Abstract
CD8 T cells are among the most vigorous soldiers of the immune system that fight viral infections and cancer. CD8 T cell development, maintenance, activation and differentiation are under the tight control of multiple transcriptional and post-transcriptional networks. Over the last two decades it has become clear that non-coding RNAs (ncRNAs), which consist of microRNAs (miRNAs) and long ncRNAs (lncRNAs), have emerged as global biological regulators. While our understanding of the function of specific miRNAs has increased since the discovery of RNA interference, it is still very limited, and the field of lncRNAs is just starting to blossom. Here we will summarize our knowledge on the role of ncRNAs in CD8 T cell biology, including differentiation into memory and exhausted cells.
Keywords: miRNA, lncRNA, naïve, effector, CTL, exhaustion, memory
Introduction
CD8 T cells protect the host organism by eliminating virally infected and cancerous cells, yet CD8 T cells must first acquire cytotoxic function. Prior to antigen encounter, naïve CD8 T cells are transcriptionally inactive and are maintained in a quiescent ‘resting’ state, in which they undergo minimal proliferation and rely primarily on oxidative phosphorylation, as they have very low metabolic demands. Maintenance of the quiescent state in the absence of infection or cancer is critical both for the long-term preservation of naïve cells, and to prevent unwarranted inflammation, including autoimmune diseases. Upon antigen recognition, naïve CD8 T cells increase the use of both glycolysis and oxidative phosphorylation to support the rapid proliferation of antigen specific cells, and the acquisition of effector functions. Although the majority of the responding cells die via apoptosis upon clearance of the invading pathogen or tumor, in an event known as contraction, a small number of cells will survive and differentiate into long-lived memory cells which provide life-long antigen-specific protection to the host. However, this differentiation program is often coopted by the immunosuppressive environments established during cancer and chronic viral infection, diverting responding CD8 T cells into the exhausted state. CD8 T cell exhaustion is characterized primarily by a gradual loss of effector function and eventually compromised survival. It has been recognized that this population is heterogeneous and contains less differentiated progenitors that are required for the repopulation of exhausted cells1, 2, 3. Although the presence of these cells in pathological conditions is viewed beneficial to the organism as they protect from immune-related pathology during a chronic immune response4, 5, exhausted CD8 T cells fail to form the highly protective memory pool. Accordingly, a more comprehensive understanding of the molecular mechanisms which orchestrate CD8 T cell differentiation, and support the survival of CD8 T cells at each stage is of great interest.
Here, we will review the post-transcriptional regulation of gene expression mediated by non-coding RNAs (ncRNAs) during CD8 T cell development and differentiation. NcRNAs include small non-coding RNAs called microRNAs (miRNAs), and long non-coding RNAs (lncRNAs). Specifically, this review will focus on miRNAs as they have been the most extensively studied. Although there is much less known regarding lncRNAs, we will also highlight the emerging evidence suggesting an important role for lncRNAs in CD8 T cells as well.
miRNA mediated regulation of gene expression
The most well-studied ncRNAs are miRNAs- short ~22 nucleotide (nt) RNAs that inhibit gene expression by sequence-specific Watson Crick base pairing to target mRNAs. Although it has been reported that up to 1,193 miRNAs have been identified in mice6, and 2,588 in humans7, 8, the use of stringent functional and molecular parameters during in silico analysis suggests the actual number of miRNAs is closer to 475 and 519 in mouse and human, respectively6. The genomic organization of miRNAs is an important factor which contributes to the tissue and context specific expression of these regulatory RNAs9. The expression of genes encoding miRNAs can be controlled by their own promoters or by the promoters of other genes. Interestingly, some promoters of miRNA encoding genes may control expression of one miRNA gene, or several10, 11. The latter are referred to as clustered miRNAs.
The generation of mature miRNAs is a multi-step process that has been well described in great detail previously8. For this review, it is important to know that the last step of miRNA biogenesis involves processing of the pre-miRNA intermediate precursor by the endonuclease Dicer to generate a fully mature double stranded miRNA, one strand of which is loaded into RNA-Induced Silencing Complex (RISC). The RISC contains the Argonauate-2 protein which binds to the miRNA and is critical for its mRNA-silencing function.
miRNAs bind their mRNA targets using a highly conserved motif in the 2–8 nt position in the 5’ end of the miRNA, which is termed the seed region. miRNAs are grouped together in ‘families’ based on the shared use of the same seed sequence for targeting mRNAs6. These miRNAs are generally distinguished by the placement of a letter following the miRNA name, and are referred to as ‘sister’ miRNAs (let-7a, let-7b, let-7c and etc.). Typically, the region of the mRNA to which the miRNA seed sequence binds is located in the 3’UTR, although non-canonical binding sites within the open reading frame of mRNAs have been described12, 13, 14. The degree of sequence complementarity between the miRNA seed sequence and the target mRNA determines the mechanism by which the expression of the mRNA is prevented. If a miRNA seed sequence is a perfect match to the target sequence in the mRNA, the poly-A tail is degraded, leading to destabilization of the mRNA8. Rather, if a miRNA binds with less-than-perfect complementarity, ribosomal progression is blocked by the RISC. The net result of both of these mechanisms is inhibition of gene expression at the post-transcriptional level.
Evidence of RNA interference in CD8 T cell immunity
It is important to note that miRNA-mediated RNA interference is a very ancient process that evolved at the dawn of multicellular organisms. Therefore, it is not surprising that many miRNAs and their targets co-evolved together and are ultra-conserved among different animals. Furthermore, many miRNA genes (e.g. let-7) were duplicated multiple times during evolution, suggesting a vital importance for such miRNAs in the regulation of various biological processes. It is also interesting to mention the peculiar co-appearance of some miRNAs and the adaptive immunity in early vertebrate animals. As such, it is likely that miRNA-mediated regulation is widespread and highly conserved throughout the adaptive immune system6. The significance of the contribution of RNA interference to T cell immunity was first observed by T cell specific deletion of the miRNA biogenesis enzyme Dicer, using Lck driven Cre. These mice had a smaller thymus, with reduced CD4 and CD8 T cell numbers in the thymus, despite no defects in the proportion of CD4 and CD8 single positive cells, and the dramatic loss of mature T cells in the periphery15, 16. Interestingly, CD8 T cell specific deletion of Dicer caused cells to respond more rapidly to T cell receptor (TCR) stimulation, ultimately resulting in compromised differentiation into short-lived effector cells (SLECs), and a failure to survive contraction and seed the memory pool17, 18, 19. MiRNA profiling in naïve, effector, and memory CD8 T cells demonstrated that miRNAs are differentially expressed in different CD8 T cell states20. Since then, several miRNAs have been implicated in controlling the development, maintenance and differentiation of CD8 T cells.
miRNAs in the development and maintenance of naïve CD8 T cells
Efficient CD8 T cell responses in the periphery are only possible if mature CD8 T cells are successfully generated in the thymus. The first evidence elucidating the role of specific miRNAs in CD8 T cells came from the pioneering work of the lab of David Bartel, where the ectopic expression of miR-181 in hematopoietic stem cells blocked the development of CD8 T lymphocytes21. It was later shown that miR-181 targets multiple phosphatases, negative regulators of TCR-signaling, suggesting a potential positive role in thymic selection22. It is well documented that the compromised proliferation or survival of immature thymocytes may negatively affect the numbers of mature T cells. Depletion of miR-142 in T cell precursors triggered upregulation of the expression of the cell cycle inhibitor, Cdkn1 resulting in severe lymphopenia due to the suppressed proliferation of thymocytes and mature T cells23. Recent publication demonstrated that miRNA cluster miR17–92 is critical for IL-7 signaling and therefore the survival of developing thymocytes24. Consistent with reports of Dicer-deficient mice, it was suggested that miRNAs may also be directly involved in lineage commitment and thymic selection by silencing of co-receptor genes and lineage specifying transcription factors Runx3 and Thpok, thus influencing productive CD8 T cell development25. However, the true impact of miRNAs in the development of mature αβ T cells in the thymus remains largely unknown.
Upon egress from the thymus, mature naïve CD8 T cells populate secondary lymphoid organs where they are retained for the lifetime of the host until they encounter their cognate antigen. In the adult organism, the fitness and longevity of naïve CD8 T cells become critical to preserve the diversity of the TCR repertoire because of the rapid decline of T cell development due to age-related thymic involution26. It has been shown that the maintenance of naïve T cells depends primarily on tonic TCR signaling through self-peptide:MHC interactions, and the cytokine IL-726, 27, 28. These signals keep T cells in a quiescent state resulting in low metabolism and minimal proliferation.
Investigation of specific miRNAs identified in the miRNA signature of CD8 T cells20, suggested the regulatory potentials of miRNAs in T cell maintenance (FIG-1 shows only non-coding RNAs that have been demonstrated experimentally to be involved in the maintenance and differentiation of CD8 T cells). Although only a fraction of such miRNAs have been characterized, it is reasonable to predict that the main function of miRNAs that are highly expressed in naïve T cells, and suppressed upon activation, is to maintain the quiescent phenotype of naïve cells by promoting survival while restricting cell activation and proliferation. It has been shown that the most highly expressed miRNAs29 in naïve T cells, such as miR-15018, 20, 30, 31, 32, miR-2620, 33, 34, miR-15/1620, 35, miR-142–3p/miR-142–5p20, 33, 36, 37, 38, miR-34231, miR-3020, miR-18131, 36, miR-10134 and the let-7 family14 of miRNAs are downregulated after activation, through an as of yet unknown mechanism.
Figure-1.
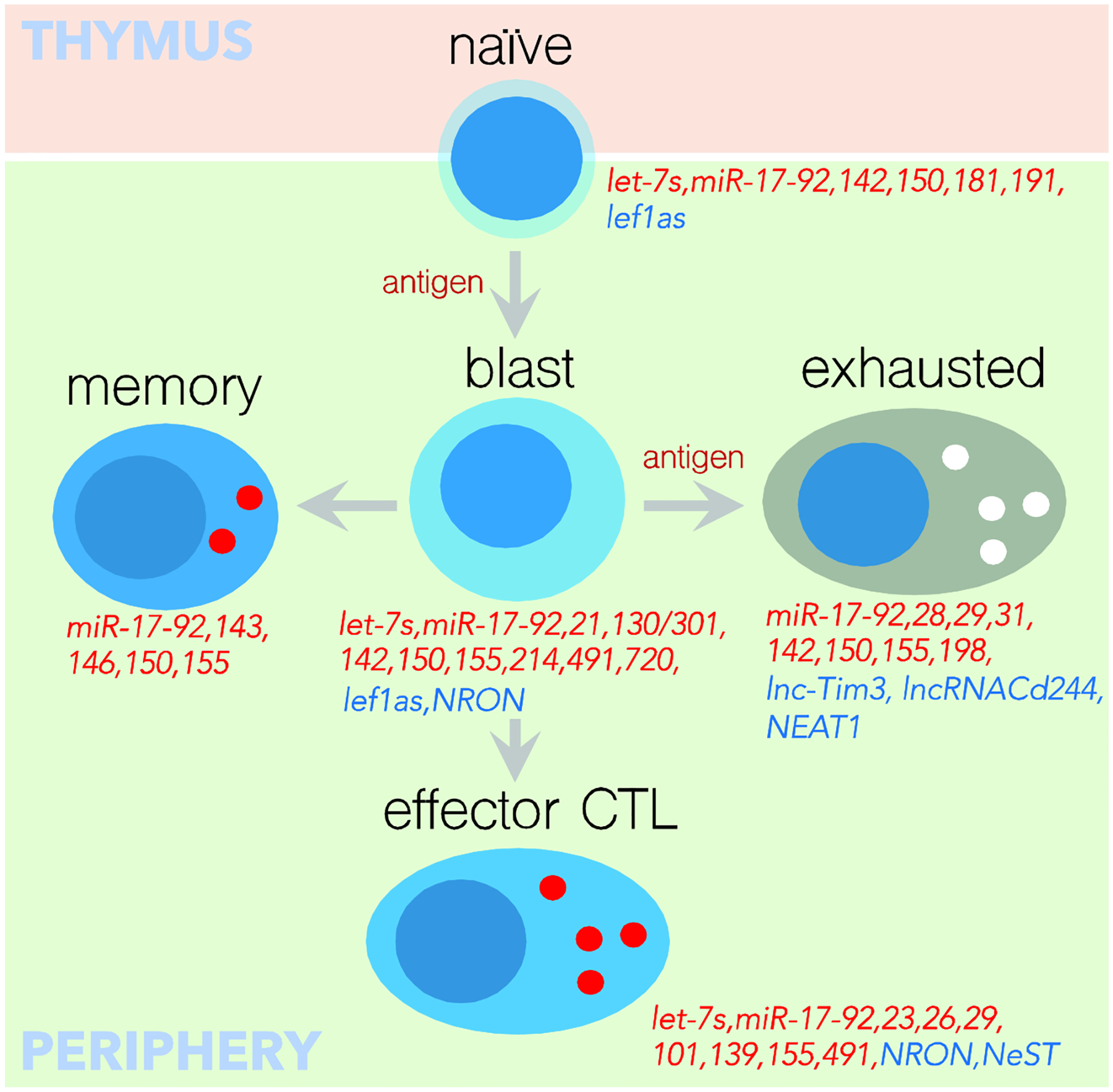
Validated miRNAs (in red) and lncRNAs (in blue) that play a role in CD8 T cell development and differentiation.
It was shown that miRNAs positively regulate the quiescent state of naïve cells by promoting IL-7 cytokine signaling and preventing cell activation by tonic TCR/MHC signals. For example, homeostatic cytokine signaling was shown to be dependent on miR-19139, which is expressed in both thymocytes and mature T cells. Specifically, miR-191 targets insulin receptor substrate 1 (IRS1), an antagonist of STAT5 activation that in turn is necessary for IL-7 signaling. Thus, upon T cell specific deletion of miR-191, aberrant IRS1 over-expression led to compromised STAT5 signaling and therefore gradual loss of lymphocytes, including naïve CD8 T cells. Perhaps the most notable miRNAs demonstrated to play a role in the maintenance of naïve CD8 T cells is the let-7 family of miRNAs, which are abundantly expressed in naïve CD8 T cells14, 20, 40, 41. Loss of let-7 miRNA expression in naïve CD8 T cells resulted in upregulation of the activation marker CD44 and heightened expression of the IL-2 receptor beta chain, CD12214. Moreover, let-7 deficiency facilitated the entry of naive cells into the cell cycle, ultimately increasing the rate of proliferation14, 40. Consistent with this report, fetal derived T cells expressing the protein Lin28B, which inhibits let-7 expression, have a hyperactivated phenotype42. Not only is let-7 expression necessary to maintain the quiescent state of naïve CD8 T cells, but also to support the long term survival of CD8 T cells, as indicated by rampant apoptosis of naïve CD8 T cells and lymphopenia in let-7 deficient mice40. The transcripts which let-7 miRNAs target to maintain naïve CD8 T cell quiescence and survival are not yet clear.
It is also important to limit the spontaneous activation of naïve CD8 T cells in the absence of cognate antigen and costimulation, as this may result in anergy43. In fact, miR-150 was demonstrated to suppress such activation of naïve CD8 T cells by inhibiting TMEM20 expression to prevent the accumulation of intracellular calcium and activation of downstream signaling cascades that drive cells into an anergic dysfunctional state44.
The activation of CD8 T cells is dependent on modulation of miRNA expression
The activation of naïve CD8 T cells through TCR-peptide:MHC recognition, costimulation, and cytokines leads to the initiation of several signal transduction cascades. These signaling events trigger T cell reprograming on both chromatin and transcriptional levels, which is driven by transcription factors (e.g. RUNX3, AP-1, NF-AT, NF-kB, Notch, MYC, JAK/STAT) in combination with chromatin remodeling complexes. Interestingly, signaling through the TCR drastically changes the expression of many miRNAs during initial T cell activation. In contrast to the vast majority of miRNAs that are expressed in naïve T cells and are downregulated by TCR signaling, the miR-17–92 cluster31, miR-22120, 31, miR130/30119, 20, miR-22220, 31, miR-2120, 31, 45, 46, miR-14633, 46, 47, 48, miR-2949, miR-3150, miR −193a50, miR-32050, miR −13231, 50, miR-101b50, miR-29850, miR-731, 50, miR-34550, miR-15531, 51, miR-34a52 and miR-720 are directly upregulated through TCR stimulation13, 53, 54, 55 which suggests they may positively regulate and support the activation of CD8 T cells. For example, the rapid upregulation of CD69 on activated T cells is critical for the retention of antigen specific T cells in lymphoid organs during priming, whereas its expression must be downregulated to facilitate effector cell egress to the site of inflammation. It has been reported that downregulation of CD69 is controlled by the miR-130/301 family of miRNAs that are gradually induced upon TCR-signaling19. Moreover, miR-155 limits the expression of the inhibitor of AKT, SHIP1, suppressor of cytokine signaling-1, SOCS1 and the protein tyrosine phosphatase Ptpn2 to amplify TCR and common gamma chain mediated cytokine signals53, 56, 57, 58, to improve effector CD8 T cell survival and function56, 59, 60. During T cell activation, costimulatory signaling through AKT is amplified by miR-21 that increases IFN-g production and consequently improves the anti-tumor function of cytotoxic T lymphocytes (CTLs) in vivo61.
The downstream effect of the afore mentioned transcriptional programs is a rapid proliferative burst, termed clonal expansion, and the acquisition of effector function, both of which significantly increase the demand for biomacromolecules. Accordingly, these transcriptional networks also change the metabolism of the cell by initiating glycolysis and increasing rates of oxidative phosphorylation62, 63. Thus, the initiation of these programs is essential for successful CD8 T cell differentiation. In fact, more evidence indicating that the early events of T cell activation may regulate the subsequent differentiation and fate of effector CD8 T cells has recently emerged64, 65, 66, 67, 68, 69.
Clonal expansion ensures that there are sufficient numbers of antigen specific CD8 T cells to clear an infection, and is initiated first by entry into the cell cycle, which can be observed by exit from the G1 phase into S phase. The phosphatase Cdc25a, and a complex formed by Ccnd2 and Cdk6 is essential for progression into S phase. The let-7 miRNAs target the mRNA of all three of these proteins, and failure to downregulate let-7 severely impairs CD8 T cell proliferation14. In addition, the cell cycle regulator Cdk4 is a direct target of miR-491, such that when miR-491 is overexpressed, CD8 T cell proliferation is inhibited70. The transcriptional network which drives proliferation downstream of these factors, is also regulated by miRNAs. MiR-720 restrains CD8 T cell proliferation by repressing the AP-1 family member FOSB13. Several negative regulators of the cell cycle are also targeted by miRNAs to facilitate clonal expansion, including the transcription factors E2f7 and E2f8, which are direct targets of miR-14237. Cluster of miRNAs, miR-17–92 is induced by NFkB-signaling through TCR-stimulation and inhibits expression of two tumor suppressors, PTEN the negative regulator of TCR-mediated activation and pro-apoptotic molecule Bim71. Overexpression of the miR-17–92 cluster in T cells resulted in lymphoproliferative disease and rampant autoimmunity. It was reported that another miRNA, miR-214, which is induced by co-stimulatory signals, can also accelerate the proliferation of CD8 T cells by targeting PTEN72. To further facilitate clonal expansion, activated CD8 T cells become less dependent on the homeostatic cytokines IL-7 and IL-15 and more dependent on the growth factor IL-273, 74. In fact, activated CD8 T cells upregulate the expression of CD25, the alpha-subunit of high affinity IL-2 receptor, and downregulate miR-150, which has been reported to decrease expression of CD2518.
To accommodate the immediate increase in bioenergetic demands required to support clonal expansion, CD8 T cells modify their metabolism, not only by increasing rates of oxidative phosphorylation, but also initiating glycolysis62, 63. In fact, let-7 works as a molecular brake of this “metabolic switch” by targeting several of the enzymes and transporters involved in glycolysis14. Importantly, the expression of many of these genes is induced by the transcription factor MYC. Although it plays a central role in metabolism, the tight regulation of MYC is essential for T cell survival, as prolonged expression in some cases is associated with hyperproliferation in leukemia cancers75, while in others it can lead to apoptosis76. Accordingly, MYC has been shown to be a direct target of both let-7 and miR-720 in CD8 T cells13, 14. mTOR is a metabolic hub that also facilitates the metabolic switch in activated CD8 T cells. Interestingly, one of the consequences of PTEN suppression by miR-17–92 cluster, is an indirect increase of mTOR activity54, 55.
Increased metabolism is also necessary to produce effector molecules, including cytotoxic proteins that mediate CD8 T cell killing of target cells. The acquisition of cytotoxic function is driven by a complex network of transcription factors including Eomes, T-bet and Blimp-1. It has been noted that many of the genes involved in CD8 T cell differentiation may not be accessible at first, and the chromatin will need to be remodeled before a transcription factor can bind to the promoter. Downregulation of two miRNAs miR-26a and mir-101 upon T cell activation, has been described to promote generation of polyfunctional effector cells by de-repressing the histone methyltransferase and transcriptional repressor, EZH2 which positively regulates Notch signaling by suppressing its transcriptional inhibitors NUMB and FBXW734.
Once the chromatin has been made accessible, transcription factors can then bind target gene promoters and initiate their transcriptional programs. Upon TCR engagement, upregulation of miR-155 has been reported to enhance T-bet expression by targeting the SHIP-1 phosphatase to drive IFN-g production while restraining type I interferon signaling and thus inducing STAT5 activation during anti-tumor and anti-viral responses77, 78, 79, 80. Consistent with these observations, mice deficient in miR-155 have impaired responses to both bacterial and acute viral infections, and in anti-tumor responses, while over expression of miR-155 enhanced anti-tumor responses by inhibiting SOCS1 and SHIP1, and therefore promoting chromatin remodeling through upregulation of PRC2-associated factor Phf1956, 57, 81. A very elegant study performed by the Rudensky group took the issue even further by generating knockin mice with the mutated miR-155 site in the 3’UTR region of the SOCS1 gene53. Interestingly, they demonstrated that miR-155-dependent regulation of SOCS1 in CD8 T cells is necessary for CD8 T cell responses during persistent chronic infection, and is dispensable during acute infection, suggesting that miR-155 regulates the function of CD8 T cells in acute viral infection through other targets.
Interestingly, several miRNAs were described as negative regulators of this program, and thus their downregulation is essential for efficient cytotoxic responses. MiR29, and the let-7 miRNAs directly target Eomes and T-bet, consequently inhibiting expression of Granzymes, Perforin, and IFN-g14, 82, 83. Moreover, miR-23a inhibits Blimp-1 to suppress cytotoxic function84. In addition, the effector molecules themselves may also be regulated by miRNAs. It has been shown that miR-29 may also directly inhibit IFN-g expression85, while miR-139–3p specifically targets perforin18, adding another layer of regulatory complexity. In addition, it was observed that miR-491 can inhibit IFN-g production, although no mechanism was described70. It is important to mention that many processes that control the activation and differentiation of CD8 T cells have a profound impact on the formation of memory exhaustion suggesting potential importance of miRNAs regulating them in CD8 T cell fate (FIG-1).
miRNAs drive formation of the memory pool
Transition through the effector stage of CD8 T cell differentiation and survival during contraction is essential for the proper development of a functional memory pool. This includes changing cytokine dependence back to the homeostatic cytokines, IL-7 and IL-15, as they promote long term survival28, 74. Mir-146a has been shown to be important for the resolution of the T cell immune response by inhibiting IL-2 production through AP-147, 48, 86. It has also been suggested to be important for surviving contraction as it acts as an anti-apoptotic factor by directly targeting and suppressing the expression of FADD, Fas-associated death domain gene47. Mir-143 was also demonstrated to decrease cell apoptosis in a HER2 chimeric antigen receptor-T (CAR-T) cell model87. Another important component of memory CD8 T cell survival and function is the switch back to slow proliferation and a reduction in glycolytic metabolism. For example, re-expression of miR-143 in memory T cells facilitates metabolic reprograming by directly targeting GLUT1 and suppressing glycolysis87. Conversely, miRNAs such as miR-17/92 are silenced in memory T cells, leading to inhibition of mTOR signaling through upregulation of upstream inhibitors of this pathway and therefore a reduction of effector-like metabolism55. Another TCR-induced miRNA, miR-155, has a similar pattern of expression to miR-17/92 and was shown to be important in suppressing the differentiation of memory T cells. Specifically, miR-155 deficient T cells were demonstrated to preferentially differentiate into the extremely long-lived central memory subset88. This would be consistent with observations that high miR-155 expression is associated with T cell exhaustion. The role of miR-150 in CD8 memory lymphocytes is less clear. One report suggested that miR-150 is required for the function of memory T cells upon restimulation in vitro and in vivo29. However, later TCR-mediated downregulation of miR-150 has been reported to promote memory CD8 T cell differentiation and recall responses by derepressing the transcription factors FOXO1A and cMyb, which are direct targets of miR-150, and are responsible for the expression of Eomes and the pro-survival factors Bcl2 and Bclxl89, 90. The importance of cMyb regulation was further emphasized in a recent publication, where cMyb was implicated in differentiation of memory stem cells by antagonizing the expression of transcription factor Zeb2 that is required for terminal differentiation of CD8 T cells91. To fully understand the role of different miRNAs in memory formation more miRNAs and their targets need to be tested using multiple genetic models.
miRNAs in CD8 T cell exhaustion
Our knowledge of the role of miRNAs in exhausted CD8 T cells is extremely limited, but without a doubt could have important clinical implications for manipulating the immune responses, inhibiting autoimmunity and improving therapies against cancer and chronic infection. CD8 T cell exhaustion occurs in pathological conditions, when antigen persists, driving up expression of inhibitory receptors which suppress CD8 T cell function when engaged43, 92. T cells use this negative feedback mechanism to prevent hyperactivation, which may result in a cytokine storm and tissue pathology. Accordingly, although exhausted T cells come at the expense of clearance of the antigen, and are relatively fragile, the maintenance of exhausted T cells is important to prevent tissue damage caused by prolonged inflammation during chronic infection.
It has been shown that TCR-induced miR-155 is highly expressed in terminally exhausted T cells, and its overexpression leads to increased expression of inhibitory receptors. Importantly, miR-155 also supports the maintenance of T cells during chronic infection by facilitating lymphocyte survival53, 93. The inflammatory milieu associated with these infections includes type I interferons. The expression of miR-31 that is upregulated in activated CD8 T lymphocytes through NFAT, drives cell dysfunction by increasing sensitivity to type I interferons, in addition to upregulating the expression of inhibitory receptors50. Furthermore, CD8 T cells isolated from the immuno-suppressive tumor microenvironment of patients with renal cell carcinoma, have elevated expression of miR-29 and miR-198 which compromises cytokine signaling and survival through direct inhibition of JAK3 and Mcl1 expression94. Alternatively, some miRNAs may have the ability to inhibit exhaustion. In fact, in a mouse melanoma model, miR-28 was downregulated in exhausted T cells and was proposed to increase cytokine production of activated T cells by directly binding the 3’UTR of mRNAs of inhibitory receptors PD-1, Tim-3 and Btla95. Mir-150 has also been demonstrated to directly target PD-1, as well as CTLA429 suggesting a potential role in the regulation of exhausted T cells. Moreover, bone marrow chimeras generated from miR-142 KO bone marrow cells yielded fewer ‘exhausted’ T cells upon transfer into lymphoreplete hosts37. Since the role of only a few miRNAs have been tested (FIG-1), it is clear that more work needs to be done in order to fully characterize the impact of miRNAs in the differentiation of exhausted and memory T cells.
lncRNA mediated regulation of gene expression
In contrast to miRNAs, lncRNAs are greater than 200 nt in their mature form. Although also transcribed by RNAPII and processed with a 5’ cap, and a polyA tail, the genomic organization of lncRNAs is somewhat more complex than that of miRNAs and has important consequences for the function of the lncRNA96. Intergenic lncRNAs require their own promoter to be expressed as they are typically at least 1 kb away from a coding region while intronic lncRNAs are spliced out from the coding regions of the gene in which they are located. There are bidirectional lncRNAs that are positioned head-to-head with an adjacent gene, and coopt the use of that promoter in a bidirectional manner. As a result of these types of genomic organization, lncRNAs often overlap with protein-coding loci and serve to regulate the expression of these genes97, 98. The regulatory functions of lncRNAs extend beyond complementary base pairing to target transcripts of coding genes to inhibit expression. LncRNAs can function either in the cytoplasm or the nucleus, and can bind DNA, RNA, and proteins96, 99. The lncRNAs which are retained in the nucleus have been reported to serve as scaffold RNAs within nuclear bodies, to interact with ribonucleoprotein complexes to modify histone complexes, and in association with DNA to create R-loops that directly regulate transcription96, 99, 100, 101, 102. The most common types of lncRNAs are the cytoplasmic natural antisense transcripts (NATs) which are complementary to mRNA transcripts96, 103. These lncRNAs can function either in cis, in which the lncRNA and the target gene are located in the same genomic region, or in trans, where the lncRNA is expressed in a distinct genomic region than the target gene104.
lncRNAs in CD8 T cell biology
Interestingly, approximately 25% of the genes expressed in mouse and human CD8 T cells are lncRNAs, suggesting a functional importance for these genes105. It has even been suggested that there may be as many lncRNA genes as there are protein coding genes in the mammalian genome105. Actually, many antisense lncRNAs have been found to be highly homologous to protein coding genes and may have arisen due to duplication events106. The majority of these lncRNAs are expressed simultaneously with the mRNAs of protein coding genes to which they bind98. For example, the transcription factor Lef1 has two isoforms, a short isoform (dominantly negative) which is expressed in naïve CD8 T cells, and a long isoform (transcriptional enhancer) which is expressed in activated CD8 T cells. Antisense Lef1 ncRNA, Lef1as, specifically overlaps with the longer isoform and is more highly expressed in naïve cells suggesting its suppressive role98. The field of lncRNAs in CD8 T cell differentiation and function is still very young. Only several lncRNAs have been identified in various subsets of CD8 T cells, and the function of most of them is not known107, 108 (FIG-1). For example, Malat1 is highly expressed in T cell subsets, but its depletion had no effect on CD8 T cell responses to LCMV109. One of the best studied lncRNAs in CD8 T cells is NRON (non-coding RNA repressor of NFAT), which was first found to modulate NFAT localization in a Jurkat cell line110. In fact, NRON is included in the ribonucleoprotein complex that, in addition to kinases, keeps NFAT phosphorylated, and thus out of the nucleus110. Consistent with this role, siRNA mediated knockdown of NRON results in increased IL-2 production from Jurkat cell line111. Moreover, NRON was found to be reduced in cytomegalovirus-driven ageing CD8 T cells in elderly patients, such that uncontrolled NFAT activity may contribute to the accumulation of this population of CD8 T cells112. Importantly, lncRNAs with positive regulatory functions have also been observed in CD8 T cells. NeST, one of the first described lncRNA in T cells113, improves CD8 T cell responses to bacterial and viral infections by acting as an ‘enhancer-like’ lncRNA by binding to the WDR5 component of the H3K4 methylase complex to open chromatin at the Ifng locus, thus promoting its expression114.
Some lncRNAs have also been implicated in exhaustion of CD8 T cells. For example, the lncRNA lnc-Tim3 is highly expressed in the tumor infiltrating lymphocytes (TILs) of hepatocellular carcinoma (HCC) patients. It was found that lnc-Tim3 binds to Tim3 to prevent its interaction with Bat3, leading to the accumulation of inactive Lck and therefore the disruption of TCR-signal115. Moreover, free Bat3 facilitates the nuclear translocation of acetyltransferase p300 and subsequent recruitment of p53 and RelA, leading to cell cycle arrest and potentially a survival signal in exhausted CD8 T cells115, 116. Further, 2B4 expression in CD8 T cells from TB (tuberculosis) patients correlates with expression of lncRNACd244117. LncRNACd244 recruits EZH2 to the promoters of Ifng and Tnfa to establish a closed chromatin state, thus preventing their expression and contributing to T cell dysfunction. It is interesting to mention that some expressed lncRNAs in CD8 T cells overlap with annotated miRNAs (miR-22, miR-142 and miR-17–92 cluster), suggesting an unexplored role of lncRNAs in the regulation of these miRNAs during CD8 T cell differentiation.
Concluding remarks
Although much progress has been made in identifying and understanding the role ncRNAs play in CD8 T cell differentiation and function, there is much that remains to be learned. How the expression of these ncRNAs is of particular importance. Is everything regulated through TCR and costimulatory signals? Or can cytokine signaling have direct consequences on miRNA expression? There is even a possibility ncRNAs to regulate other ncRNAs. It has been reported that the lncRNA NEAT1 is upregulated in CD8 T cells from the hepatocellular carcinoma patients, and that NEAT1 inhibits miR-155 to reduce CD8 T cell cytotoxicity and survival118. The continued study of ncRNAs in CD8 T cells will have important outcomes not only for our understanding of CD8 T cell biology, and RNA biology, but also for the development of novel therapeutic strategies to improve disease.
Highlights.
MicroRNAs and lncRNAs regulate CD8 T cell development and differentiation
Non-coding RNAs guide CD8 T cell differentiation into effector CTLs, memory or exhausted lymphocytes
The most studied microRNAs in CD8 T cell biology are let-7, miR-17–92, miR-150, miR-155
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blackburn SD, Shin H, Freeman GJ & Wherry EJ Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc Natl Acad Sci U S A 105, 15016–15021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn SD et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10, 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paley MA et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wherry EJ T cell exhaustion. Nat Immunol 12, 492–499 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Zajac AJ et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188, 2205–2213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP Metazoan MicroRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwal V, Bell GW, Nam JW & Bartel DP Predicting effective microRNA target sites in mammalian mRNAs. Elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha M & Kim VN Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Treiber T, Treiber N & Meister G Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 20, 5–20 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A & Tuschl T New microRNAs from mouse and human. RNA 9, 175–179 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Jeon K, Lee JT, Kim S & Kim VN MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21, 4663–4670 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai EC Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30, 363–364 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Wang Y et al. Regulation of T cell function by microRNA-720. Sci Rep 5, 12159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells AC et al. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobb BS et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med 201, 1367–1373 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muljo SA et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med 202, 261–269 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumann FM, Yuzefpolskiy Y, Sarkar S & Kalia V Dicer Regulates the Balance of Short-Lived Effector and Long-Lived Memory CD8 T Cell Lineages. PLoS One 11, e0162674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trifari S et al. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci U S A 110, 18608–18613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N & Bevan MJ Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci U S A 107, 21629–21634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H et al. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One 2, e1020 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CZ, Li L, Lodish HF & Bartel DP MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Li QJ et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129, 147–161 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Mildner A et al. MicroRNA-142 controls thymocyte proliferation. Eur J Immunol 47, 1142–1152 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Regelin M et al. Responsiveness of Developing T Cells to IL-7 Signals Is Sustained by miR-17 approximately 92. J Immunol 195, 4832–4840 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Rupp LJ et al. The microRNA biogenesis machinery modulates lineage commitment during alphabeta T cell development. J Immunol 193, 4032–4042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldrath AW & Bevan MJ Selecting and maintaining a diverse T-cell repertoire. Nature 402, 255–262 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Kimura MY et al. IL-7 signaling must be intermittent, not continuous, during CD8(+) T cell homeostasis to promote cell survival instead of cell death. Nat Immunol 14, 143–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surh CD & Sprent J Homeostasis of naive and memory T cells. Immunity 29, 848–862 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Smith NL, Wissink EM, Grimson A & Rudd BD miR-150 Regulates Differentiation and Cytolytic Effector Function in CD8+ T cells. Sci Rep 5, 16399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cobb BS et al. A role for Dicer in immune regulation. J Exp Med 203, 2519–2527 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grigoryev YA et al. MicroRNA regulation of molecular networks mapped by global microRNA, mRNA, and protein expression in activated T lymphocytes. J Immunol 187, 2233–2243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuchen S et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32, 828–839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monticelli S et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol 6, R71 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao E et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat Immunol 17, 95–103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gagnon JD & Ansel KM MicroRNA regulation of CD8 + T cell responses. Non-coding RNA Investigation 3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bronevetsky Y et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med 210, 417–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y et al. Mature T cell responses are controlled by microRNA-142. J Clin Invest 125, 2825–2840 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y et al. PU.1-dependent transcriptional regulation of miR-142 contributes to its hematopoietic cell-specific expression and modulation of IL-6. J Immunol 190, 4005–4013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lykken EA & Li QJ The MicroRNA miR-191 Supports T Cell Survival Following Common gamma Chain Signaling. J Biol Chem 291, 23532–23544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pobezinskaya EL et al. Survival of Naive T Cells Requires the Expression of Let-7 miRNAs. Front Immunol 10, 955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pobezinsky LA et al. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat Immunol 16, 517–524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J et al. Fetal and adult progenitors give rise to unique populations of CD8+ T cells. Blood 128, 3073–3082 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schietinger A & Greenberg PD Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol 35, 51–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim TD et al. MicroRNA-150 modulates intracellular Ca (2+) levels in naive CD8(+) T cells by targeting TMEM20. Sci Rep 7, 2623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carissimi C et al. miR-21 is a negative modulator of T-cell activation. Biochimie 107 Pt B, 319–326 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Wang L et al. Regulation of T lymphocyte activation by microRNA-21. Mol Immunol 59, 163–171 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Curtale G et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 115, 265–273 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Yang L et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med 209, 1655–1670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandiran K et al. Notch1 primes CD4 T cells for T helper type I differentiation through its early effects on miR-29. Mol Immunol 99, 191–198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moffett HF et al. The microRNA miR-31 inhibits CD8(+) T cell function in chronic viral infection. Nat Immunol 18, 791–799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haasch D et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol 217, 78–86 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Hart M et al. miR-34a: a new player in the regulation of T cell function by modulation of NF-kappaB signaling. Cell Death Dis 10, 46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu LF et al. A Single miRNA-mRNA Interaction Affects the Immune Response in a Context- and Cell-Type-Specific Manner. Immunity 43, 52–64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salaun B et al. Differentiation associated regulation of microRNA expression in vivo in human CD8+ T cell subsets. J Transl Med 9, 44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu T et al. Temporal expression of microRNA cluster miR-17–92 regulates effector and memory CD8+ T-cell differentiation. Proc Natl Acad Sci U S A 109, 9965–9970 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dudda JC et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity 38, 742–753 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lind EF, Elford AR & Ohashi PS Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol 190, 1210–1216 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Yang J et al. Unexpected positive control of NFkappaB and miR-155 by DGKalpha and zeta ensures effector and memory CD8+ T cell differentiation. Oncotarget 7, 33744–33764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ji Y & Gattinoni L miR-155 releases the brakes on antitumor T cells. Oncoimmunology 4, e1026533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji Y et al. miR-155 augments CD8+ T-cell antitumor activity in lymphoreplete hosts by enhancing responsiveness to homeostatic gammac cytokines. Proc Natl Acad Sci U S A 112, 476–481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He W et al. MiR-21 is required for anti-tumor immune response in mice: an implication for its bi-directional roles. Oncogene 36, 4212–4223 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Geltink RIK, Kyle RL & Pearce EL Unraveling the Complex Interplay Between T Cell Metabolism and Function. Annu Rev Immunol 36, 461–488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward PS & Thompson CB Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang JT et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315, 1687–1691 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Kaech SM & Ahmed R Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol 2, 415–422 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kakaradov B et al. Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol 18, 422–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.King CG et al. T cell affinity regulates asymmetric division, effector cell differentiation, and tissue pathology. Immunity 37, 709–720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verbist KC et al. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532, 389–393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D et al. The Transcription Factor Runx3 Establishes Chromatin Accessibility of cis-Regulatory Landscapes that Drive Memory Cytotoxic T Lymphocyte Formation. Immunity 48, 659–674 e656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu T et al. MicroRNA-491 regulates the proliferation and apoptosis of CD8(+) T cells. Sci Rep 6, 30923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao C et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol 9, 405–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jindra PT, Bagley J, Godwin JG & Iacomini J Costimulation-dependent expression of microRNA-214 increases the ability of T cells to proliferate by targeting Pten. J Immunol 185, 990–997 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaech SM et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 4, 1191–1198 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Schluns KS, Kieper WC, Jameson SC & Lefrancois L Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol 1, 426–432 (2000). [DOI] [PubMed] [Google Scholar]
- 75.King B et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chou C et al. c-Myc-induced transcription factor AP4 is required for host protection mediated by CD8+ T cells. Nat Immunol 15, 884–893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dickey LL, Worne CL, Glover JL, Lane TE & O’Connell RM MicroRNA-155 enhances T cell trafficking and antiviral effector function in a model of coronavirus-induced neurologic disease. J Neuroinflammation 13, 240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gracias DT et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol 14, 593–602 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hope JL et al. The Transcription Factor T-Bet Is Regulated by MicroRNA-155 in Murine Anti-Viral CD8(+) T Cells via SHIP-1. Front Immunol 8, 1696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huffaker TB et al. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J Biol Chem 292, 18530–18541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji Y et al. miR-155 harnesses Phf19 to potentiate cancer immunotherapy through epigenetic reprogramming of CD8(+) T cell fate. Nat Commun 10, 2157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steiner DF et al. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity 35, 169–181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wissink EM, Smith NL, Spektor R, Rudd BD & Grimson A MicroRNAs and Their Targets Are Differentially Regulated in Adult and Neonatal Mouse CD8+ T Cells. Genetics 201, 1017–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin R et al. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest 124, 5352–5367 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma F et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol 12, 861–869 (2011). [DOI] [PubMed] [Google Scholar]
- 86.Huffaker TB et al. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep 2, 1697–1709 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang T et al. miR-143 Regulates Memory T Cell Differentiation by Reprogramming T Cell Metabolism. J Immunol 201, 2165–2175 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Tsai CY, Allie SR, Zhang W & Usherwood EJ MicroRNA miR-155 affects antiviral effector and effector Memory CD8 T cell differentiation. J Virol 87, 2348–2351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ban YH et al. miR-150-Mediated Foxo1 Regulation Programs CD8(+) T Cell Differentiation. Cell Rep 20, 2598–2611 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Chen Z et al. miR-150 Regulates Memory CD8 T Cell Differentiation via c-Myb. Cell Rep 20, 2584–2597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gautam S et al. The transcription factor c-Myb regulates CD8(+) T cell stemness and antitumor immunity. Nat Immunol 20, 337–349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McLane LM, Abdel-Hakeem MS & Wherry EJ CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 37, 457–495 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Stelekati E et al. Long-Term Persistence of Exhausted CD8 T Cells in Chronic Infection Is Regulated by MicroRNA-155. Cell Rep 23, 2142–2156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gigante M et al. miR-29b and miR-198 overexpression in CD8+ T cells of renal cell carcinoma patients down-modulates JAK3 and MCL-1 leading to immune dysfunction. J Transl Med 14, 84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Q et al. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget 7, 53735–53750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yao RW, Wang Y & Chen LL Cellular functions of long noncoding RNAs. Nat Cell Biol 21, 542–551 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Guttman M et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pang KC et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol 182, 7738–7748 (2009). [DOI] [PubMed] [Google Scholar]
- 99.Wang KC & Chang HY Molecular mechanisms of long noncoding RNAs. Mol Cell 43, 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chedin F Nascent Connections: R-Loops and Chromatin Patterning. Trends Genet 32, 828–838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guttman M et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spitale RC, Tsai MC & Chang HY RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6, 539–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Katayama S et al. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566 (2005). [DOI] [PubMed] [Google Scholar]
- 104.Ransohoff JD, Wei Y & Khavari PA The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19, 143–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hudson WH et al. Expression of novel long noncoding RNAs defines virus-specific effector and memory CD8(+) T cells. Nat Commun 10, 196 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Milligan MJ & Lipovich L Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front Genet 5, 476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fu Y, Gao K, Tao E, Li R & Yi Z Aberrantly Expressed Long Non-Coding RNAs In CD8(+) T Cells Response to Active Tuberculosis. J Cell Biochem 118, 4275–4284 (2017). [DOI] [PubMed] [Google Scholar]
- 108.Hrdlickova B et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med 6, 88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yao Y et al. Long noncoding RNA Malat1 is not essential for T cell development and response to LCMV infection. RNA Biol 15, 1477–1486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Willingham AT et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309, 1570–1573 (2005). [DOI] [PubMed] [Google Scholar]
- 111.Sharma S et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A 108, 11381–11386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang YH, Yu XH, Qu GJ, Qiao FF & Han H Reduced expression of the lncRNA NRON is a potential hallmark of the CMV-amplified CD8+ T cell accumulations commonly seen in older humans. Exp Gerontol 115, 46–54 (2019). [DOI] [PubMed] [Google Scholar]
- 113.Vigneau S, Rohrlich PS, Brahic M & Bureau JF Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol 77, 5632–5638 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gomez JA et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152, 743–754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ji J et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis 9, 478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Das M, Zhu C & Kuchroo VK Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev 276, 97–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Y et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A 112, E3883–3892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yan K et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8(+)T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int J Biochem Cell Biol 110, 1–8 (2019). [DOI] [PubMed] [Google Scholar]


