the formation of nucleosome-depleted, transcriptionally active promoter regions (NDRs)1,2. The essential Saccharomyces cerevisiae SWI/SNF complex RSC3 contains 16 subunits, including the ATP-dependent DNA translocase Sth14,5. RSC removes nucleosomes from promoter regions6,7 and positions the specialized +1 and –1 nucleosomes that flank NDRs8,9. Here, we present the cryo-EM structure of RSC in complex with a nucleosome substrate. The structure reveals that RSC forms five protein modules and suggests key features of the remodelling mechanism. The body module serves as a scaffold for the four flexible modules that we call DNA-interacting, ATPase, arm and ARP modules. The DNA-interacting module binds extra-nucleosomal DNA and is involved in the recognition of promoter DNA elements8,10,11 that influence RSC functionality12. The ATPase and arm modules sandwich the nucleosome disc with their ‘SnAC’ and ‘finger’ elements, respectively. The translocase motor of the ATPase module engages with the edge of the nucleosome at superhelical location +2. The mobile ARP module may modulate translocase-nucleosome interactions to regulate RSC activity5. The RSC-nucleosome structure provides a basis for understanding NDR formation and the structure and function of human SWI/SNF complexes that are frequently mutated in cancer13.
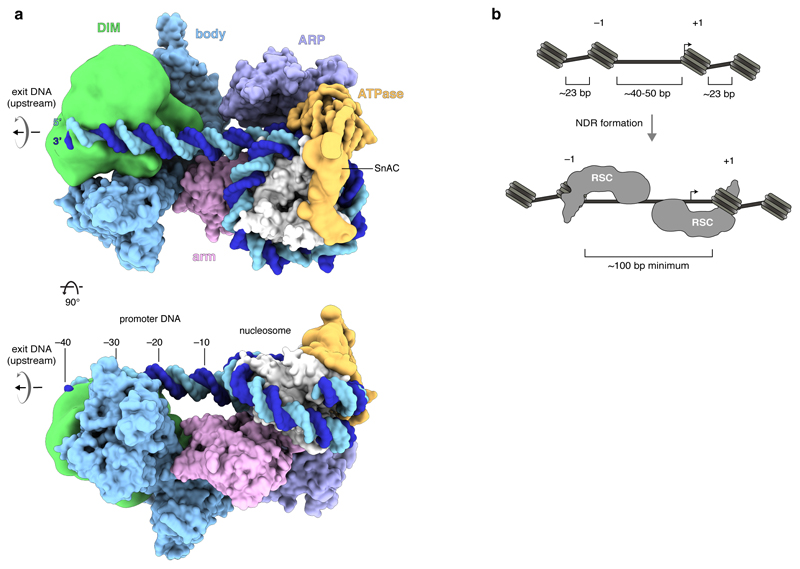
We isolated endogenous RSC from the yeast Saccharomyces cerevisiae by affinity purification of the tagged subunit Rsc2 (Extended Data Figure 1a) (Methods). A RSC-nucleosome complex was assembled in the presence of the ATPase transition state analogue ADP-BeF3 (Extended Data Figure 1b). Cryo-EM analysis resulted in a reconstruction at ~15 Å resolution that revealed the nucleosome, four turns of DNA exiting from one side of the nucleosome, and five RSC modules that we refer to as ATPase, ARP, body, arm, and DNA-interaction module (DIM) (Figure 1a; Extended Data Figure 2). Focussed 3D classification enabled modelling of the nucleosome, the associated ATPase, and the ARP module (Methods). We also subjected the free RSC complex to cryo-EM analysis. This resulted in structures of the body and arm modules at resolutions of 3.6 Å and 3.8 Å, respectively, which we could fit into the density of the RSC-nucleosome complex (Extended Data Figures 3, 4a-c). This led to a structural model of the RSC-nucleosome complex that only lacks the DIM module and agrees with lysine-lysine crosslinking information (Extended Data Figures 1c, d, Supplementary Tables 1, 2).
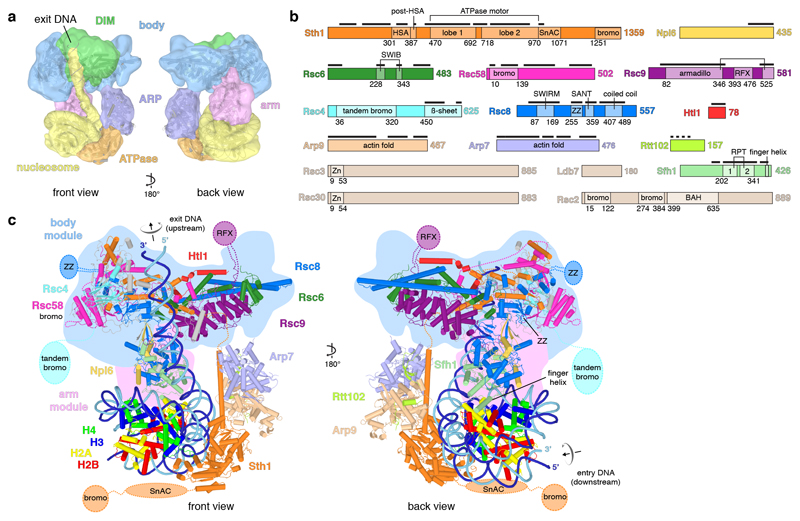
Figure 1. RSC-nucleosome complex structure.
a. Two views of the low pass-filtered cryo-EM density reveal the overall architecture. The five RSC modules are in different colours. The nucleosome with exit DNA is in yellow. DIM, DNA-interaction module.
b. RSC subunit domain architecture. Domain boundaries marked with residue numbers. Black bars indicate modelled regions. HSA, helicase-SANT-associated; SnAC, Snf2 ATP coupling; bromo, bromodomain; armadillo, armadillo repeat fold; RFX, DNA-binding RFX-type winged-helix; SWIRM, Swi3 Rsc8 Moira; ZZ, ZZ-type zinc finger; SANT, Swi3 Ada N-Cor TFIIIB; Zn, Zn(2)-C6 fungal-type zinc finger; RPT, repeat; BAH, bromo-adjacent homology.
c. Cartoon representation. Unassigned elements in grey. Mobile domains depicted schematically. Arrows indicate directionality of DNA translocation.
RSC structure
The structure reveals the intricate architecture of RSC (Figure 1, Supplementary Video 1). The body module contains subunits Rsc4, Rsc6, Rsc8, Rsc9, Rsc58, Htl1, and the N-terminal region of Sth1 (Extended Data Figure 5a, Extended Data Table 1). The ARP module is flexibly tethered to the body and comprises the helicase-SANT associated (HSA) region of Sth1, the actin-related proteins Arp7 and Arp9, and subunit Rtt102. The C-terminal region of Sth1 extends from the HSA region and forms the ATPase module (Extended Data Figure 6a). The arm module protrudes from the body and contains subunit Sfh1 and parts of Rsc8, Npl6, and Rsc9 (Extended Data Figure 5a). The arm and body modules are tightly connected by two copies of Rsc8 that adopt different structures (Extended Data Figure 6b). The N-terminal SWIRM domains of the two Rsc8 copies reside in the arm, whereas the SANT domains and one of the ZZ zinc finger domains reside in the body, as do the long C-terminal helices.
RSC also contains six domains that are implicated in interactions with histone tails. The N-terminal bromodomain in Rsc58 locates to the surface of the body (Extended Data Figure 6c). The five other domains are mobile, and include a bromodomain in Sth1, two bromodomains in Rsc2, a BAH domain in Rsc2 that binds histone H314, and a tandem bromodomain in Rsc4 that interacts with acetylated H3 tails, in particular acetylated lysine K1415,16. RSC also contains five putative DNA-binding domains, of which four are mobile. These include the zinc finger domains in subunits Rsc3 and Rsc30, an RFX domain in subunit Rsc9, and a ZZ finger domain in one of the two Rsc8 subunits. In summary, RSC consists of five modules and nine flexibly connected domains, of which some are involved in substrate selection.
ATPase and translocation
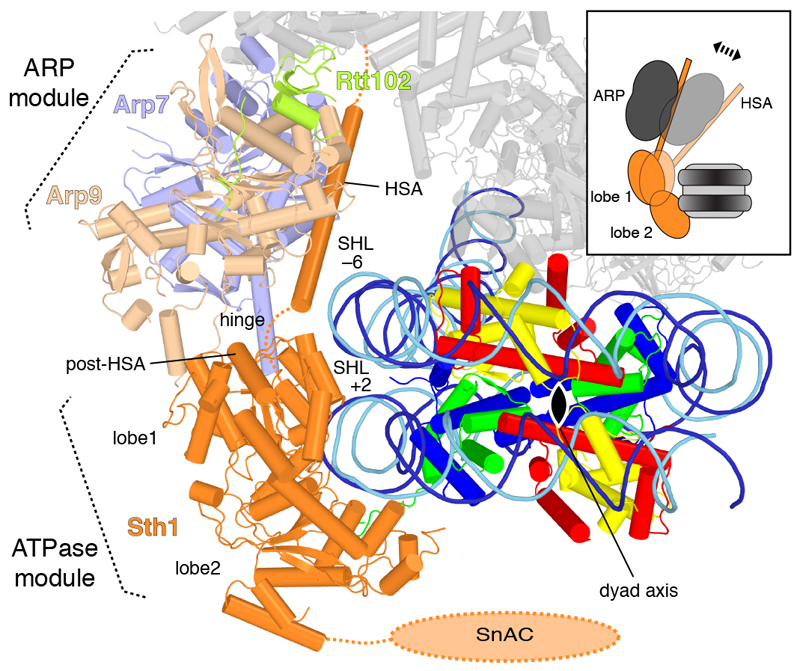
RSC engages in multivalent interactions with its substrate, contacting both DNA and histones (Figure 1). The ATPase and arm modules interact with the nucleosome, whereas the DIM module engages with DNA exiting from the nucleosome. The ATPase module binds the edge of the nucleosome, contacting both DNA gyres in a conformation poised for translocation activity (Extended Data Figure 6d). The two lobes of the ATPase motor domain contact one gyre at superhelical location (SHL) +2 and adopt the same relative orientation as in the structure of the related Snf2 ATPase bound to a nucleosome17. The N-terminal ATPase lobe 1 also binds the second DNA gyre around SHL –6 (Figure 2), a location where the N-terminal tail of histone H3 protrudes (Extended Data Figure 4d). Considering the known directionality of the translocase18, we arrive at the model that the RSC ATPase motor pumps DNA towards the nucleosome dyad and along the octamer surface in the exit direction, which corresponds to the upstream direction of transcription, thus liberating more promoter DNA.
Figure 2. RSC ATPase and ARP modules.
View along the nucleosome dyad (black oval). View as in Fig. 1c, right, but rotated by 45° around a horizontal axis.
The ARP module couples RSC ATPase activity to DNA translocation and regulates the remodelling activity5,19. Our results suggest that this regulation involves changes in the position of the mobile ARP module that influence the conformation and mobility of the ATPase lobe 1 and its interactions with both DNA gyres (Figure 2). These changes are likely transmitted through the hinge region between the HSA region and lobe 1 that includes the ‘post-HSA’ region of Sth1. Mutations of the post-HSA region increase ATPase activity and DNA translocation, suggesting that the hinge acts as a throttle for the ATPase4,5,20. The ARP module adopts a defined position in the RSC-nucleosome complex, but it is mobile in the free RSC structure. We propose that the position of the mobile ARP module influences the motility of the bilobal ATPase motor and thereby controls RSC translocation activity (Figure 2).
Nucleosome sandwiching and sliding
The structure also shows that RSC contacts the nucleosome disc not only at the edge, but also binds both of its faces, effectively sandwiching the histone octamer. The SnAC domain in subunit Sth1 binds the outer face of the histone octamer, whereas the arm module binds the inner face (Figure 3a). The SnAC domain in the SWI/SNF homologue Snf2 is important for remodelling in vivo and biochemical data suggested that it acts as a histone anchor that is required for nucleosome sliding21. The strength of the SnAC-histone octamer contact may be influenced by the N-terminal tail of histone H4, which binds at the interface of the SnAC and ATPase motor of Sth1 (Figure 3a). Since histone acetylation can impair octamer transfer by RSC to the histone chaperone Nap122, histone acetylation may modulate the sandwiching contacts, maybe influencing nucleosome eviction or sliding.
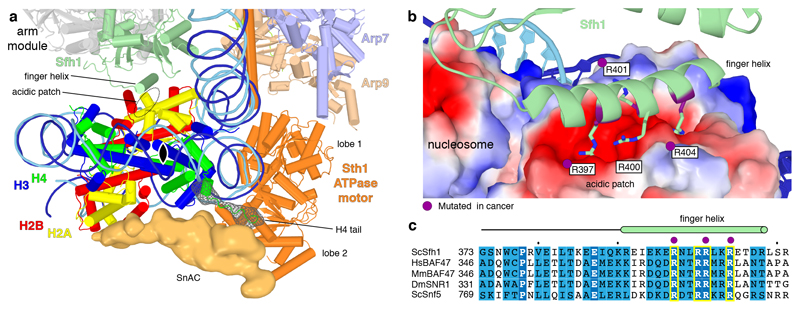
Figure 3. RSC sandwiches the nucleosome.
a. RSC-nucleosome interactions viewed along the nucleosome dyad (black oval). On the outer face of the histone octamer, densities for the Sth1 SnAC domain and the histone H4 tail are shown as an orange surface and a green mesh, respectively. On the inner face, the arm module and Sfh1 finger helix are depicted.
b. Interaction of the Sfh1 finger helix with the acidic patch of the inner face of the histone octamer (surface representation coloured by electrostatic charge; red, negative; blue, positive). Conserved arginine residues are depicted. Residues mutated in cancer (Methods) highlighted in purple.
c. Sequence alignment of the finger helix region (green cylinder) in S. cerevisiae’s (Sc) Sfh1 with its homologs H. sapiens (Hs) BAF47, M. musculus (Mm) BAF47, D. melanogaster (Dm) SNR1 and ScSnf5. Invariant and conserved residues highlighted in dark and light blue, respectively. Yellow boxes contain arginine residues shown in (b). Purple dots mark residues mutated in cancer (Methods).
Binding of the arm module to the inner face of the histone octamer is mediated by an exposed ‘finger’ helix, which resides in the C-terminal region of subunit Sfh1 (Figure 3a, Extended Data Figures 1d, 4e). The finger helix contains four arginine residues (R397, R400, R401 and R404) that contact the acidic patch of the octamer. Three of these arginines are known to be mutated in human cancers (Figure 3b), pointing to the functional significance of the finger helix-acidic patch interaction. The finger helix and its arginine residues are highly conserved in Sfh1 homologs throughout eukaryotes (Figure 3c). The SnAC domain is also conserved over species and between SWI/SNF complexes21, suggesting that the sandwiching interactions are formed by all SWI/SNF family complexes.
Nucleosome and DNA recognition
The arm module and its finger helix may also contribute to substrate selection. RSC preferentially recognizes nucleosomes that contain the histone variant H2A.Z23. Such nucleosomes show a more extended acidic patch24 and may have increased affinity for the basic RSC finger. The arm module may also contact the unique C-terminal tail of H2A.Z that protrudes near Sfh1 (Extended Data Figure 4d). The observed arm-octamer interaction may also explain why ubiquitination of histone H2B counteracts RSC function25. The ubiquitin moiety is linked to H2B residue K123 (human K120) and, although flexible, can adopt a position that sterically interferes with the arm-octamer interaction (Extended Data Figure 4f).
RSC not only binds the nucleosome, but also DNA that exits from it (Extended Data Figure 7a). The DIM module contacts exiting DNA ~20–40 bp upstream of SHL –7 of the nucleosome. This is in agreement with RSC protecting ~50 bp of extra-nucleosomal DNA from nuclease digestion26. The DIM-DNA contact also explains how RSC recognizes specific DNA elements that are enriched in promoters8,10–12. According to our cryo-EM and crosslinking results, the DIM contains parts of RSC subunits Rsc2, Rsc3 and Rsc30 (Extended Data Figure 1d). Rsc3 and Rsc30 recognize a CGCG DNA element located upstream of the transcription start site, probably via their zinc cluster domains10.
NDR formation
The results further elucidate the formation of NDRs. In S. cerevisiae, the DNA linker length between two nucleosomes is only ~23 bp on average27. Steric considerations predict that RSC can bind to DNA as observed in our structure only at sites in chromatin where the length of the DNA linking two nucleosomes is at least 40–50 bp (Extended Data Figure 7b). This may explain why RSC is targeted preferentially to promoter regions, which are intrinsically nucleosome-depleted, and also why two closely spaced nucleosomes impair binding of the RSC-related complex SWI/SNF28.
NDR formation involves sliding of both flanking nucleosomes away from the NDR center6. The RSC-nucleosome structure can describe RSC action on both, the +1 and the –1 nucleosome. In the latter case, DNA exits in downstream direction, rather than upstream, the ATPase engages with SHL –2, rather than SHL +2, and DNA translocation slides the nucleosome upstream, rather than downstream. Provided that RSC remains bound to both flanking nucleosomes after remodelling, a minimum NDR size of ~100 bp would result (Extended Data Figure 7b). However, larger NDRs can be formed when RSC evicts a nucleosome8.
SWI/SNF family and cancer
The RSC structure is a good model for its human counterpart PBAF. PBAF contains subunits homologous to Sth1, Rsc6, Rsc8, Sfh1, Arp7 and Arp9, and putative counterparts of subunits Rsc2, Rsc4, and Rsc9 (Extended Data Table 1). Projection of the homologous regions onto the RSC structure reveals that the ATPase, ARP and arm modules and a large part of the body are conserved in PBAF (Extended Data Figure 5b). Although PBAF apparently lacks the DIM, it contains 12 putative DNA-binding domains that may mediate DNA recognition (Extended Data Table 1). Due to these similarities, the RSC structure can be used to locate protein sites in PBAF that are mutated in human cancers (Extended Data Figure 5b). Most mapped mutations are predicted to destabilize protein folds. However, mutations are particularly enriched within the ATPase, ARP and arm modules that surround and contact the nucleosome, suggesting that they cause functional defects. Sequence and domain conservation further suggest that the architecture of the RSC-related yeast SWI/SNF complex and its human counterpart BAF are also similar (Extended Data Table 1, Extended Data Figure 8). Comparison of our structure with nucleosome complex structures of other families of chromatin remodelers reveals strong differences (Extended Data Figure 5c).
When our manuscript was under review, three related structures were reported, two structures of RSC-nucleosome complexes29,30 and a structure of the yeast SWI/SNF-nucleosome complex31. Together, these structures provide a basis for understanding the multiple functions of SWI/SNF family remodelers in chromatin biology.
Methods
Preparation of RSC complex
The yeast Saccharomyces cerevisiae contains two isoforms of RSC that comprise either the subunit Rsc1 or its homologue Rsc232 (Extended Data Table 1). We isolated the Rsc2-containing 16 subunit isoform. The RSC2-TAP-HIS3 yeast strain (YSC1177-YLR357W) was purchased from the Dharmacon TAP-tagged open reading frame (ORF) library. A colony from a YPD agar plate was used to prepare a 2 L pre-culture in YPD medium with 50 µg/mL ampicillin sodium salt and 12.5 µg/mL tetracyclin-hydrochloride with OD600 of 1.6. Cells were fermented from OD600 ~0.006 to OD600 ~10 in 250 L of 3% YEP broth (w/v, Formedium) supplemented with 2% glucose, 50 g/L ampicillin sodium salt and 12.5 g/L tetracyclin-hydrochloride. The pellet was resuspended in cold 2x lysis buffer (100 mM HEPES pH 7.6, 20% glycerol (v/v), 1.4 M KAc, 2 mM MgCl2, 2 mM DTT and 3x protease inhibitor (100x: 0.028 mg/mL leupeptin, 0.137 mg/mL pepstatin A, 17 mg/mL PMSF, 33 mg/mL benzamidine)), frozen in liquid nitrogen to pea-sized granules and stored at –80 °C.
RSC was purified based on the TAP-tag purification strategy4,33,34, with several modifications. All purification procedures were performed at 4 °C unless stated otherwise. 600 g yeast granules were lysed by cryo-milling (Spex Freezer/Mill 6875D) and stored at –80 °C. Yeast powder was thawed at 30 °C, diluted with 100 mL 1x lysis buffer and cleared by centrifugation (25,200 xg). The supernatant was incubated for 6 h with 10 mL IgG Sepharose 6 Fast Flow resin (GE Healthcare) pre-equilibrated in lysis buffer. The resin was recovered by centrifugation (3,200 xg) and washed with 100 mL elution buffer A (50 mM K-HEPES, pH 7.6, 150 mM KAc, 10% glycerol (v/v), 3 mM CaCl2, 1 mM imidazole, 1 mM DTT, 0.5x protease inhibitor). IgG resin was resuspended in 10 mL elution buffer A, mixed with 2 mL calmodulin resin (Agilent Technologies) pre-equilibrated in elution buffer A, and supplied with catalytic amounts of TEV protease. The resin was washed with 100 mL elution buffer A without protease inhibitors, and protein was eluted with 50 mL elution buffer B (3 mM EGTA instead of 3 mM CaCl2). Elution was applied to a HiTrap Q 1 mL HP column (GE Healthcare) pre-equilibrated with Q-150 buffer (50 mM HEPES pH 7.6, 150 mM KAc, 10% glycerol, 1 mM DTT) and washed with 10 CV Q-150 buffer. Protein was eluted with a linear gradient from 0 – 100 % buffer Q-1500 (1.5 M KAc instead of 150 mM KAc) over 50 CV. RSC-containing fractions were concentrated, dialysed overnight to 50 mM HEPES pH 7.6, 150 mM KAc, 10% glycerol, 5 mM MgCl2,1 mM DTT, and immediately used for cryo-EM sample preparation. Typical yields were 0.2 – 0.3 mg from 600 g yeast granules.
Preparation of nucleosome substrates
Xenopus leavis histones were expressed and purified as described35,36. Briefly, histones were expressed recombinantly in E. coli cells and purified as inclusion bodies using a Dounce tissue grinder (Sigma-Aldrich). Histones were aliquoted, flash-frozen, lyophilised, and stored at –80 °C. For octamer preparation, lyophilised histones were resuspended in unfolding buffer (20 mM HEPES pH 7.5, 7 M guanidinium hydrochloride, 10 mM DTT) to a concentration of 3 mg/mL. Histones H2A, H2B, H3 and H4 were combined at a molar ratio of 1.2:1.2:1:1 and dialysed against two times 2 L of refolding buffer (10 mM HEPES pH 7.5, 2 M NaCl, 1 mM EDTA, 2.5 mM DTT) for a total of 12 h at 4 °C. The sample was concentrated and applied to a Superdex 200 Increase 10/300 size exclusion column pre-equilibrated with refolding buffer. Peak fractions were pooled and frozen in liquid nitrogen at a concentration of 1.34 mg/mL.
DNA fragments for nucleosome reconstruction were prepared by PCR as described37. gBlock DNA (IDT) containing the 145-bp Widom 601 sequence38 with a 55 bp extension at the 5’-end and a 37 bp extension at the 3’-end was used as a template together with two primers (forward: TCATTACCCAGCCCGCCTAG, reverse: CCTACGGACCGGATATCTTCCCTG). Reactions were pooled (42 mL) and DNA products recovered by phenol-chloroform-extraction. DNA was resuspended in MilliQ water and applied to a Superose 6 Increase 10/300 size exclusion chromatography column pre-equilibrated in gel filtration buffer (20 mM HEPES pH 7.5, 200 mM NaCl, 1 mM EDTA). Peak fractions were pooled, concentrated ten times, and stored at –20 °C.
Nucleosome reconstitution was performed as described36, with minor modifications. DNA and histone octamers were mixed at a 1:1.2 molar ratio in reconstitution buffer (20 mM HEPES pH 7.5, 1 mM EDTA, 2 mM DTT) containing 2 M NaCl and incubated for 30 min on ice. Sample was transferred to a Slide-A-Lyzer 3.5K MWCO MINI device and gradient-dialysed from 500 ml high salt reconstitution buffer against 2 L of low salt reconstitution buffer (20 mM NaCl) for 22 h. After a heat shift for 30 min at 50 °C, the sample was recovered and immediately used for complex formation.
RSC-nucleosome complex formation
Newly prepared RSC complex was mixed with ADP-BeF3 at a final concentration of 1 mM and incubated on ice for 30 min. A 1.6-fold molar excess of the nucleosome substrate was added, the mixture incubated for 15 min at 30 °C and transferred back on ice. RSC-nucleosome complex was cross-linked using the GraFix method39. The sample was applied to a gradient generated from a 10% sucrose light solution (10% sucrose (w/v), 50 mM HEPES pH 7.6, 150 mM KAc, 5% glycerol (v/v), 2 mM MgCl2, 1 mM DTT, 0.5 mM ADP-BeF3) and a 25% sucrose heavy solution (25% sucrose (w/v) instead of 10%) containing 0.2% glutaraldehyde crosslinker with a BioComp Gradient Master 108 (BioComp Instruments). Centrifugation was carried out for 16 h at 32,000 rpm in a SW 60 Ti swinging-bucket rotor (Beckmann) at 4 °C. 200 µL fractions were collected and quenched with aspartate (pH 7.5) at a final concentration of 50 mM. Fractions containing RSC-nucleosome complex were dialysed for 8 h at 4 °C to 20 mM HEPES pH 7.6, 150 mM KAc, 1% glycerol (v/v), 3 mM MgCl2, 1 mM DTT and applied to cryo-EM grids.
Cryo-EM analysis of RSC-nucleosome complex
RSC-nucleosome complex was absorbed to a thin carbon film before plunge freezing as described40, with minor modifications. A small, thin (~3.1 nm) carbon film was floated from the mica sheet onto a 50 µL drop of sample and incubated for 2 – 3 min. The carbon film was recovered with copper R2/1 or R3.5/1 grids (Quantifoil) and vitrified by plunge-freezing in liquid ethane using a Vitrobot Mark IV (FEI) operated at 4 °C and 100% humidity.
Electron micrographs were acquired on an FEI Titan Krios G2 transmission election microscope operated at 300 keV in EFTEM mode, equipped with a Quantum LS 967 energy filter (Gatan), zero loss mode, 30 eV slit width, and a K2 Summit direct electron detector (Gatan) in counting mode. Automated data acquisition was done using the FEI EPU software package at a nominal magnification of 130,000x, resulting in a calibrated pixel size of 1.05 Å/px. Micrographs for the two datasets were collected at a dose rate of 4.78 e–/Å2/s over 10 s resulting in a total dose of 47.8 e–/Å2, and at a dose rate of 5.67 e–/Å2/s over 8 s resulting in a total dose of 45.4 e–/Å2, respectively. Both datasets were dose fractionated over 40 frames.
Dose weighting, CTF estimation and motion correction were carried out during data collection using Warp41. Automated particle picking by Warp resulted in 112,657 particles from the first dataset (4404 micrographs) and 1,119,875 particles from the second dataset (19,415 micrographs). Particle coordinates were exported, combined, extracted and processed using RELION 3.042. Removal of bad particles through global 3D classifications with a negative stain reconstruction of the RSC complex as reference resulted in high-quality particles that could be refined to an overall map of the RSC remodeler together with the nucleosome (map 1) at a resolution of ~15 Å (Extended Data Figure 2). The reported resolution value is based on visual inspection and comparison with lowpass-filtered high resolution structures (e.g. the nucleosome) because the software-generated values were overestimated for the density in this resolution range. Further processing of the particles revealed great flexibility and dynamics which could not be resolved by focused 3D classifications and refinements.
The particles corresponding to the RSC-nucleosome map were re-extracted centred on the nucleosome with a box mainly including the nucleosome and the Sth1 ATPase module. Global 3D classification resulted in a good class that revealed the Sth1 ATPase subunit bound to the nucleosome. Focused 3D refinement and postprocessing with automatic B-factor determination in RELION excluding the ATPase density provided a nucleosome map (map 2) at a resolution of 3.6 Å (gold-standard Fourier shell correlation 0.143 criterion) and a B-factor of –155 Å2 (Extended Data Figure 2). Improvement of the ATPase density turned out to be very difficult and showed its highly dynamic nature in this sample. A strategy of focused 3D classification without image alignment on the ATPase part, followed by a global 3D refinement and additional focused 3D classification on the ATPase-nucleosome density led to the best results. A focused 3D classification and postprocessing with automatic B-factor determination in RELION resulted in an overall resolution of the ATPase-nucleosome map (map 3) of 4.3 Å (FSC 0.143 criterion) and B-factor of –186 Å2 (Extended Data Figure 2). Final focused maps were combined using the Frankenmap tool distributed with Warp (map 7) (Extended Data Figure 2). Masks encompassing the regions of interest were created with UCSF Chimera43 and RELION.
Cryo-EM analysis of the free RSC complex
Freshly purified RSC complex was mixed with ADP-BeF3 to a final concentration of 1 mM and incubated for 15 min on ice. BS3 (bis(sulfosuccinimidyl(suberate))) cross-linker (Thermo Fischer Scientific) was added to a final concentration of 1 mM, incubated on ice for 30 min before quenching with Tris-HCl, pH 7.5, and ammonium bicarbonate at a final concentration of 100 mM and 20 mM, respectively. After size exclusion chromatography using a Sepharose 6 Increase 3.2/300 column (GE Healthcare) pre-equilibrated in gel filtration buffer (50 mM HEPES pH 7.6, 150 mM KAc, 4 mM MgCl2, 1 mM DTT), peak fractions were immediately applied to cryo-EM grids. 4 µL of sample were applied to glow-discharged (Pelco easiGlow) R2/2 gold grids (Quantifoil). Grids were blotted and vitrified as described above.
Cryo-EM data was collected as described above, with small modifications. The energy filter slit width was set to 20 eV. Micrographs for the two 0° tilt datasets were collected at a dose rate of 4.88 e–/Å/s for 8 s resulting in a total dose of 39 e–/Å2 and at a dose rate of 5.02 e–/Å2/s over 9 s resulting in a total dose of 45.2 e–/Å2, respectively, and fractionated over 40 frames. The third, 25° tilted dataset was acquired in 44 frames at a dose rate of 4.99 e–/Å2/s for 11 s resulting in a total dose of 54.9 e–/Å2.
Pre-processing and particle picking were carried out as described above and resulted in 363,824 particles from the first dataset (3284 micrographs), 170,028 particles from the second dataset (1216 micrographs) and 475,168 particles from the tilted dataset (3158 micrographs). Particles were processed with global 3D classifications using RELION 342 and a negative stain reconstruction of the RSC complex as a first reference to obtain an improved initial reference. All 1,009,020 particles were newly extracted and bad particles were sorted out in multiple rounds of global 3D classifications in combination with global 3D refinements. The best resulting class was refined with a mask excluding the flexible DNA-interaction module (DIM). Particles corresponding to this reconstruction were subjected to CTF refinement and Bayesian polishing in RELION. Using focused 3D refinements, the maps for the arm module (map 4), and body1 (map 5) and body2 (map 6) submodules were further improved. Postprocessing with automatic B-factor determination in RELION resulted in overall resolutions of 3.8 Å, 3.6 Å and 3.6 Å, respectively, and B-factors of –136 Å2, –103 Å2 and –100 Å2, respectively (Extended Data Figure 3). Final focused maps were combined with Warp (map 8). Postprocessing with automatic B-factor determination in RELION resulted in overall resolutions of 3.6 Å and a B-factor of –106 Å2 (Extended Data Figure 3).
Structural modelling
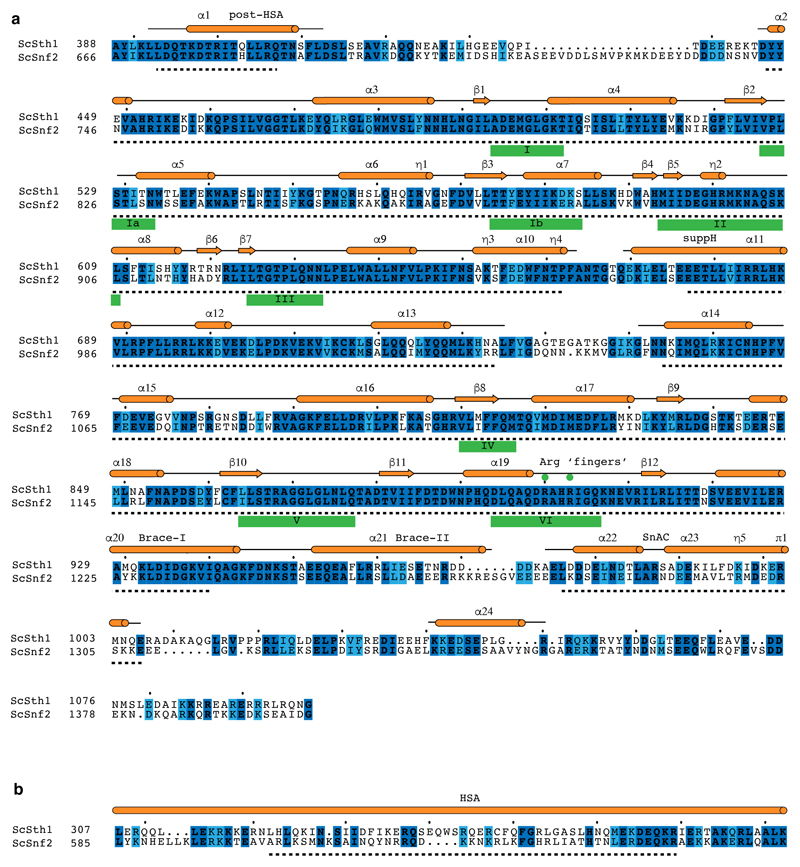
Previous electron microscopy studies revealed the overall shape of RSC, but did not allow for any molecular modeling44–46. To enable molecular modelling based on our data, we first used the combined cryo-EM map 7 aligned to map 1 for model building of the Sth1 ATPase domain bound to the nucleosome. The final map 7 was created with the local resolution tool from RELION and a B-factor of –150 Å2. The structure of the yeast Snf2 bound to the nucleosome in the ADP-BeF3 state (PDB code 5Z3U)17 was used as basis for modelling. Published data together with the close homology between Sth1 and Snf2 (Extended Data Figure 8) suggest that Sth1 also binds at SHL +2. The remodeler and the nucleosome part were fitted separately. The Xenopus laevis histones and Widom 601 sequence of PDB 5Z3U were the same as used in our study. The nucleosome structure was rigid-body fitted into our cryo-EM map in UCSF Chimera43 and the entry side DNA and histone tails trimmed according to the density in COOT47. DNA sequence information was not resolved and assumed to match the 5Z3U template. Due to lower resolution, amino acid side chains of residues 15 – 22 of H4 (chain B) were stubbed in COOT. The nucleosome structure was flexibly fitted using Namdinator48 and real space refined in PHENIX49 with secondary structure restraints (including base paring and base stacking restraints).
High conservation of amino acids between Sth1 and Snf2 ATPase domains (Extended Data Figure 8) allowed for generation of a Sth1 homology model with Rosetta50,51. The homology model was trimmed according to the density in COOT, Brace-II helix was removed, and amino acid side chains were stubbed owing to the lower resolution of the map area before rigid-body docking using UCSF Chimera. Additional real space refinement with secondary structure restraints (including base paring and base stacking restraints) was performed in PHENIX. Density for the Sth1 SnAC domain was clearly observed at lower contour level. However, the local resolution was not sufficient to build a model for the SnAC structure or to further investigate its interactions with the nucleosome. The overhanging exit side DNA was modelled by generating a bend B-DNA following the density in map 1 using 3D-DART52. The DNA duplex was connected to the nucleosomal Widom 601 DNA and geometry optimized with base pairing and base stacking restraints in PHENIX.
Map 1 allowed for the rigid-body docking of the crystal structure of the ARP module bound to the Snf2 HSA region (PDB code 4I6M)19 using UCSF Chimera. The amino acid residues of the Snf2 HSA helix were mutated to the ones from Sth1 according to sequence alignment (Extended Data Figure 8) starting at the C-terminus, ignoring gaps, and stubbed. The model for the Sth1 HSA helix is thus an extrapolation based on the strong α-helical secondary structure prediction and the register might differ slightly19. We next aligned the focused refined maps 4 – 6 and the combined map 8 to map 1 and used them for model building of the RSC core. SWISS-MODEL53,54 was used to generate homology models for the Rsc58 N-terminal bromodomain (PDB code 3LJW)55, the Rsc6 SWIB domain (PDB code 1UHR), the Rsc8 SWIRM (PDB code 2FQ3)56, SANT (PDB code 2YUS) and ZZ zinc finger domains (PDB code 1TOT)57, the Rsc9 armadillo-like domain (PDB code 4V3Q)58 and the Sfh1 RPT1 and RPT2 domains (PDB code 6AX5). The homology models were rigid-body placed using UCSF Chimera43 and manually adjusted and re-build in COOT47.
The quality of the maps allowed for de novo building of the other model parts (Supplementary Table 1). Modelling was guided and validated by BS3 cross-linking data visualized with xVis59 and secondary structure predictions performed with Quick2D60 and PSIPRED61,62. Amino acid residues connecting the domains of the two Rsc8 subunits could not be modelled. For clarification, they were placed into a single chain (chain L) clustered by proximity. The Sfh1 C-terminal finger helix was built into the density of map 7. A poly-alanine model was placed into density that could not be assigned to any RSC subunit (chain X). Bulky amino acid side chain density in the maps 4 – 8 enabled us to assign the sequence registers, however in some regions register shifts cannot be entirely excluded. The modelled RSC subunits Rsc4, Rsc58, Rsc6, Rsc8, Rsc9, Npl6, Htl1, Sfh1 and Sth1 (residues 48 – 297) together with the poly-alanine chain were applied to several rounds of real space refinement and geometry optimisation using PHENIX49, and flexible fitting with Namdinator48 against the combined map 8. MolProbity63 was used to flip and optimise Asn, Gln and His side chains. The C-terminal finger helix of Sfh1 was real space refined with PHENIX against map 7. The final structure displayed excellent stereochemistry as shown by MolProbity (Supplementary Table 2, map7 (ATPase + nucleosome), map8 (RSC core)). The slightly worse model statistics of the full structure arise from outliers in the ARP module crystal structure that was rigid-body docked due to insufficient density information. Figures were created using PyMol64,UCSF Chimera43 and UCSF ChimeraX65. The angular distribution plots were generated using the AngularDistribution tool distributed with Warp41.
The RSC structure and density assignments are consistent with a large amount of biochemical and genetic data, including the known requirement of the Rsc4 C-terminal region for cell growth15, the interaction between Rsc6 and Rsc866, the lethality of Rsc58 truncation67, and the known interaction between Rsc3 and Rsc3068. Sites of missense mutations found in human cancers were derived from the cBio cancer genomics portal (cBioPortal)69,70 and mapped onto the RSC structure for residues that are identical in its human counterpart PBAF using MSAProbs60,71. MSAPobs and Aline72 were used to map conservation between RSC and PBAF.
Preparation of crosslinking samples for mass spectrometry
RSC-nucleosome complex was prepared as described above. The crosslinking reaction was performed with BS3 (bis(sulfosuccinimidyl(suberate))) crosslinker (Thermo Fischer Scientific) at a final concentration of 1 mM on ice for 30 min before quenching with Tris-HCl, pH 7.5, and ammonium bicarbonate at a final concentration of 100 mM and 20 mM, respectively. The crosslinked sample was applied to a 10% – 25% sucrose gradient as described above (no glutaraldehyde in the heavy solution) and protein containing fractions were pooled (~800 µL, ~50 µg complex) and applied to in-solution digest. 150 µL of urea buffer (8 M urea, 50 mM NH4HCO3 pH 8) and 60 µL 0.1 M DTT (in 50 mM NH4HCO3 pH 8) were added to reduce the sample for 30 min at 37 °C, 300 rpm. The sample was alkylated with 60 µL 0.4 M iodoacetamide (in 50 mM NH4HCO3 pH 8) for 30 min at 37 °C, 300 rpm in the dark. The reaction was quenched by addition of 60 µL 0.1 M DTT (in 50 mM NH4HCO3 pH 8). The sample was digested for 30 min at 37 °C with 0.5 µL Pierce Universal Nuclease (250 U/µl) in presence of 1 mM MgCl2. The final sample volume was adjusted to 1200 µL with 50 mM NH4HCO3 pH 8 resulting in a final urea concentration of 1 M. Trypsin digest was performed overnight at 37 °C with 2.5 µg trypsin (Promega, V5111). Tryptic peptides were desalted with C18 spin columns (Harvard Apparatus 74-4601), lyophilized and dissolved in 30% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid. The peptide mixture was separated on a Superdex Peptide 3.2/300 (GE Healthcare) column run at 50 µl/min with 30% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid. Crosslinked species are enriched by size exclusion chromatography based on their higher molecular weight compared to linear peptides. Therefore 50 µL fractions were collected from 1.0 mL post-injection. Fractions from 1.0 – 1.6 mL post-injection were dried in a speed-vac and dissolved in 5% (v/v) acetonitrile, 0.05% (v/v) trifluoroacetic acid and subjected to LC-MS/MS.
LC-MS/MS analysis and crosslink identification
LC-MS/MS analyses were performed on a Q Exactive HF-X hybrid quadrupole-orbitrap mass spectrometer (Thermo Scientific) coupled to a Dionex Ultimate 3000 RSLCnano system. The peptide mixtures from in-solution digest were loaded on a Pepmap 300 C18 column (Thermo Fisher) at a flow rate of 10 µL/min in buffer A (0.1 % (v/v) formic acid) and washed for 3 min with buffer A. The sample was separated on an in-house packed C18 column (30 cm; ReproSil-Pur 120 Å, 1.9 µm, C18-AQ; inner diameter, 75 µm) at a flow rate of 300 nL/min. Sample separation was performed over 120 min using a buffer system consisting of 0.1 % (v/v) formic acid (buffer A) and 80 % (v/v) acetonitrile, 0.08 % (v/v) formic acid (buffer B). The main column was equilibrated with 5 % B, followed by sample application and a wash with 5 % B. Peptides were eluted by a linear gradient from 15 – 48 % B. The gradient was followed by a wash step at 95 % B and re-equilibration at 5 % B. Eluting peptides were analyzed in positive mode using a data-dependent top 30-acquisition methods. MS1 and MS2 resolution were set to 120,000 and 30,000 FWHM, respectively. Precursors selected for MS2 were fragmented using 30 % normalized, higher-energy collision-induced dissociation (HCD) fragmentation. Allowed charge states of selected precursors were +3 to +7. Further MS/MS parameters were set as follows: isolation width, 1.4 m/z; dynamic exclusion, 10 sec; max. injection time (MS1/MS2), 60 ms / 200 ms. The lock mass option (m/z 445.12002) was used for internal calibration. All measurements were performed in duplicates. The .raw files of all replicates were searched by the software pLink 2, version 2.3.173 against a customized protein database containing the expressed proteins and protein-protein crosslinks were filtered with 1 % FDR. Crosslinks appearing less than three times were excluded to increase confidence and plotted using xVis59 and xiNET74.
Extended Data
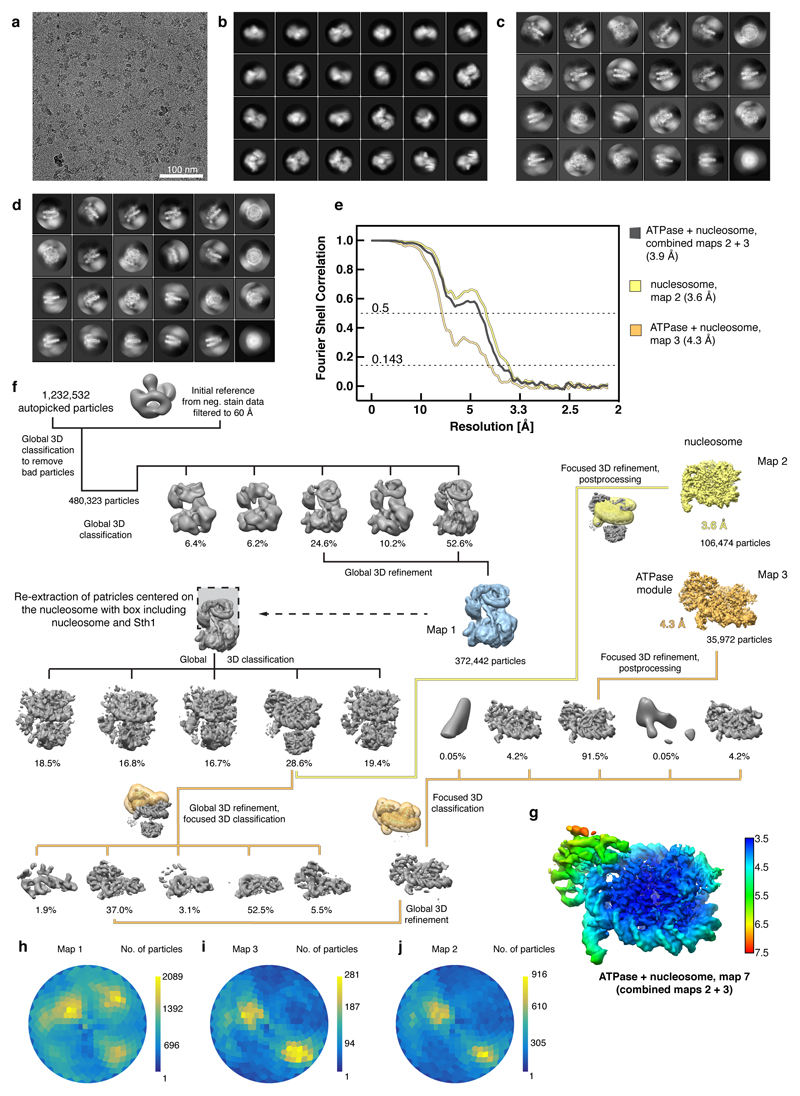
Extended Data Figure 1. Preparation and characterization of RSC-nucleosome complex. Related to Figure 1.
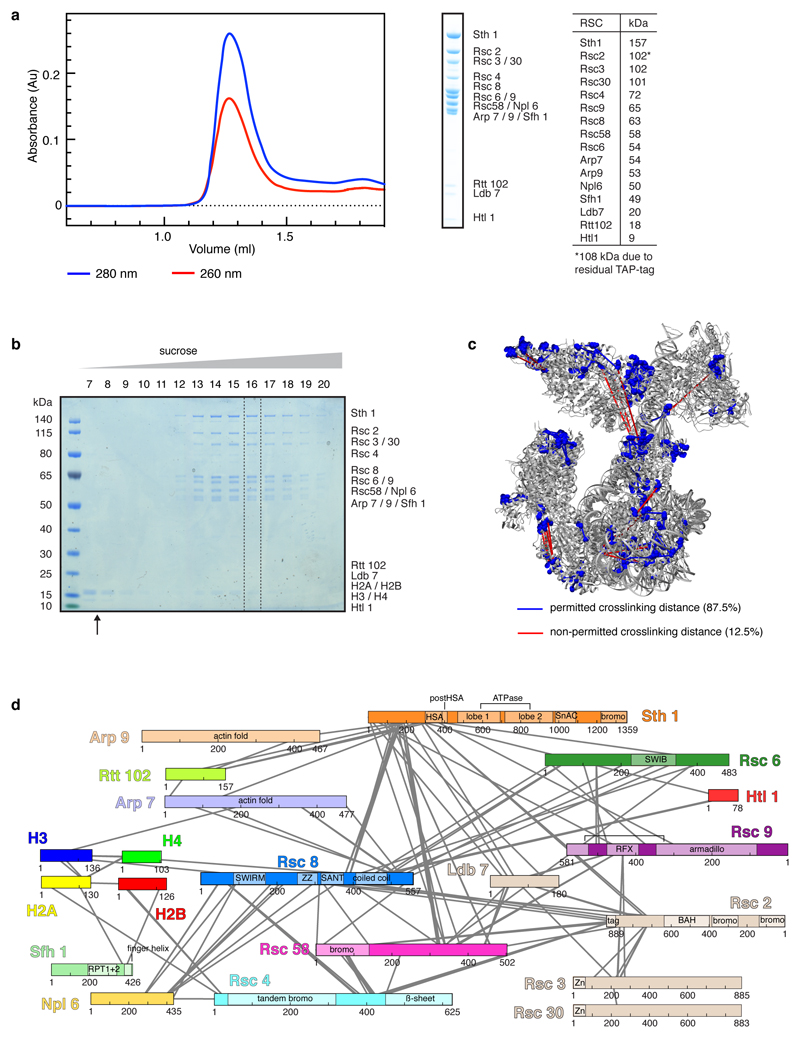
a. Preparation of endogenous Rsc2-containing isoform of the RSC complex from S. cerevisiae. Analysis of purified RSC by size-exclusion chromatography and SDS-PAGE showed high purity and homogeneity with stoichiometric subunits as assessable by Coomassie stain. Subunit identity was confirmed by mass spectrometry. The table shows the expected molecular weights of the RSC subunits. For gel source data, see Supplementary Figure 1.
b. Assembly of the RSC-nucleosome complex. SDS-PAGE analysis of the fractions 7 – 20 of a 10 – 25% sucrose gradient ultracentrifugation. Complex formation was successful as demonstrated by the co-migration of histones with the RSC complex. The unbound over-stoichiometric nucleosomes only migrated to fractions 7 and 8 (black arrow). Fraction 16 in the presence of crosslinker was used for cryo-EM grid preparation (dashed box).
c. Location of crosslinking sites mapped onto the structure. BS3 crosslinks that appeared at least in triplicates were mapped onto the RSC-nucleosome structure. Lysine residues involved in the crosslinking network are shown as blue spheres and crosslinked residues are connected with lines indicating permitted (blue) and non-permitted (red) crosslinking distances. 87.5% of the mapped crosslinks are within the permitted crosslinking distance which was set to 30 Å. The remaining 12.5% of non-permitted crosslinks likely reflect ambiguity caused by the presence of two identical Rsc8 subunits in the structure as well as flexibility of the complex in buffer or arise from technical errors.
d. Crosslinking network between subunits of the RSC-nucleosome complex. Subunits are coloured as in Figure 1. Crosslinks with a score above 2.5 are shown. A comprehensive list of crosslinks can be found in the Supplementary Data 1. Cross-linking mass spectrometry experiments were performed in duplicates with similar results.
Extended Data Figure 2. Cryo-EM analysis of the RSC-nucleosome complex. Related to Figures 1 – 3.
a. Representative cryo-EM micrograph of the RSC-nucleosome complex shows homogeneously distributed individual particles.
b-d. 2D class averages of the RSC-nucleosome complex (b), the ATPase-nucleosome subcomplex (c) and the nucleosome subcomplex (d).
e. Fourier shell correlation plots reveal the overall resolutions of the cryo-EM reconstructions.
f. Cryo-EM processing workflow for the reconstructions of the RSC-nucleosome complex, the ATPase-nucleosome subcomplex, and the nucleosome subcomplex. Particle distribution after 3D classifications is indicated below the corresponding map. The final maps are shown in colours. The masks used for focused classifications and refinements are colour coded corresponding to the final maps they were used for. Views are generally rotated by 180° with respect to Figure 1c, left.
g. Local resolution estimation of the combined ATPase-nucleosome map as implemented in RELION42. We note that the resolution of the peripheral area with the ATPase module is overestimated.
h-j. Angular distribution plot for all particles contributing to the final reconstructions of the RSC-nucleosome complex (h), the ATPase-nucleosome subcomplex (i) and the nucleosome subcomplex (j).
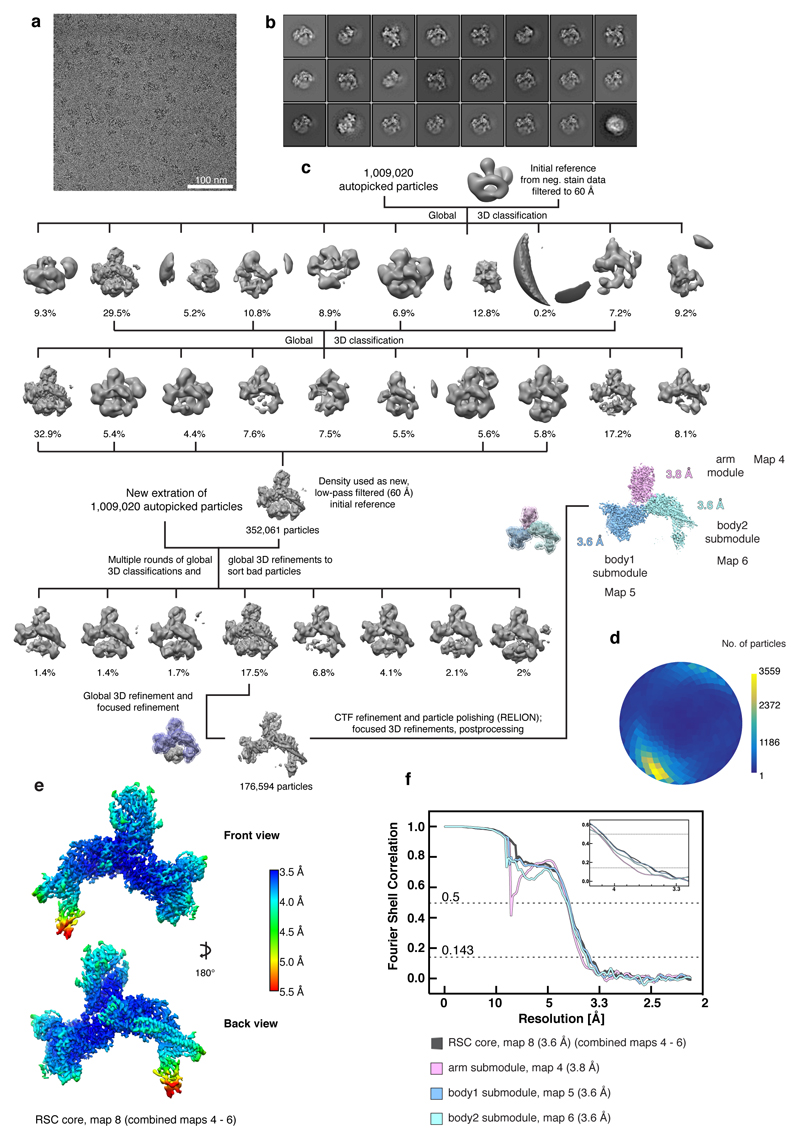
Extended Data Figure 3. Cryo-EM analysis of the free RSC complex. Related to Figures 1 – 3.
a. Representative cryo-EM micrograph of the free RSC complex shows homogeneously spaced individual particles.
b. 2D class averages of the free RSC complex.
c. Cryo-EM processing workflow for the reconstruction of the free RSC complex. Particle distribution after 3D classifications is indicated below the corresponding map. The final maps after focused 3D refinement and masks are depicted in colour. Views are generally rotated by 180° with respect to Figure 1c, right.
d. Angular distribution plot for all particles contributing to the final reconstruction of the free RSC complex.
e. Two views of the combined RSC core map coloured according to the local resolution as implemented in RELION42.
f. Fourier shell correlation plots of the maps used for model building of the RSC core complex.
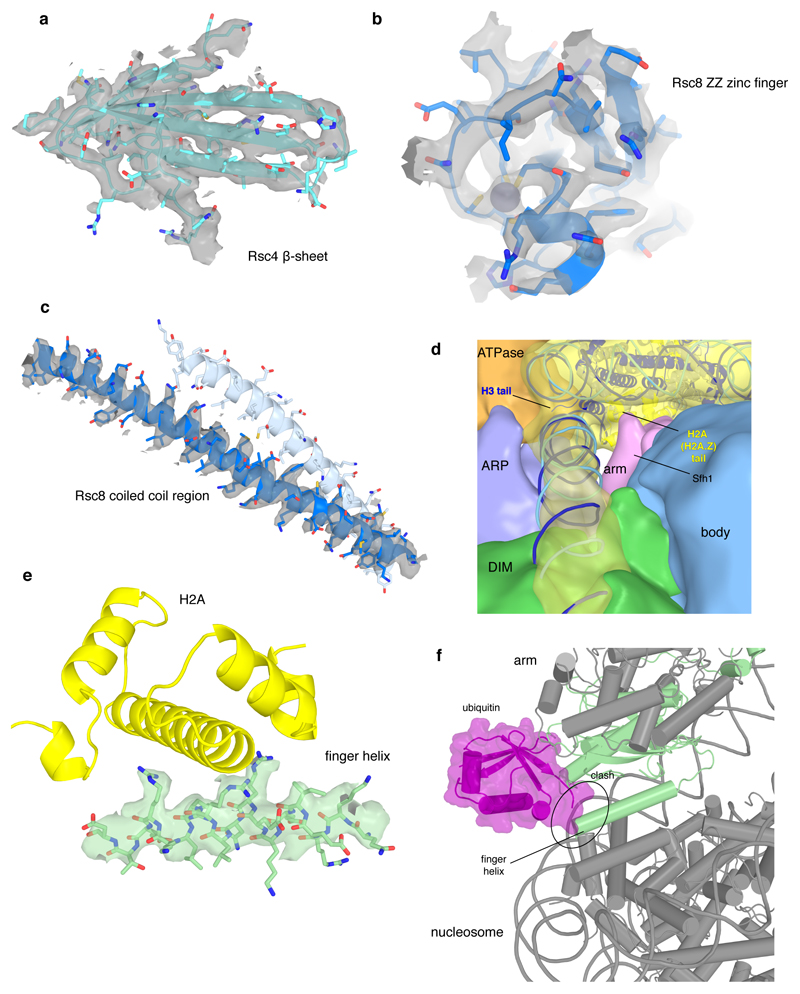
Extended Data Figure 4. Cryo-EM densities for selected RSC regions. Related to Figures 1 – 3.
a-c. Examples of map quality. Close-up of the Rsc4 β-sheet shows clear separation of individual strands (a). The high quality of the map for the ZZ zinc finger of Rsc8 allowed backbone tracing and placement of side chains as well as for the zinc ion (b). Coiled coil helices of the two Rsc8 subunits with density for one helix (c).
d. View along the exit DNA in the direction of the nucleosome showing the low pass-filtered maps for the modules ATPase, ARP, DIM, arm, body, and the nucleosome. At the site where the H2A C-terminal tail protrudes from the nucleosome near Sfh1, low resolution density connecting the arm module and the nucleosome is visible. Density bridging form the ARP module to the exit DNA close to the H3 histone tail can be observed.
e. Density representing the finger helix (green) at the acidic patch of the nucleosome (indicated by H2A in yellow). Side chain density is visible for conserved arginine residues.
f. Interaction of RSC with the nucleosome is sterically impaired by the flexibly bound ubiquitin moiety at H2B lysine 123 (human K120). The Sfh1 finger helix and the ubiquitin moiety (ubiquitylated nucleosome PDB code 6NOG)75 overlap after superposition of nucleosomes.
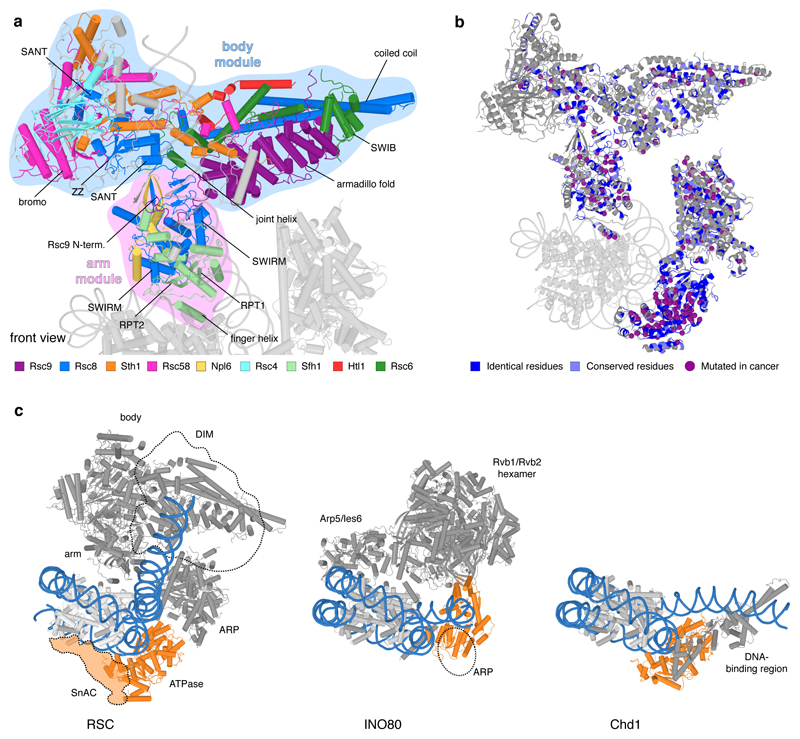
Extended Data Figure 5. Structure of RSC body and arm modules, cancer mutations and remodeler families. Related to Figure 1.
a. Cartoon representation of RSC core viewed as in Figure 1. Important structural elements are labelled.
b. Conservation between SWI/SNF complexes RSC (yeast) and PBAF (human). Residues that are identical (blue) or conserved (light blue) in human PBAF highlighted on the RSC structure (grey). Purple spheres depict identical residues that show missense mutations in various cancers (Methods).
c. Comparison of overall structure of RSC with complexes of INO80 (yeast INO8076) and CHD (yeast CHD177) families. ATPase motor domains are shown in orange, DNA in blue. With regard to the INO80 family, the ATPase of the SWR1 complex also binds SHL +278, whereas the ATPase of the INO80 complex binds SHL –676,79. The ARP module of INO80 contacts exit DNA, which is not the case in RSC. The INO80 complex also contacts both faces of the histone octamer76, resembling the sandwiching interactions made by RSC on a topological level. With respect to the CHD family, the ATPase motor of yeast Chd1 also binds SHL +2, but its DNA-binding region engages with exit DNA near the nucleosome, leading to a different DNA trajectory77,80. With respect to the ISWI family, the ATPase motor binds SHL +281, but other interactions have not been structurally resolved (not shown).
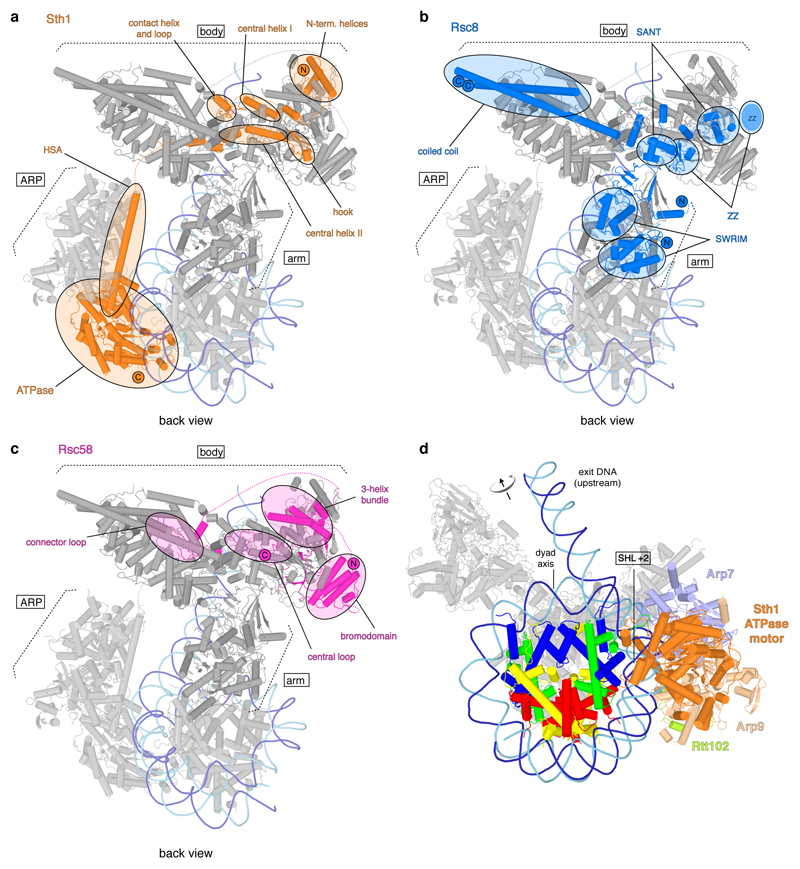
Extended Data Figure 6. Course of polypeptide chains of architectural subunits Sth1, Rsc8, Rsc58 and ATPase-nucleosome interactions. Related to Figure 1.
a. Back view of RSC. The Sth1 subunit of RSC starts with its N-terminus in the body module and tracks through it turning around with a contact helix and loop. Forming the central helix I, the hook and the central helix II it folds back and forth tightly interweaving the body module before it exits with its HSA region through the ARP module to build the ATPase module.
b. RSC with the domains of the two Rsc8 subunits highlighted in blue. Both Rsc8 start N-terminal with their SWIRM domains in the arm module where they support the two repeat domains of Sfh1 in a similar manner. They then follow distinct paths through the arm towards the body module where they contribute with both their SANT and ZZ zinc finger domains. Here the two domains of each subunit form different contacts with various interactions partners and whereas one ZZ zinc finger domain is tightly packed at the body and DNA-interaction module interface, the other seems to extend from the body, presumably as additional interaction surface. Both Rsc8 subunits unite again with their C-terminal long helices in a coiled coil fold on the opposite side of the body module.
c. Rsc58 N-terminal bromodomain attaches to the top of the body module. Then, Rsc58 follows an interwound path through the body module via the central and connector loop. It turns back docking to the body with a 3-helix bundle and stabilizing the module with its C-terminal end.
d. Contacts of Sth1 ATPase motor (orange) with the nucleosome. View as in Fig. 1c, left, but rotated by 45° around a horizontal axis. Arrows indicate directionality of DNA translocation.
Extended Data Figure 7. DNA recognition and NDR formation. Related to Figure 1.
a. Space-filling RSC-nucleosome structure with DIM (green) and SnAC (orange) densities. View on the top as in Fig. 1c, left, but rotated by 90° around the vertical and horizontal axis. Arrows indicate directionality of DNA translocation. Number of upstream DNA base pairs relative to SHL –7 is provided.
b. Schematic of a promoter before (top) and after (bottom) RSC remodelling shows NDR formation by sliding the flanking –1 and +1 nucleosomes away from the NDR center. Arrows indicate the transcription start site.
Extended Data Figure 8. Sequence alignments for the Sth1 ATPase domain and HSA region. Related to Figures 1 – 3.
a. Sequence alignment of the S. cerevisiae Sth1 ATPase domain to the homologous Snf2 ATPase domain of the same organism. Secondary structure elements are represented in orange according to the cryo-EM structure of the Snf2 ATPase (PDB entry 5Z3U)17. Residues modelled in the Snf2 structure are topped by a back line with helical regions shown as cylinders and sheet regions as arrows. The Sth1 residues modelled in this work are indicated with a black dashed line below. ATPase motifs are underlined. Invariant residues are coloured in dark blue and conserved residues in light blue. The alignment was generated with MSAProbs71 within the MPI Bioinformatics Toolkit60 and visualized using ESPript82.
b. Sequence alignment of the HSA regions from S. cerevisiae homologues Sth1 and Snf2. Illustration and generation of the alignment as in (a).
Extended Data Table 1. Subunit composition of RSC and related chromatin remodelling complexes.
Assignment to the structural modules based on the S. cerevisiae structure of RSC presented in this work. Subunits occurring together in the complex are separated by comma, a slash indicates the use of one of the subunits. Subunits that could not be assigned to a module by homology are listed below. The PBAF subunit BAF20083 likely corresponds to Rsc9 because it comprises an armadillo repeat fold53, and the BAF180 subunit comprises regions that resemble Rsc2 and Rsc484. Only the small RSC subunits Rsc58, Rtt102 and Htl1 lack obvious counterparts. PBAF subunits contain 12 DNA-binding domains located in subunits BAF180 (HMG box)85, BAF200 (AT-rich domain, two C2H2 zinc fingers, RFX domain)83, BAF57 (HMG box)86 and BCL11A/B (six C2H2 zinc fingers)87. The BAF subunit BAF250a is predicted to contain five armadillo repeats88, and is likely the counterpart of Rsc9.
| Module | S. cerevisiae | S. pombe | D. melanogaster | H.sapiens | ||||
|---|---|---|---|---|---|---|---|---|
| RSC | SWI/SNF | RSC | SWI/SNF | PBAP | BAP | PBAF | BAF | |
| ATPase module | Sth1 | Snf2 | Snf21 | Snf22 | BRM | BRM | BRG1 | BRG1/BRM |
| Arp module | Arp9 | Arp9 | Arp9 | Arp9 | β-actin | β-actin | β-actin | β-actin |
| Arp7 | Arp7 | Arp42 | Arp42 | BAP55 | BAP55 | BAF53A/B | BAF53A/B | |
| Rtt102 | Rtt102 | |||||||
| arm module | Sfh1 | Snf5 | Sfh1 | Snf2 | SNR1 | SNR1 | BAF47 | BAF47 |
| Npl6 | Swp82 | Rsc7 | Snf59 | |||||
| body module | Rsc6 | Swp73 | Ssr3 | Ssr3 | BAP60 | BAP60 | BAF60A/B/C | BAF60A/B/C |
| Rsc9 | Swi1 | Rsc9 | Soli | BAP 170 | OSA | BAF200 | BAF250A/B | |
| Htl1 | ||||||||
| Rsc58 | Rsc58 | |||||||
| Rsc4 | Rsc4 | Polybromo | BAF 180 | |||||
| Rsc2 / Rsc1 | Rsc1 | |||||||
| DNA-interaction module | Rsc3 | |||||||
| Rsc30 | ||||||||
| Scaffold | Rsc8, Rsc8 |
Swi3, Swi3 |
Ssr1, Ssr2 |
Ssrl, Ssr2 |
MOR, MOR |
MOR, MOR |
BAF 155, BAF 170 |
BAF 155, BAF 170 |
| Ldb7 | ||||||||
| BAP111 | BAP111 | BAF57 | BAF57 | |||||
| Snf11 | ||||||||
| Snf6 | ||||||||
| Taf14 | Tfg3 | |||||||
| Snf30 | ||||||||
| SAYP | BAF45A | BAF45A/B/C | ||||||
| BRD7 | BRD9 | |||||||
| BCL11A/B | BCL11A/B | |||||||
| BCL7A/B/C | BCL7A/B/C | |||||||
| SS18/SS18L1 | ||||||||
Supplementary Material
Supplementary Information is available for this paper.
Acknowledgements
We thank current and former members of the Cramer Laboratory, including S. Osman, G. Kokic, P. Seweryn, S. Schilbach, S. Neyer, and H. Hillen. F.R.W. was supported by a Boehringer Ingelheim Fonds PhD fellowship. H.U. was supported by the Deutsche Forschungsgemeinschaft (SFB860). P.C. was supported by the Deutsche Forschungsgemeinschaft (SFB860, SPP1935, EXC 2067/1-390729940), the European Research Council Advanced Investigator Grant TRANSREGULON (grant agreement No 693023), and the Volkswagen Foundation.
Footnotes
Data availability statement
The coordinate file for the RSC-nucleosome complex structure was deposited with the Protein Data Bank with accession code 6TDA. The cryo-EM density maps used for model building were all deposited with the Electron Microscopy Data Base (EMDB) with accession code EMD-10465.
Author Contributions
F.R.W. carried out all experiments and data analysis unless stated otherwise. C.D. assisted with data collection and model building. A.S. and H.U. carried out crosslinking and mass spectrometry analysis. H.W. helped with nucleosome biochemistry. D.T. helped with cryo-EM data processing. P.C. designed and supervised the project. F.W. and P.C. wrote the manuscript, with input from all authors.
Additional Information
Reprints and permissions information is available at www.nature.com/reprints.
Competing interests
The authors declare no competing financial or non-financial interests.
References
- 1.Lorch Y, Kornberg RD. Chromatin-remodeling for transcription. Q Rev Biophys. 2017;50:e5. doi: 10.1017/S003358351700004X. [DOI] [PubMed] [Google Scholar]
- 2.Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns BR, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 4.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clapier CR, et al. Regulation of DNA Translocation Efficiency within the Chromatin Remodeler RSC/Sth1 Potentiates Nucleosome Sliding and Ejection. Mol Cell. 2016;62:453–461. doi: 10.1016/j.molcel.2016.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krietenstein N, et al. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell. 2016;167:709–721 e712. doi: 10.1016/j.cell.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein-Brill A, Joseph-Strauss D, Appleboim A, Friedman N. Dynamics of Chromatin and Transcription during Transient Depletion of the RSC Chromatin Remodeling Complex. Cell Rep. 2019;26:279–292 e275. doi: 10.1016/j.celrep.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubik S, et al. Nucleosome Stability Distinguishes Two Different Promoter Types at All Protein-Coding Genes in Yeast. Mol Cell. 2015;60:422–434. doi: 10.1016/j.molcel.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Ramachandran S, Zentner GE, Henikoff S. Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res. 2015;25:381–390. doi: 10.1101/gr.182618.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badis G, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorch Y, Maier-Davis B, Kornberg RD. Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev. 2014;28:2492–2497. doi: 10.1101/gad.250704.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubik S, et al. Sequence-Directed Action of RSC Remodeler and General Regulatory Factors Modulates +1 Nucleosome Position to Facilitate Transcription. Mol Cell. 2018;71:89–102 e105. doi: 10.1016/j.molcel.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 13.Pulice JL, Kadoch C. Composition and Function of Mammalian SWI/SNF Chromatin Remodeling Complexes in Human Disease. Cold Spring Harb Symp Quant Biol. 2016;81:53–60. doi: 10.1101/sqb.2016.81.031021. [DOI] [PubMed] [Google Scholar]
- 14.Chambers AL, Pearl LH, Oliver AW, Downs JA. The BAH domain of Rsc2 is a histone H3 binding domain. Nucleic Acids Res. 2013;41:9168–9182. doi: 10.1093/nar/gkt662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasten M, et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanDemark AP, et al. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, et al. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature. 2019;567:409–413. doi: 10.1038/s41586-019-1029-2. [DOI] [PubMed] [Google Scholar]
- 18.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 19.Schubert HL, et al. Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler. Proc Natl Acad Sci U S A. 2013;110:3345–3350. doi: 10.1073/pnas.1215379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szerlong H, et al. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen P, et al. The SnAC domain of SWI/SNF is a histone anchor required for remodeling. Mol Cell Biol. 2013;33:360–370. doi: 10.1128/MCB.00922-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorch Y, Maier-Davis B, Kornberg RD. Histone Acetylation Inhibits RSC and Stabilizes the +1 Nucleosome. Mol Cell. 2018;72:594–600 e592. doi: 10.1016/j.molcel.2018.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cakiroglu A, et al. Genome-wide reconstitution of chromatin transactions reveals that RSC preferentially disrupts H2AZ-containing nucleosomes. Genome Res. 2019;29:988–998. doi: 10.1101/gr.243139.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 25.Materne P, et al. Histone H2B ubiquitylation represses gametogenesis by opposing RSC-dependent chromatin remodeling at the ste11 master regulator locus. Elife. 2016;5 doi: 10.7554/eLife.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brahma S, Henikoff S. RSC-Associated Subnucleosomes Define MNase-Sensitive Promoters in Yeast. Mol Cell. 2019;73:238–249 e233. doi: 10.1016/j.molcel.2018.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dechassa ML, et al. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel AB, et al. Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. bioRxiv. 2019 doi: 10.1101/804534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Y, et al. Structure of the RSC complex bound to the nucleosome. Science. 2019 doi: 10.1126/science.aay0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Y, Reyes AA, Malik S, He Y. Cryo-electron microscopy structure of a nucleosome-bound SWI/SNF chromatin remodeling complex. bioRxiv. 2019 doi: 10.1101/805184. [DOI] [Google Scholar]
- 32.Cairns BR, et al. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 33.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 34.Lorch Y, Kornberg RD. Isolation and assay of the RSC chromatin-remodeling complex from Saccharomyces cerevisiae. Methods Enzymol. 2004;377:316–322. doi: 10.1016/S0076-6879(03)77019-0. [DOI] [PubMed] [Google Scholar]
- 35.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 36.Dyer PN, et al. Reconstitution of Nucleosome Core Particles from Recombinant Histones and DNA. Methods in Enzymology. 2003;375:23–44. doi: 10.1016/S0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 37.Maskell DP, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 39.Kastner B, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 40.Stark H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 2010;481:109–126. doi: 10.1016/S0076-6879(10)81005-5. [DOI] [PubMed] [Google Scholar]
- 41.Tegunov D, Cramer P. Real-time cryo-EM data pre-processing with Warp. bioRxiv. 2018 [Google Scholar]
- 42.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Asturias FJ, Chung WH, Kornberg RD, Lorch Y. Structural analysis of the RSC chromatin-remodeling complex. Proc Natl Acad Sci U S A. 2002;99:13477–13480. doi: 10.1073/pnas.162504299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaban Y, et al. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leschziner AE, et al. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci U S A. 2007;104:4913–4918. doi: 10.1073/pnas.0700706104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kidmose RT, et al. Namdinator - automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ. 2019;6:526–531. doi: 10.1107/S2052252519007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raman S, et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77(Suppl 9):89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Dijk M, Bonvin AM. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 2009;37:W235–239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bienert S, et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45:D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charlop-Powers Z, Zeng L, Zhang Q, Zhou MM. Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell Res. 2010;20:529–538. doi: 10.1038/cr.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Da G, et al. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci U S A. 2006;103:2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Legge GB, et al. ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J Mol Biol. 2004;343:1081–1093. doi: 10.1016/j.jmb.2004.08.087. [DOI] [PubMed] [Google Scholar]
- 58.Reichen C, et al. Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects. Acta Crystallogr D Struct Biol. 2016;72:168–175. doi: 10.1107/S2059798315023116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grimm M, Zimniak T, Kahraman A, Herzog F. xVis: a web server for the schematic visualization and interpretation of crosslink-derived spatial restraints. Nucleic Acids Res. 2015;43:W362–369. doi: 10.1093/nar/gkv463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmermann L, et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 61.Buchan DWA, Jones DT. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019;47:W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 63.Williams CJ, et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015. [Google Scholar]
- 65.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treich I, Ho L, Carlson M. Direct interaction between Rsc6 and Rsc8/Swh3, two proteins that are conserved in SWI/SNF-related complexes. Nucleic Acids Res. 1998;26:3739–3745. doi: 10.1093/nar/26.16.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taneda T, Kikuchi A. Genetic analysis of RSC58, which encodes a component of a yeast chromatin remodeling complex, and interacts with the transcription factor Swi6. Mol Genet Genomics. 2004;271:479–489. doi: 10.1007/s00438-004-0999-3. [DOI] [PubMed] [Google Scholar]
- 68.Angus-Hill ML, et al. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- 69.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y, Schmidt B, Maskell DL. MSAProbs: multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities. Bioinformatics. 2010;26:1958–1964. doi: 10.1093/bioinformatics/btq338. [DOI] [PubMed] [Google Scholar]
- 72.Bond CS, Schuttelkopf AW. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 73.Yang B, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 74.Combe CW, Fischer L, Rappsilber J. xiNET: cross-link network maps with residue resolution. Mol Cell Proteomics. 2015;14:1137–1147. doi: 10.1074/mcp.O114.042259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Worden EJ, Hoffmann NA, Hicks CW, Wolberger C. Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell. 2019;176:1490–1501 e1412. doi: 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eustermann S, et al. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018;556:386–390. doi: 10.1038/s41586-018-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Farnung L, Vos SM, Wigge C, Cramer P. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature. 2017;550:539–542. doi: 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willhoft O, et al. Structure and dynamics of the yeast SWR1-nucleosome complex. Science. 2018;362 doi: 10.1126/science.aat7716. [DOI] [PubMed] [Google Scholar]
- 79.Ayala R, et al. Structure and regulation of the human INO80-nucleosome complex. Nature. 2018;556:391–395. doi: 10.1038/s41586-018-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sundaramoorthy R, et al. Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. Elife. 2018;7 doi: 10.7554/eLife.35720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yan L, Wu H, Li X, Gao N, Chen Z. Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat Struct Mol Biol. 2019;26:258–266. doi: 10.1038/s41594-019-0199-9. [DOI] [PubMed] [Google Scholar]
- 82.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan Z, et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xue Y, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nicolas RH, Goodwin GH. Molecular cloning of polybromo, a nuclear protein containing multiple domains including five bromodomains, a truncated HMG-box, and two repeats of a novel domain. Gene. 1996;175:233–240. doi: 10.1016/0378-1119(96)82845-9. [DOI] [PubMed] [Google Scholar]
- 86.Wang W, et al. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci U S A. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Satterwhite E, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–3420. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
- 88.Sandhya S, Maulik A, Giri M, Singh M. Domain architecture of BAF250a reveals the ARID and ARM-repeat domains with implication in function and assembly of the BAF remodeling complex. PLoS One. 2018;13:e0205267. doi: 10.1371/journal.pone.0205267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.