Abstract
Cell surface translocation of the chaperone glucose regulated protein 78 kDa (GRP78) is a key event that promotes cancer cell survival during endoplasmic reticulum stress. Here, we identify Gα-Interacting Vesicle Associated Protein (GIV) - an enhancer of pro-survival signaling during ER stress - as a binding partner of GRP78. We show that GIV and GRP78 interact in an ER stress-dependent manner through their respective carboxyl terminal domains and that GIV aids in the localization of GRP78 to the plasma membrane. Kaplan-Meier analysis of disease-free survival in cancer patients shows poor prognosis for patients with high expression of both GIV and GRP78, further suggesting a vital role of these two proteins in enhancing cancer cell viability.
Keywords: GIV/Girdin, GRP78, ER stress, Unfolded protein response
INTRODUCTION
Endoplasmic reticulum (ER) chaperones play vital roles in maintaining normal ER function. The 78 kDa glucose regulated protein (GRP78), also known as Immunoglobulin Binding Protein (BiP), is one of the best characterized of these chaperones1,2. GRP78 is regarded as the master regulator of ER function as it plays important roles in multiple events including protein folding and assembly, targeting of misfolded proteins for ER-associated protein degradation (ERAD), and sensing of the increased misfolded protein load in the ER (termed ER stress) to activate the unfolded protein response (UPR)3,4. The UPR is an evolutionarily conserved signaling response that works to alleviate the stress by temporarily shutting down general protein translation and increasing ERAD, and selectively enhancing the expression of chaperone proteins such as GRP78 to facilitate correct protein folding5–7.
Traditionally, GRP78 is regarded as an ER luminal protein due to the KDEL retention motif at its carboxyl terminus8. However, in the past several years, multiple studies have reported GRP78 in other subcellular locations including the cytosol, mitochondria, and cell surface9–14. Of these locations, localization to the cell surface is believed to be one of the pivotal events that aid in enhanced cell survival, angiogenesis, invasion, and chemoresistance in cancer cells15. Cell surface GRP78 has been shown to interact with a number of proteins, such as Cripto and β1-Integrin, to promote cell survival and proliferation through activation of the phosphatidylinositol 3-kinase (PI3K)-Akt pathway16–19. However, the exact mechanism by which GRP78 reaches the cell surface and activates PI3K-Akt signaling remains elusive.
Recently, our laboratory identified Gɑ-Interacting Vesicle Associated Protein (GIV), aka Girdin, as a key protein that promotes cell survival during ER stress20. GIV, a prototypical member of the family of guanine nucleotide exchange modulators (GEMs), has been extensively studied for its role in the activation of heterotrimeric G protein signaling downstream of multiple cell surface receptors21,22. It has been shown to regulate numerous cellular processes including cell migration, autophagy, and intracellular vesicular trafficking23. Our recent work added UPR to the repertoire of cellular functions of GIV by showing that it mediates activation of the pro-survival PI3K-Akt pathway and dampens the induction of the pro-apoptotic transcription factor, CCAAT/enhancer binding protein homologous protein (CHOP), thus enhancing cell survival in the face of ER stress20. Despite these insights, the molecular mechanism by which GIV senses ER stress and couples survival signaling to ER stress remained unknown. Here, we show that GIV interacts with GRP78 in an ER stress-dependent manner. We mapped the interacting domains on GIV and GRP78 and provide evidence that GRP78 is not restricted to the ER under ER stress conditions. It can localize to other subcellular locations including the cytosol and plasma membrane with the latter being dependent on GIV. Further, using a Boolean analysis of human colorectal, liver, and breast cancer transcriptomes, we show that high level of GIV in tumors with high GRP78 expression is associated with lower disease-free survival across all cohorts analyzed. Together, our work provides evidence for a collaborative role for GIV and GRP78 in ensuring cell survival under ER stress conditions.
MATERIALS AND METHODS
Materials and Reagents:
All reagents used were of analytical grade. Antibodies against GIV (catalog #133371, #393757), GRP78 (catalog #13968, #376768), and glutathione S-transferase (GST; catalog #5399) were obtained from Santa Cruz Biotechnology, Dallas, TX. Antibodies against calnexin (catalog #2679), epidermal growth factor receptor (EGFR; catalog #4267), total Akt (tAkt; catalog #4691), phospho-Akt (pAkt-S473; catalog #4060), cleaved poly ADP-ribose polymerase (c-PARP; catalog #5625), CHOP (catalog#2895), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; catalog #5174), actin (catalog #3700), and tubulin (catalog #2128) were from Cell Signaling Technology, Danvers, MA. Anti-hexa-histidine (catalog #ab18184) was from Abcam, Cambridge, MA. Goat anti-rabbit and goat anti-mouse IRDye 680RD and IRDye 800CW secondary antibodies for immunoblotting were from Li-COR Biosciences, Lincoln, NE. Goat anti-rabbit Alexafluor Plus 647 and goat anti-mouse Alexafluor Plus 488 secondary antibodies for immunofluorescence were from Invitrogen, Carlsbad, CA. ProLong Gold Antifade Mountant with DAPI was from ThermoFisher Scientific, Waltham, MA.
Cell Culture and Transfection:
HeLa and Cos7 cells (American Type Culture Collection) were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Hyclone, Logan, UT) supplemented with 10% Fetal Bovine Serum (FBS; Hyclone) and 1X Penicillin, Streptomycin, and Glutamine (PSG; Corning, Manassas, VA). HeLa scrambled shRNA (Scr-shRNA) control and HeLa GIV-shRNA cell lines were a kind gift from Dr. Mikel Garcia-Marcos (Boston University, MA)24. These cell lines were maintained in DMEM supplemented with 10% FBS, 1X PSG, and 1 μg/mL Puromycin (Gibco, Grand Island, NY). Cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C. Dulbecco’s phosphate buffered saline (DPBS) and trypsin (0.25%) containing 2.21mM EDTA were from Hyclone and Corning, respectively. Transfections were performed using TransIT®-LT1 reagent (Mirus Bio, Madison, WI) following the manufacturer’s instructions. ER stress was triggered by incubating cells with medium containing tunicamycin (10 μg/mL; Sigma) for desired time-points. The control cells for tunicamycin treatment were incubated in medium containing dimethylsulfoxide (DMSO). The GST-GRP78 constructs were a kind gift from Dr. Sue Firth (University of Sydney, Australia)25.
Immunoblotting:
Samples were separated using standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to Immobilon™ FL PVDF membranes (EMD Millipore). The recipes for buffers and transfer conditions for GIV were as previously described26. Infrared imaging with two-color detection and quantification of blots were performed according to the manufacturer’s protocols using the Odyssey Fc imaging system (Li-COR Biosciences). All immunoblotting images were processed and assembled using Image Studio Lite (Li-COR Biosciences), Adobe Photoshop, and Adobe Illustrator software.
Recombinant Protein Expression and Purification:
Plasmids encoding His6-GIV-CT (residues 1660–1870), GST-GRP78 (residues 1–654), GST-GRP78-NT (residues 1–340), and GST-GRP78-CT (residues 341–654) fusion constructs were used to express these proteins in the E. coli BL21-DE3 strain. Cells were grown in Luria Bertani broth containing the appropriate antibiotic at 37°C to early log phase (OD600 ~0.6–0.8) and protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 25°C for 1h for GST-GRP78 constructs, or at 37°C for 2h for His6-GIV-CT. Affinity purification of solubilized proteins was carried out using glutathione-agarose resin and HisPur Cobalt Resin per the manufacturer’s (ThermoFisher Scientific) protocol.
GST Pull-Down Assays:
For in vitro pull-down assays, equimolar amounts (0.2 μM) of bacterially expressed and purified His6-GIV-CT was incubated with either GST-GRP78, GSTGRP78-NT, GST-GRP78-CT or GST alone immobilized on glutathione-agarose beads in binding buffer (50 mM Tris·HCl pH 7.4, 150 mM NaCl, 1 mM CaCl2, 2 mM DTT, 0.5% Triton X-100, protease inhibitors) for 16–18h. The in cellulo pull-down assays were performed as previously described27. Briefly, lysates of cells expressing GST-GIV constructs were incubated with glutathione-agarose beads for 16–18h at 4°C. The proteins bound to the beads were washed, eluted off, and analyzed by immunoblotting.
Differential Centrifugation:
HeLa cells grown on 10 cm plates to 90–95% confluency were treated with either tunicamycin or DMSO for 6 h, trypsinized, washed with DPBS, and incubated with hypotonic extraction buffer (10 mM HEPES, pH 7.8, 25 mM potassium chloride, 1 mM EGTA, protease and phosphatase inhibitors) for 20 min. The hypotonic extraction buffer was removed, and the cells were resuspended in isotonic extraction buffer (10 mM HEPES, pH 7.8, 250 mM sucrose, 25 mM potassium chloride, 1 mM EGTA, protease and phosphatase inhibitors) and lysed with a total of 75 strokes of a Dounce homogenizer. The homogenate was centrifuged at 1,000xg for 10 min at 4°C to remove nuclei. The post nuclear supernatant (PNS) was then centrifuged for 1 h at 100,000xg at 4°C. The resulting supernatant (cytosolic fraction) and pellet (membrane fraction; resuspended in a volume of 1X isotonic extraction buffer equivalent to the cytosolic fraction) were analyzed by immunoblotting.
Cell Surface Protein Labeling:
Scr-shRNA and GIV-shRNA cells grown on 10 cm plates to 90–95% confluency were treated with tunicamycin or DMSO for 6h. Cell surface proteins were then separated from intracellular proteins using the Pierce™ Cell Surface Protein Isolation Kit (ThermoScientific) per the manufacturer’s protocol. Briefly, cell surface proteins were biotinylated, the labeling reaction was quenched before the cells were lysed, and the biotinylated fraction was separated from the unlabeled fraction through affinity purification with a streptavidin-conjugated resin. The labeled and unlabeled fractions were analyzed by immunoblotting.
Proximity Ligation Assay:
HeLa cells seeded on glass coverslips were grown to 60–70% confluency, washed three times with PBS, and fixed for 25 min at room temperature with 4% (vol/vol) paraformaldehyde (Alfa Aesar, Haverhill, MA). After washing with PBS, cells were permeabilized for 20 min and processed for proximity ligation assay (PLA) using the Duolink in situ PLA kit (Sigma-Aldrich; catalog #DUO92101) according to the manufacturer’s instructions. PLA dots were quantified using the DuoLink image tool from the manufacturer.
Immunofluorescence Microscopy:
Cells grown on coverslips were fixed, blocked, and permeabilized, followed by sequential incubations for 1h each with primary and secondary antibodies. Cells were mounted on glass slides using the ProLong Gold Antifade Mountant with DAPI. Images were taken using the Olympus Fluoview 1000 confocal laser scanning system mounted on an inverted microscope (Olympus IX-81) with either the 40X oil immersion UPLFLN (NA 1.30, WD 0.20 mm) or the 60X oil immersion PLAPON (NA 1.42, WD 0.15 mm) objective. The images were analyzed using ImageJ.
Analysis of gene expression data:
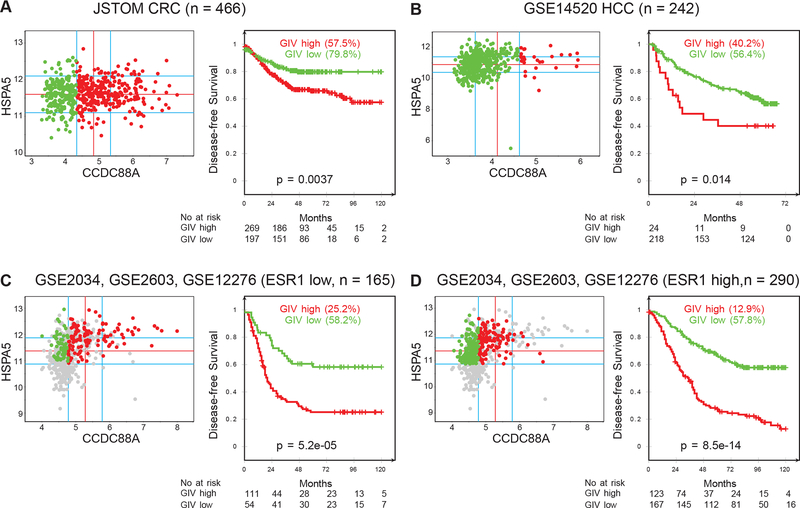
Gene expression data from three different cancer types (colorectal cancer, liver cancer, breast cancer) were collected from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) website28,29. The colorectal cancer dataset was prepared by pooling data from GSE13067, GSE14333, GSE17538, GSE31595, GSE37892, GSE33113, and normalizing them together using Robust Multi-chip Average (RMA) algorithm. Similarly, the breast cancer dataset was pooled from GSE2034, GSE2603 and GSE12276. GSE14520 (n = 242) was used for the liver cancer dataset. Patient survival data were carefully annotated for Kaplan-Meier analysis. To derive optimal cut-off values of gene expression levels, they were ordered from low to high and a rising step function was computed to define a threshold by StepMiner algorithm30. A noise margin of +/− 0.5 around StepMiner threshold was used to soften or harden the actual threshold. Almost all colorectal cancer and liver cancer samples were observed to have high levels of GRP78 (HSPA5) expression. Therefore, GIV high and low values were employed to perform Kaplan-Meier analyses using R statistical software (R version 3.4.4 2018-03-15). For the breast cancer dataset, both HSPA5 high and low values were observed. Therefore, GIV high samples were compared to GIV low within HSPA5 high samples in breast cancer separated independently by Estrogen receptor 1 (ESR1) high and low samples.
Statistical Analysis:
The densitometric analysis was performed using the Image Studio software (Li-COR Biosciences). All graphical data were prepared and statistical analyses were performed using GraphPad Prism. Statistical significance between the difference of means was calculated either by a one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test or by a paired student’s t-test. *p<0.05; **p<0.01, ***p<0.001, ****p <0.0001.
RESULTS
GIV and GRP78 interact during ER stress
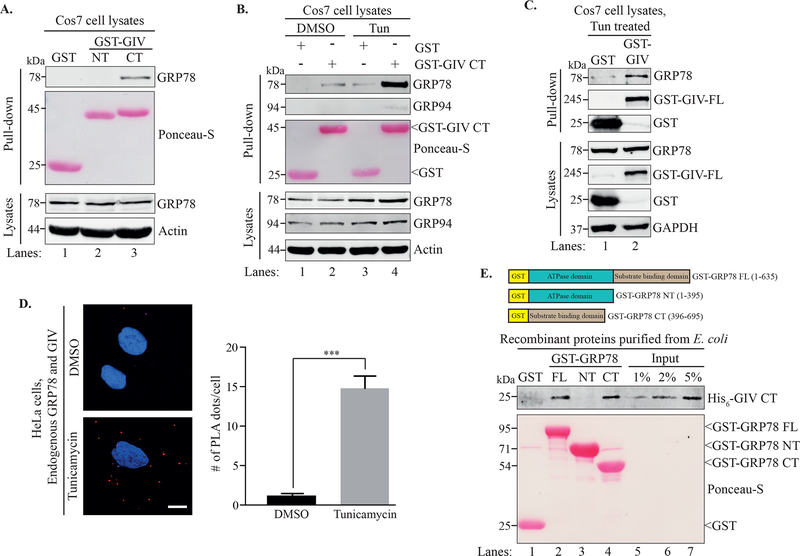
Recent work from our laboratory showed that GIV plays a crucial role in enhancing cell survival during ER stress by activating the PI3K-Akt pathway and by downregulating the pro-apoptotic response of the UPR20. Since GRP78 is another well-known contributor to survival in cells undergoing ER stress, we asked whether GIV and GRP78 interact with each other. Because we previously determined that the expression of the C-terminal region of GIV (residues 1660–1870) is sufficient to rescue the viability of GIV-depleted cells during ER stress20, we hypothesized that the C-terminus of GIV might mediate the protein-protein interactions during the UPR. To this end, we performed GST pull-down assays on cleared lysates of Cos7 cells transiently expressing GST-GIV-CT. As shown in Figure 1A, endogenous GRP78 was pulled down with GST-GIV-CT (lane 3) whereas neither GST alone nor GST-GIV N-terminus (NT; residues 1–220) showed any interaction with GRP78 (Figure 1A, lanes 1 and 2), suggesting that GRP78 specifically interacted with GIV-CT. We next wished to see whether induction of ER stress would affect the GIV•GRP78 interaction. Cos7 cells transiently expressing GST alone or GST-GIV-CT were either treated with tunicamycin or DMSO (vehicle control) for 6h. Tunicamycin induces ER stress by blocking N-linked glycosylation, a critical post-translational modification involved in the processing and maturation of newly synthesized proteins in the ER, resulting in the accumulation of unprocessed proteins in the ER lumen. The lysates of these cells were then subjected to a GST pull-down. As shown in Figure 1B, treatment with tunicamycin greatly enhanced the interaction of GST-GIV-CT with GRP78 as compared to the DMSO control (compare lanes 2 and 4). On the other hand, GRP94, another ER luminal chaperone, showed very minimal interaction with GST-GIV-CT under similar conditions (Figure 1B), implying specificity of the GIV•GRP78 interaction. Together, these results demonstrated that GIV-CT can bind GRP78 and that this interaction is promoted in an ER stress-dependent manner. We confirmed that full length GIV also binds to GRP78 by performing a similar pull-down on cell lysates expressing GST-tagged full length GIV (GST-GIV-FL; residues 1–1870). The cells were incubated with tunicamycin for 6h prior to lysis. As shown in Figure 1C, GST-GIV-FL was able to pull-down GRP78, indicating that full length GIV and GRP78 interact during ER stress.
Figure 1. GRP78 and GIV interact in an ER stress-dependent manner.
A. A GST pull-down assay was performed using lysates of Cos7 cells expressing GST, GST-GIV-NT or GST-GIVCT. B. Cos7 cells expressing GST or GST-GIV-CT were treated with either DMSO or tunicamycin for 6 h before a GST pull-down was performed. Ponceau S staining shows the GST-tagged proteins from the pull-downs (A, B). C. Cos7 cells expressing either GST or GST-GIVFL were treated with tunicamycin followed by lysis and a GST pull-down. The pull-downs (A-C) were analyzed for binding to GRP78 and the immunoblots of the lysates confirm expression of the indicated proteins. D. HeLa cells treated with either DMSO or tunicamycin were fixed and analyzed for in situ interaction between endogenous GIV and GRP78 by performing a proximity ligation assay (PLA). The PLA dots (red) per cell were quantified from a total of 25–30 cells per experiment (right) from 3 independent experiments. Data are presented as mean ± S.E.M. ***p <0.001. Scale bar = 10 μm. E. Top. The schematic shows the different GST-tagged GRP78 constructs used for the experiment. Bottom. Equimolar amounts (0.2 μM) of bacterially expressed and purified GST-GRP78 protein constructs immobilized on glutathione agarose beads were incubated with purified His6-GIV-CT overnight. The pull-down samples were analyzed for His6-GIV-CT binding. Lanes 5–7 show the % input control for His6-GIV-CT used in the binding. Ponceau S staining shows GST-GRP78 constructs.
Next we asked if the GIV•GRP78 interaction that we observed using cell lysates in pull-down assays also occurred between the endogenous proteins in intact cells subjected to ER stress. To test this, we performed a proximity ligation assay (PLA), which allows in situ detection of protein-protein interactions with single molecule resolution. As shown in Figure 1D, we observed ER stress-dependent interaction between endogenous GIV and GRP78 in HeLa cells as indicated by the significantly increased (~7–8 fold) average incidences of interaction per cell upon tunicamycin treatment. Together with the in cellulo pull-down experiments, these findings confirmed that the interaction between GIV and GRP78 occurs in cells under ER stress conditions.
To determine if the interaction between GIV and GRP78 is direct, and if so, which domain of GRP78 may interact with GIV, we next performed in vitro binding assays between bacterially expressed and purified His6-GIV-CT and either full length or individual domains of bacterially expressed and purified GST-GRP78. As shown in Figure 1E, GST-GRP78-FL was able to pull-down His6-GIV-CT indicating that the interaction observed between the two recombinant proteins is likely direct (lane 2). For the individual domains, GST-GRP78-CT, but not GSTGRP78-NT was able to pull down His6-GIV-CT (lanes 3 and 4), suggesting that the interaction between GIV and GRP78 is mediated through the substrate binding C-terminal domain of GRP78.
GRP78 co-localizes with GIV during ER stress
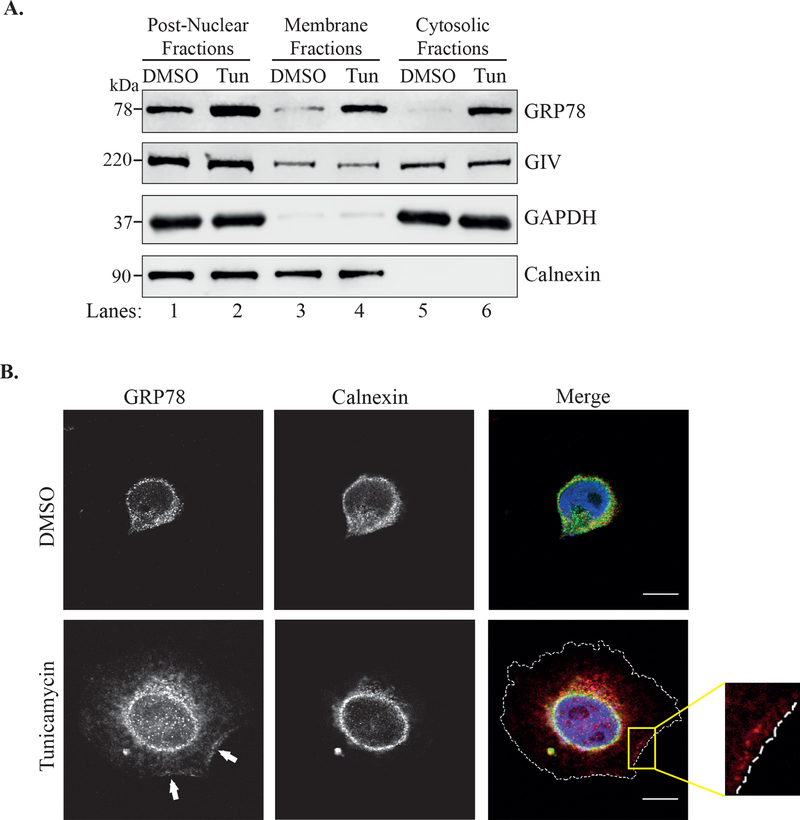
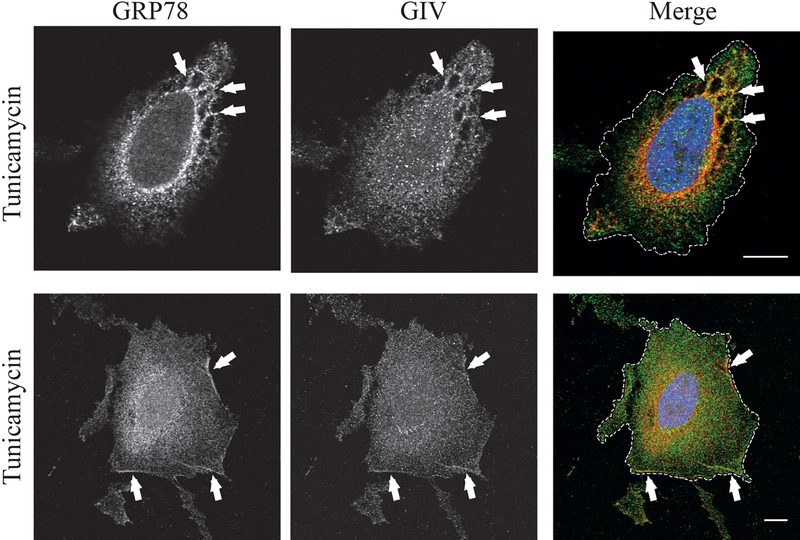
Under normal homeostatic conditions, GRP78 resides within the ER8. GIV, on the other hand, is a cytosolic protein that can peripherally associate with membranes31,32. Because these two proteins reside in different cellular compartments under normal conditions, we next wished to identify the subcellular location where these two proteins interact during ER stress. To do this, we first performed differential centrifugation to fractionate cell lysates into cytosolic and membrane fractions. As shown in Figure 2A, GIV partitioned to the membrane as well as cytosolic fractions under both control and ER stress conditions. GRP78 on the other hand, partitioned mostly to the membrane fraction under control (DMSO) conditions, but showed an equal distribution to both membrane and cytosolic fractions when treated with tunicamycin (Figure 2A). These data suggest that during ER stress, in addition to being present in the ER, GRP78 can also localize to the cytosol, which is where the GIV•GRP78 interaction could potentially take place. We also performed immunofluorescence microscopy to visualize the sub-cellular location of GRP78 in ER stressed cells. As shown in Figure 2B, in cells treated with DMSO, GRP78 co-localized with the ER membrane protein calnexin (upper panels). Upon treatment with tunicamycin, GRP78 staining appeared stronger as expected due to increased expression (Figure 2B, lower panels). It showed a broader localization pattern surpassing that of calnexin and could also be visualized along the plasma membrane (lower panels, white arrows and inset). When we co-stained tunicamycin treated cells for GIV and GRP78, the two proteins appeared to co-localize inside the cell on internal membranes (Figure 3, top panels) as well as at the plasma membrane (bottom panels).
Figure 2. GRP78 localization is not restricted to the ER during ER stress.
A. Lysates of HeLa cells treated with either DMSO or tunicamycin for 6h were separated into cytosolic and membrane fractions by differential centrifugation. The fractions were analyzed by immunoblotting. GAPDH is used as a cytosolic marker and Calnexin as an ER marker. B. HeLa cells treated with tunicamycin or DMSO for 6h were fixed and stained for GRP78 (red), calnexin (green) and DAPI (blue). Cells were visualized by confocal microscopy. The arrows point to the localization of GRP78 along the cell periphery. The cell periphery is outlined by the dashed white line in the bottom, merged panel. The inset shows enlargement of the boxed region. Scale bar = 10 μm.
Figure 3. GRP78 co-localizes with GIV in tunicamycin treated cells.
A. HeLa cells treated with tunicamycin were fixed and stained for GRP78 (red), GIV (green), and DAPI (blue). The cells were visualized by confocal microscopy. The arrows point to the co-localization along the cell periphery. The cell periphery is outlined by the dashed white line in the merged images. Scale bar = 10 μm.
ER stress-induced cell surface localization of GRP78 is mediated by GIV
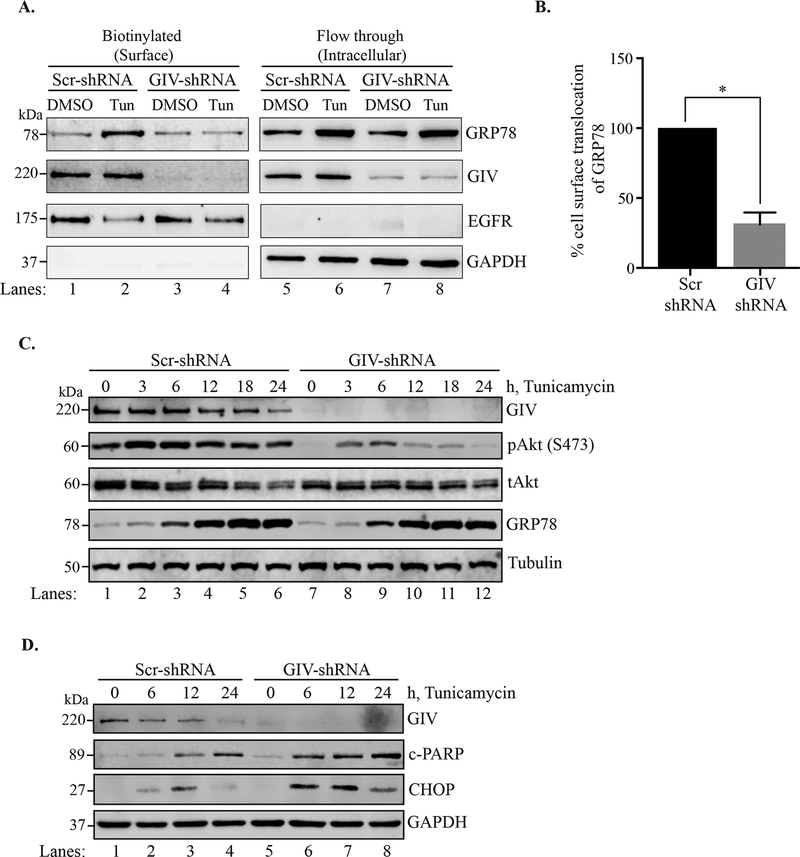
Multiple studies have shown that GRP78 is able to localize to the cell surface in numerous types of cancer15,33–37. Because we observed co-localization between the two proteins at the plasma membrane, we next determined whether the localization of GRP78 to the cell surface is influenced by GIV. We performed a cell surface protein biotinylation assay using Sulfo-NHS-SS-Biotin to biotinylate the exposed primary amines of cell surface proteins in control (Scr-shRNA) and GIV-depleted (GIV-shRNA) HeLa cells. Cells were incubated with either DMSO or tunicamycin for 6h before biotinylation was performed. As shown in Figure 4A, ER stress led to enhanced level of GRP78 in the cell surface protein fraction in Scr-shRNA cells (compare lanes 1 and 2). However, in the GIV-shRNA cells, GRP78 translocation to the cell surface upon ER stress was impaired (Figure 4A, compare lanes 3 and 4) and was significantly less than that observed in Scr-shRNA cells (Figure 4B), suggesting that GIV plays a role in the translocation of GRP78 to the cell surface.
Figure 4. ER stress induced cell surface localization of GRP78 and Akt signaling is reduced in GIV depleted cells.
A. Scr-shRNA and GIV-shRNA cells were treated with tunicamycin or DMSO for 6h following which the cell surface proteins were biotinylated and separated from the non-biotinylated fraction through affinity purification using the streptavidin-agarose resin. The biotinylated (cell surface proteins) and flow-through (intracellular) fractions were then analyzed by immunoblotting. EGFR served as a cell surface protein control, and GAPDH as an intracellular protein control. B. The % cell surface localization of GRP78 was calculated by densitometric analysis. The Scr control was plotted to 100% and the relative % localization in GIV-shRNA cells was calculated. n=3. Errors bars represent ± S.E.M. *p <0.05. C. and D. Scr-shRNA and GIV-shRNA cells were ER stressed using tunicamycin for the indicated time-points before lysis. The cell lysates were analyzed by immunoblotting.
The cell surface localization of GRP78 has been previously shown to be important for activation of the PI3K-Akt pathway35. Because we previously demonstrated that GIV mediates the activation of the PI3K-Akt pathway in ER stressed HeLa cells20 and because translocation of GRP78 to the cell surface requires GIV (Figure 4A and B), we predicted that GIV-depleted cells will have reduced Akt activation even upon prolonged ER stress which markedly increases expression of GRP78. To test this, we treated Scr-shRNA and GIV-shRNA cells with tunicamycin for 0, 3, 6, 12, 18, and 24h and analyzed the cell lysates by immunoblotting. As shown in Figure 4C, Scr-shRNA cells showed increased activation of Akt (as confirmed by blotting for pAkt-S473) peaking at 3–6h and sustained over the entire course of treatment (lanes 1–6). The GIV-shRNA cells on the other hand, showed a very modest level of Akt activation initially (3–6h) which declined thereafter even though GRP78 level rose comparable to the control cells (lanes 7–12). Consistent with dampened pro-survival signaling, the GIV-shRNA cells also show enhanced pro-apoptotic response as determined by the levels of c-PARP and CHOP (Figure 4D). Together, these results suggested that GIV may have little or no impact on the transcription/translation/stability of GRP78 in response to UPR; instead, the primary and crucial role that it performs is that of facilitating the localization of GRP78 to the plasma membrane from where sustained activation of the pro-survival PI3K-Akt signals may be initiated.
High expression of GIV and GRP78 is associated with shorter disease-free survival in patients with colorectal, liver, and breast cancers
High expression of GRP78 has previously been correlated to a poor prognostic outcome for patients affected with many different cancers38. To determine if this is exacerbated by a concomitant high expression of GIV, we carried out a comparative gene expression analysis to assess the effect of high vs low expression of GIV on the disease-free survival of patients with colorectal, liver, and breast cancer expressing high levels of GRP78 (see methods). Samples were divided into “low” and “high” subgroups with regard to GRP78 (HSPA5) and GIV (CCDC88A) gene expression levels using the StepMiner algorithm, implemented within the hierarchical exploration of gene-expression microarrays online (HEGEMON) software39 (Figure 5). Kaplan-Meier analyses of the disease-free survival showed that among patients whose tumors had high GRP78 expression, concomitant expression of GIV at high levels correlated to a significantly poorer prognosis compared to those with low GIV (Figure 5). This trend was true for all four groups tested – colorectal cancer (Figure 5A), liver cancer (Figure 5B), breast cancer with low estrogen receptor 1 (ESR1) expression (Figure 5C), and breast cancer with high ESR1 expression (Figure 5D). These analyses suggested that cancers with high expression of GRP78 and GIV expression are more robust, resulting in lower disease-free survival of the affected patients. In addition to the diverse cohorts (mixed population of patients with different treatments and various stages) described above, we also analyzed a selected group of colorectal cancer patients all of whom received cytotoxic chemotherapy (Figure S1). These samples were subgrouped based on high vs low expression of both GRP78 and GIV. Our analysis indicated that while the disease-free survival in patients expressing high levels of both proteins was significantly reduced compared to those expressing high levels of GRP78 alone, the outcome of patients expressing low GRP78 was unchanged regardless of the expression level of GIV (Figure S1). This analysis suggested that GIV may likely be involved in promoting chemoresistance in tumors experiencing ER stress and UPR (increased GRP78 expression), but not in tumors with low or no ER stress/UPR (low GRP78 expression).
Figure 5. Concurrent high expression of GIV and GRP78 correlates with poor prognosis for disease free survival of colorectal, liver, and breast cancer patients.
A. Expression pattern of CCDC88A (GIV) was analyzed in colorectal cancer (CRC) dataset (JSTOM, n = 466, pooled data from GSE13067, GSE14333, GSE17538, GSE31595, GSE37892, GSE33113). StepMiner threshold minus 0.5 was used for CCDC88A. Almost all CRC have high levels of HSPA5 (GRP78) expression. B. Expression pattern of CCDC88A was analyzed in hepatocellular carcinoma (HCC) dataset (GSE14520, n = 242). StepMiner threshold plus 0.5 was used for CCDC88A. Almost all HCC have high levels of HSPA5 expression. C, D. Gene expression data from three different datasets (GSE2034, GSE2603, GSE12276) were combined to create the Breast Cancer database (n = 572). Duplicate samples were carefully removed and all data were normalized together. The dataset mainly contains breast cancer patients with distant metastases such as bone, lung and brain. StepMiner threshold minus 0.5 was used for ESR1, HSPA5, CCDC88A. ESR1 low patients (C, n=165) and ESR1 high patients (D, n=290) expressing high HSP5A were further categorized into groups of patients based on expression levels of CCDC88A.
DISCUSSION
GRP78 has been hailed as a master regulator of the UPR and an important player in promoting cancer survival, chemoresistance, and metastasis40–42. This study has identified GIV, another key regulator of the UPR and contributor to cell survival during ER stress, as a binding partner of GRP78 and has addressed the functional importance of this interaction in an ER stressed cell.
Our domain mapping experiments showed that these two proteins can bind directly to each other through their respective C-terminal domains (Figure 1E). The C-terminal region of GIV has been shown to interact with many proteins including (but not limited to) receptor tyrosine kinases, heterotrimeric G proteins, PI3K, Akt, and actin, serving as the hub of the signaling events that contribute to important properties of cancer cells such as survival, invasion, and metastasis21,23. GIV-CT is predicted to be intrinsically disordered, which makes it a good signaling platform that can be molded (e.g. via post-translational modifications) to serve as a binding partner for multiple effectors43. It also makes an excellent site for interaction with the C-terminal substrate binding domain of GRP78, which binds to unstructured regions on partially folded client proteins44,45. Interestingly, Gipie, another member of the GEM family has also been shown to interact with GRP78 and serve as a regulator of the UPR in endothelial cells46. However, this study suggested that GIV did not bind to GRP78, which contradicts our findings here. This difference could be due to different cell lines used, Matsuhita et al. used human umbilical vein endothelial cells (HUVECs) while we used HeLa cells as our primary model system to study the interaction between endogenous GIV and GRP78. Incidentally, HeLa cells and many other cancer cell lines do not express detectable level of Gipie46 suggesting that regulation of ER stress may be carried out by different members of the GEM family depending on the specific cell type.
Our data show that under ER stress conditions, GRP78 is also found in the cytosol (Figure 2). There are a few plausible mechanisms through which this could happen. GRP78 can be retrotranslocated from the ER lumen to the cytosol using the ERAD pathway as shown by Duriez et al.9. In this study, the authors showed that the hepatitis B virus precore protein is retrotranslocated from the ER to the cytosol using the ERAD pathway and in the process translocates GRP78 to the cytosol as well. Since ERAD is upregulated during ER stress to help restore cellular homeostasis47, it is possible for GRP78 to hitchhike with retrotranslocating misfolded proteins to the cytosol. Another mechanism could be through a global change in permeability of the ER membrane as shown by Wang et al.10. Here the authors showed that ER membrane permeability is altered in cells treated with tunicamycin and thapsigargin, which allows passage of ER luminal proteins including GRP78 and protein disulfide isomerase to the cytosol. The third possible mechanism is the translation of an alternatively spliced mRNA form of GRP78 that is missing the signal sequence as shown by Ni et al.11. The authors showed that the alternatively spliced transcript devoid of the signal peptide sequence is stabilized under conditions of ER stress leading to accumulation of cytosolic GRP78, which retains all the functional aspects of the protein. Regardless of whether one or all of the abovementioned mechanisms play a role in cytosolic localization of GRP78, the cytosol presents a likely site of interaction between GIV and GRP78.
Our co-localization results using immunofluorescence microscopy (Figure 3) indicate that these proteins can potentially also interact on internal membranes as well as the plasma membrane. It is possible that the reticular pattern of co-localization that we observed represents binding of GIV to GRP78 that is retrotranslocating out of the ER. The integrity of the ER-to-Golgi trafficking has been shown to be partially required for the cell surface translocation of GRP78 in HeLa cells14 and GIV has been shown to play a role in regulating vesicular trafficking at the Golgi32. It is possible that in addition to directly binding to the retrotranslocated GRP78 in the cytosol and mediating its localization to the cell surface by an as yet unknown mechanism, GIV may also mediate trafficking of the pool of GRP78 that escapes the ER due to saturation of the KDEL receptors upon increased expression of chaperones during ER stress. Such pool of GRP78 will fail to return to the ER and would continue along the anterograde secretory pathway, ultimately being secreted from where it can associate with the cell surface peripherally through interaction with its binding partners. Whether this is indeed the case, needs to be investigated.
Previous studies have shown cell surface expression of GRP78 in many different types of tumor cells15,33–37, however, the exact mechanism by which this translocation occurs is not well understood. It has been suggested that GRP78 must bind a substrate protein to translocate to the plasma membrane in order to promote cancer cell survival through activation of the PI3K-Akt pathway35. One study has shown that the substrate-binding activity of GRP78 is required for its cell surface translocation further suggesting that this process is mainly dependent upon GRP78 binding to client proteins14. Interestingly, antibodies targeting the C-terminus of GRP78 have been shown to curb cancer cell survival and migration through inhibition of the PI3K-Akt pathway18,36,48. Given that the interaction between GIV and GRP78 is mediated through the C-terminal domain of the latter, and that GIV depletion retards cell surface translocation of GRP78 and Akt activation, our data establish a crucial role for GIV in mediating the cytoprotective functions attributed to GRP78. The significant decline in disease-free survival in patients with tumors expressing high levels of GRP78 and GIV compared to those expressing high levels of GRP78 alone (Figures 5, S1) further supports the key role GIV plays in augmenting cancer cell survival.
Conclusions:
The results presented in this study have characterized a novel interaction between GIV and GRP78 by mapping the binding regions on both proteins; by analyzing the in situ binding between endogenous GIV and GRP78 under ER stress; by identifying the sub-cellular locations where these proteins can interact; and by determining the potential role of this ER stress-dependent binding event in facilitating localization of GRP78 to the plasma membrane. Finally, worse prognosis for the disease outcome in cancer patients concomitantly expressing high levels of these proteins further highlights the important role for these two proteins in enhancing cancer cell viability. Our current and future efforts are aimed at investigating the mechanism(s) by which GIV promotes cell surface translocation of GRP78 to enhance cell survival signals.
Supplementary Material
Acknowledgements:
We thank Drs. William Silkworth (UCLA) and Elena Grintsevich (CSULB) for their assistance with analysis of the immunofluorescence images. This work was supported by the National Institute of General Medical Sciences-National Institutes of Health (NIGMS-NIH) grant #SC2GM121246 to DB. JMN and AH were supported by the NIGMS-NIH awards #T34GM008074 and #R25GM071638, respectively. SL was supported by the CSULB BUILD program under NIGMS award numbers UL1GM118979, TL4GM118980, and RL5GM118978. PG is supported by the NIH (CA100768, CA238042, CA160911 and AI141630). DS is supported by the NIH grant #R00-CA151673, 2017 Padres Pedal the Cause/Rady Children’s Hospital Translational PEDIATRIC Cancer Research Award (Padres Pedal the Cause/RADY #PTC2017), 2017 Padres Pedal the Cause/C3 Collaborative Translational Cancer Research Award (San Diego NCI Cancer Centers Council (C3) #PTC2017). The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
ETHICAL STATEMENT:
This article does not contain any studies with human participants or animals performed by any of the authors. Publicly available tumor transcriptome data analyzed here represents deidentified information and is exempt from the need for regulatory approvals for human subject research.
Conflict of Interest: The authors declare that they have no conflict of interest.
REFERENCES
- 1.Ni M, Lee AS (2007) ER chaperones in mammalian development and human diseases. FEBS Lett. 581, 3641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M, Wey S, Zhang Y, Ye R, Lee AS (2009) Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxid Redox Signal. 11, 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casas C (2017) GRP78 at the centre of the stage in cancer and neuroprotection. Front. Neurosci 11, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendershot LM (2004) The ER function BiP is a master regulator of ER function. Mt. Sinai J. Med 71, 289–97. [PubMed] [Google Scholar]
- 5.Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–6. [DOI] [PubMed] [Google Scholar]
- 6.Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, Wang ZV, Zorzano A, Hill JA, Jaimovich E, Quest AFG, Lavandero S (2013) Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int. Rev. Cell Mol. Biol 301, 215–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karagöz GE, Acosta-Alvear D, Walter P (2019) The unfolded protein response: detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol 11, a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munro S, Pelham HRB (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907. [DOI] [PubMed] [Google Scholar]
- 9.Duriez M, Rossigno JM, Sitterlin D (2008) The hepatitis B virus precore protein is retrotransported from endoplasmic reticulum (ER) to cytosol through the ER-associated degradation pathway. J. Biol. Chem 283, 32352–32360. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Olberding KE, White C, Li C (2011) Bcl-2 proteins regulate ER membrane permeability to luminal proteins during ER stress-induced apoptosis. Cell Death Differ. 18, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni M, Zhou H, Wey S, Baumeister P, Lee AS (2009) Regulation of PERK signaling and leukemic cell survival by a novel cytosolic isoform of the UPR regulator GRP78/BiP. PLoS One 4(8), e6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun FC, Wei S, Li CW, Chang YS, Chao CC, Lai YK (2006) Localization of GRP78 to mitochondria under the unfolded protein response. Biochem. J 396, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Liu R, Ni M, Gill P, Lee AS (2010) Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J. Biol. Chem 285, 15065–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai Y-L, Zhang Y, Tseng C-C, Stanciauskas R, Pinaud F, Lee AS (2015) Characterization and mechanism of stress-induced translocation of 78-kilodalton glucose-regulated protein (GRP78) to the cell surface. J. Biol. Chem 290, 8049–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni M, Zhang Y, Lee AS (2011) Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem. J 434, 181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N, Zoubeidi A, Beraldi E, Gleave ME (2013) GRP78 regulates clusterin stability, retrotranslocation and mitochondrial localization under ER stress in prostate cancer. Oncogene 32, 1933–42. [DOI] [PubMed] [Google Scholar]
- 17.Shani G, Fischer WH, Justice NJ, Kelber JA, Vale W, Gray PC (2008) GRP78 and cripto form a complex at the cell surface and collaborate to inhibit transforming growth factor signaling and enhance cell growth. Mol. Cell. Biol 28, 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Ridder GG, Gonzalez-Gronow M, Ray R, Pizzo SV (2011) Autoantibodies against cell surface GRP78 promote tumor growth in a murine model of melanoma. Melanoma Res. 21, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y, Wey S, Wang M, Ye R, Liao CP, Roy-Burman P, Lee AS (2008) PTEN null prostate tumorigenesis and AKT activation are blocked by targeted knockout of ER chaperone GRP78/BiP in prostate epithelium. Proc. Natl. Acad. Sci. U. S. A 105, 19444–19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen P, Calderon R, Rodriguez-Ledezma Y, Araujo K, Bhandari D (2019) GIV/Girdin promotes cell survival during endoplasmic reticulum stress. Mol. Cell. Biochem 453, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Marcos M, Ghosh P, Farquhar MG (2015) GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein. J. Biol. Chem 290, 6697–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh P, Rangamani P, Kufareva I (2017) The GAPs, GEFs, GDIs and now, GEMs: new kids on the heterotrimeric G protein signaling block. Cell Cycle 16, 607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aznar N, Kalogriopoulos N, Midde KK, Ghosh P (2016) Heterotrimeric G protein signaling via GIV/Girdin: breaking the rules of engagement, space, and time. Bioassays. 38(4), 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leyme A, Marivin A, Perez-Gutierrez L, Nguyen LT, Garcia-Marcos M (2015) Integrins activate trimeric G proteins via the nonreceptor protein GIV/Girdin. J. Cell Biol. 210, 1165–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grkovic S, O’Reilly VC, Han S, Hong M, Baxter RC, Firth SM (2013) IGFBP-3 binds GRP78, stimulates autophagy and promotes the survival of breast cancer cells exposed to adverse microenvironments. Oncogene 32, 2412–20. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh P, Aznar N, Swanson L, Lo IC, Lopez-Sanchez I, Ear J, Rohena C, Kalogriopoulos N, Joosen L, Dunkel Y, Sun N, Nguyen P, Bhandari D (2016) Biochemical, biophysical and cellular techniques to study the guanine nucleotide exchange factor, GIV/Girdin. Curr. Protoc. Chem. Biol 8, 265–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhandari D, Lopez-Sanchez I, To A, Lo IC, Aznar N, Leyme A, Gupta V, Niesman I, Maddox AL, Garcia-Marcos M, Farquhar MG, Ghosh P (2015) Cyclin-dependent kinase 5 activates guanine nucleotide exchange factor GIV/Girdin to orchestrate migration-proliferation dichotomy. Proc. Natl. Acad. Sci. U. S. A 112, E4874–E4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30(1), 207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R (2005) NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 33 (Database issue), D562–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahoo D, Dill DL, Tibshirani R, Plevritis SK (2007) Extracting binary signals from microarray time-course data. Nucleic Acids Res. 35(11), 3705–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le-Niculescu H, Niesman I, Fischer T, DeVries L, Farquhar MG (2005) Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J Biol Chem. 280(23), 22012–22020. [DOI] [PubMed] [Google Scholar]
- 32.Lo IC, Gupta V, Midde KK, Taupin V, Lopez-Sanchez I, Kufareva I, Abagyan R, Randazzo PA, Farquhar MG, Ghosh P (2015) Activation of Gαi at the Golgi by GIV/Girdin imposes finiteness in Arf1 signaling. Dev Cell 33(2), 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Gronow M, Selim MA, Papalas J, Pizzo SV (2009) GRP78: a multifunctional receptor on the cell surface. Antioxid Redox Signal 11(9), 2299–2306. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XX, Li HD, Zhao S, Zhao L, Song HJ, Wang G, Guo QJ, Luan ZD, Su RJ (2013) The cell surface GRP78 facilitates the invasion of hepatocellular carcinoma cells. Biomed Res. Int 2013, 917296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Tseng CC, Tsai YL, Fu X, Schiff R, Lee AS (2013) Cancer cells resistant to therapy promote cell surface relocalization of GRP78 which complexes with PI3K and enhances PI(3,4,5)P3 production. PLoS One 8(11), e80071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Li X, Gao W, Zhou Y, Wey S, Mitra SK, Krasnoperov V, Dong D, Liu S, Li D, Zhu G, Louie S, Conti PS, Li Z, Lee AS, Gill PS (2013) Monoclonal antibody against cell surface GRP78 as a novel agent in suppressing PI3K/AKT signaling, tumor growth, and metastasis. Clin. Cancer Res. 19, 6802–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arap MA, Lahdenranta J, Mintz PJ, Hajitou A, Sarkis AS, Arap W, Pasqualini R (2004) Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell 6, 275–84. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffenbach KT, Lee AS (2011) The critical role of GRP78 in physiologic and pathologic stress. Curr. Opin. Cell Biol. 23, 150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, Zabala M, Bueno J, Neff NF, Wang J, Shelton AA, Visser B, Hisamori S, Shimono Y, van de Wetering M, Clevers H, Clarke MF, Quake SR (2011) Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol 29, 1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Lee AS (2006) Stress induction of GRP78/BiP and its role in cancer. Curr. Mol. Med 6, 45–54. [DOI] [PubMed] [Google Scholar]
- 41.Lee AS (2007) GRP78 induction in cancer: therapeutic and prognostic implications endoplasmic reticulum stress and cancer. Cancer Res 67, 3496–3505. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Bi X, Zhang G, Deng Y, Luo X, Xu L, Scherer PE, Ferdous A, Fu G, Gillette TG, Lee AS, Jiang X, Wang ZV. (2018) Glucose-regulated protein 78 is essential for cardiac myocyte survival. Cell Death Differ. 25, 2181–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin C, Ear J, Midde K, Lopez-Sanchez I, Aznar N, Garcia-Marcos M, Kufareva I, Abagyan R, Ghosh P (2014) Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol Biol Cell. 25(22), 3654–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ (1993) Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–28. [DOI] [PubMed] [Google Scholar]
- 45.Yang J, Nune M, Zong Y, Zhou L, Liu Q (2015) Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP. Structure 23, 2191–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsushita E, Asai N, Enomoto A, Kawamoto Y, Kato T, Mii S, Maeda K, Shibata R, Hattori S, Hagikura M, Takahashi K, Sokabe M, Murakumo Y, Murohara T, Takahashi M (2011) Protective role of Gipie, a Girdin family protein, in endoplasmic reticulum stress responses in endothelial cells. Mol. Biol. Cell 22(6), 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–58. [DOI] [PubMed] [Google Scholar]
- 48.Misra UK, Mowery Y, Kaczowka S, Pizzo SV (2009) Ligation of cancer cell surface GRP78 with antibodies directed against its COOH-terminal domain up-regulates p53 activity and promotes apoptosis. Mol. Cancer Ther. 8, 1350–1362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.