SUMMARY
Here, we apply quantitative chemical proteomics and untargeted lipidomics to assign a polyunsaturated fatty acid (PUFA)-specific triacylglycerol (TAG) lipase activity for diacylglycerol lipase-beta (DAGLβ) in macrophages. We demonstrate that DAGLβ but not DAGLα is expressed and active in bone marrow-derived macrophages (BMDMs) as determined by activity-based protein profiling (ABPP) analysis of SILAC BMDMs. Genetic disruption of DAGLβ resulted in accumulation of cellular TAGs composed of PUFA but not saturated/low unsaturated FA counterparts, which is recapitulated in wild-type macrophages treated with a DAGLβ-selective inhibitor. Biochemical assays with synthetic substrates confirm PUFA-TAGs as authentic DAGLβ substrates. In summary, our findings identify DAGLβ as a PUFA-specific TAG lipase in primary macrophages.
Graphical Abstract
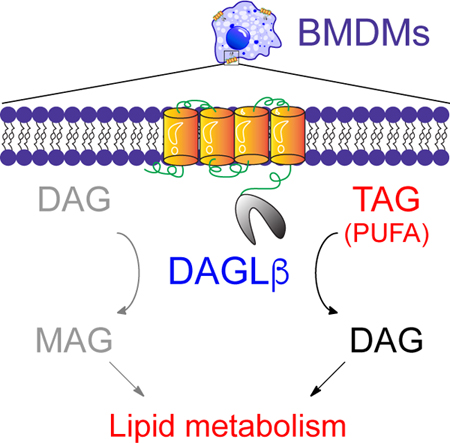
In Brief
DAGLβ-mediated metabolism was disrupted using genetic and pharmacological approaches to assign polyunsaturated fatty acid (PUFA)-esterified triglycerides as DAGLβ substrates in primary macrophages.
INTRODUCTION
Diacylglycerol lipases (DAGLs) hydrolyze arachidonic acid (AA)-esterified diacylglycerols (DAGs) to biosynthesize the principal endocannabinoid 2-arachidonylglycerol (2-AG) (Gao et al., 2010; Tanimura et al., 2010). DAGLs are members of the serine hydrolase superfamily that utilize a catalytic serine for calcium-dependent cleavage of DAGs in a sn-1 selective manner (Bisogno et al., 2003; Long and Cravatt, 2011). The function of DAGL isoforms is regulated by cell type- and tissue-specific expression in mammals. DAGLα expression is enriched in central tissues (e.g. brain and spinal cord) where this isoform regulates 2-AG involved in synaptic activity and neuroinflammation (Gao et al., 2010; Ogasawara et al., 2016; Tanimura et al., 2010; Viader et al., 2016). DAGLβ activity is enriched in macrophages (Hsu et al., 2012), microglia (Viader et al., 2016), and dendritic cells (Shin et al., 2019) where this isoform regulates 2-AG and downstream metabolic products including prostaglandins important for innate immune responses underlying peripheral and central inflammation.
To date, the substrate specificity of DAGLs have been assigned principally by targeted liquid chromatography-mass spectrometry (LC-MS) metabolomics approaches (Lee and Yokomizo, 2018). These analyses were performed with a predefined set of lipids based on insights from prior studies of substrate specificity in cells and/or animals. Targeted metabolomics has been vital for identifying DAGs, endocannabinoids, and prostaglandins as DAGLα- and DAGLβ-regulated lipids involved in mediating cell biological and animal physiological functions (Baggelaar et al., 2015; Hsu et al., 2012; Ogasawara et al., 2016; Shin et al., 2019; Viader et al., 2016). While critical for understanding DAGL biology, this targeted approach is not well-suited for global discovery of new lipids regulated by these enzymes. Consequently, the full range of substrates regulated (directly or indirectly) by DAGLs remains underexplored.
Here, we performed untargeted metabolomics to map putative lipid substrates of DAGLβ in primary macrophages. Genetic disruption of DAGLβ resulted in cellular accumulation of a rare subset of triacylglycerols (TAGs) in primary macrophages. Specifically, our metabolomics profiling identified TAGs containing polyunsaturated fatty acids (PUFAs) as putative DAGLβ substrates, which we could recapitulate by pharmacology using DAGLβ-selective inhibitors. PUFA-TAGs were confirmed as authentic DAGLβ substrates by biochemical assays using synthetic lipid standards. Collectively, our findings identify DAGLβ as a neutral lipase that is capable of hydrolyzing DAGs and TAGs with specificity that is dependent on fatty acyl composition.
RESULTS
Activity-based protein profiling (ABPP) of DAGLs in bone marrow-derived macrophages
Bone marrow-derived macrophages (BMDMs) were differentiated from bone marrow of mice using L929-supplemented media for 7 days following published procedures (Serbulea et al., 2018) (see STAR Methods for details). We chose BMDMs for our global metabolomics studies because of the ability to generate homogeneous cultures of both wild-type (WT) and DAGLβ knockout (KO) primary macrophages. To determine whether BMDMs express active DAGLα and/or DAGLβ, we performed activity-based protein profiling (ABPP (Niphakis and Cravatt, 2014)) studies with our DAGL-tailored probe HT-01 (Hsu et al., 2012). We detected prominent HT-01 labeling of a ~70 kDa protein band that was present in proteomes from Daglb+/+ but absent from Daglb−/− BMDMs (Fig S1A). We also performed HT-01-based ABPP comparisons of Dagla+/+and Dagla−/− BMDM proteomes but could not detect active DAGLα (Fig S1A). These ABPP results are in contrast with brain proteomes where we could reliably measure endogenous DAGLα and DAGLβ using HT-01 (Fig S1B). Thus, we focused the remainder of our macrophage studies on the DAGLβ isoform. In summary, gel-based ABPP profiling studies identified DAGLβ as a principal DAGL isoform expressed in BMDMs.
LC-MS metabolomics identifies PUFA-TAGs as putative DAGLβ substrates in BMDMs
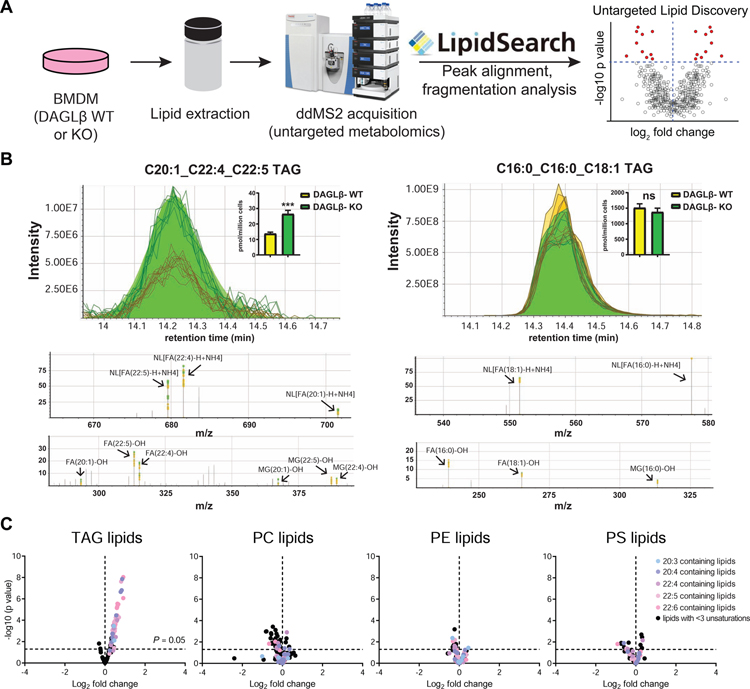
Next, we deployed an untargeted MS-metabolomics approach to gain an unbiased global evaluation of alterations in lipid composition of DAGLβ-disrupted BMDMs. Total lipids (i.e. lipidome) were extracted from Daglb+/+ and Daglb−/− BMDMs using the Folch method (Folch et al., 1957) and analyzed by liquid chromatography-mass spectrometry (LC-MS) on a high-resolution Q-Exactive Plus mass spectrometer configured for untargeted data-dependent acquisition (ddMS2, Fig 1A). Lipid precursor ions (MS1) selected for fragmentation were identified using LipidSearch™ that matches precursor (MS1) and fragment (MS2) spectra to an in-silico database containing more than 1.5 million lipid molecules and their predicted precursor and fragment ions (Taguchi et al., 2007). Identified lipids were scored based on fatty acid (FA) chain composition, which is determined from fragment ions corresponding to neutral loss of FAs, and only those with high quality (Grade A or B signifying 2–4 lipid class-specific diagnostic fragment ions and less than 5 ppm precursor mass deviation) were selected for further manual evaluation (Fig 1B; see STAR Methods for details). We detected on average ~980 lipids per BMDM sample analysis, which consisted of positive and negative ion mode LC-MS analyses. See Supplemental Dataset S1 for full list of lipids measured.
Figure 1. Untargeted lipidomics reveals PUFA-TAG accumulation in DAGLβ-disrupted BMDMs.
(A) Schematic of untargeted LC-MS lipidomic profiling platform. See STAR Methods for details. (B) Representative peak alignments of MS1 chromatograms and MS2 fragmentation spectra observed for triacylglycerol (TAG) species from LipidSearch™ and manual analyses of the raw MS data. The following diagnostic ions enable confident TAG identification: neutral loss of each fatty acyl chain (NL[FA-H+NH4]), precursor ion ([TAG+NH4]+), and fragmentation of the remaining glycerol backbone and the released fatty acyl chains (monoacylglycerol (MG)-OH and FA-OH). Inset: bar graphs showing the abundances of each TAG species from the MS1 chromatograms (Data shown represent mean ± s.e.m.; n=5 biological samples). Statistical significance was determined using a Holm-Sidak correction following a two-sided binomial test (***P < 0.001). (C) Volcano plot showing changes in abundance of lipid species detected in DAGLβ KO compared with WT BMDMs from global lipidomics analyses (n=5 biological samples). Dashed lines indicated statistical significance cutoff (horizontal; P = 0.05) and direction of fold change (vertical; left and right quadrant denote decreased and increased abundance, respectively, in KO/WT comparisons). Results show preferential accumulation of PUFA containing TAG species in DAGLβ KO vs. WT BMDM lipidomes. Phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) lipids showed negligible changes in both PUFA (colored circles) and species with low units of unsaturation (lipids with <3 unsaturation units, black circles). Statistical significance was determined using a Holm-Sidak correction following a two-sided binomial test. All data shown are representative of two independent experiments (n=2 biologically independent experiments).
From these data, we could quantitate >750 distinct species after filtering for lipids with high quality annotations of FA composition. We validated our BMDM LC-MS metabolomics approach by demonstrating that genetic disruption of DAGLβ resulted in lipid alterations that matched previous reports in peritoneal macrophages (Hsu et al., 2012). Specifically, metabolomics analysis showed accumulation of AA-esterified DAG substrates along with cellular reduction of the downstream products 2-AG and AA (Fig S2A). Interestingly, our untargeted findings revealed a striking alteration in TAG lipid profiles in DAGLβ-disrupted BMDMs. We observed statistically significant accumulation of TAG species that contained polyunsaturated fatty acids (PUFAs) in Daglb−/− compared with Daglb+/+ BMDMs (Fig 1C). Fatty acid composition of accumulated TAGs included species containing omega-6 (arachidonic acid, C20:4) and omega-3 fatty acids (docosapentaenoic acid, C22:5; Fig 1B). TAGs containing saturated or low degrees of unsaturation (<3) showed negligible differences in Daglb−/− compared with Daglb+/+ lipidomes (Fig 1C). To determine if PUFA-TAG changes were due to general alterations in macrophage lipidomes, we compared levels of phospholipids between Daglb−/− and Daglb+/+ lipidomes and observed negligible changes in phosphatidyl-ethanolamine (PE), -choline (PC), and -serine (PS) species (Fig 1C).
Differentiation of SILAC BMDMs for quantitative chemoproteomic profiling
To complement our KO findings, we used our previously reported DAGLβ inhibitor KT109 and matching negative control compound KT195 (Hsu et al., 2012) to determine whether acute blockade of DAGLβ affected TAG levels in BMDMs. We first determined by gel-based ABPP using HT-01 that treatment of BMDMs with KT109 but not KT195 resulted in concentration-dependent blockade of DAGLβ (Fig S1C). From our gel-based ABPP studies, we selected optimal treatment conditions (200 nM compounds, 4 h, 37°C) for LC-MS ABPP to determine selectivity across the serine hydrolase family.
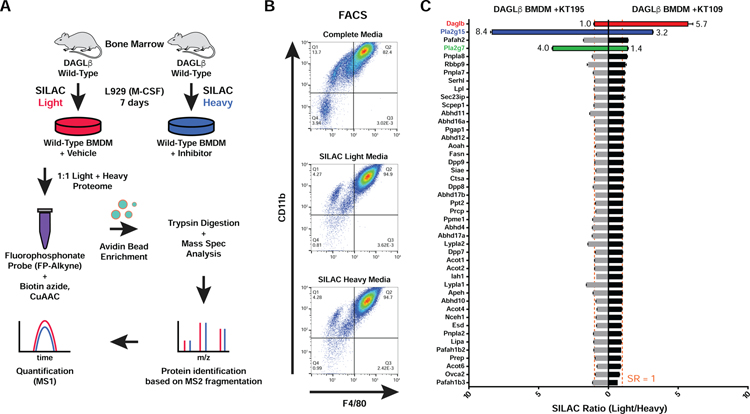
We established a quantitative proteomics strategy to assess compound potency and selectivity in inhibitor-treated primary macrophages (Fig 2). We hypothesized that isotopically-labeled lysine and arginine (i.e. SILAC (Mann, 2006)) would be efficiently incorporated into macrophage proteomes during the rapid differentiation and proliferation of bone marrow stem cells to produce SILAC light- and heavy-labeled BMDMs. For these studies, BMDMs were differentiated from bone marrow of mice using L929 supplemented media (SILAC or standard media) for 7 days as depicted in Fig 2A and as described in STAR Methods. We used fluorescence activated cell sorting (FACS) and established markers for BMDMs (F4/80 and CD11b) to compare macrophage content under standard and SILAC differentiation conditions. We observed comparable numbers of F4/80+CD11b+ macrophages in standard and SILAC media (both light and heavy) conditions, which confirmed that BMDMs differentiated under SILAC media were phenotypically similar to standard media counterparts (Fig 2B).
Figure 2. ABPP-SILAC analysis of inhibitor activity against serine hydrolases in SILAC BMDMs.
(A) Generation of SILAC primary macrophage via incorporation of isotopically-labeled amino acids during BMDM differentiation. (B) Flow cytometry analyses confirmed that BMDMs differentiated using standard or SILAC media are phenotypically equivalent as judged by analysis of macrophage cell surface markers (CD11b and F4/80). (C) ABPP-SILAC analysis of BMDMs treated with DMSO (light) or inhibitor (heavy; 200 nM of KT109 or KT195, 4 h, 37 °C) followed by labeling with FP-alkyne probe (10 μM, 2 h, 25 °C). A SILAC ratio (SR) of ~1 indicates no change in serine hydrolase activity between treatment conditions (vertical dashed line; SR = 1). Data shown represent mean ± s.e.m. All data shown are representative of two independent experiments (n=2 biologically independent experiments).
Next, we evaluated global changes in serine hydrolase activity in proteomes from DMSO vehicle-(light) compared with KT109-treated (heavy) BMDMs. After inhibitor treatment, cells were lysed, proteomes labeled with the broad-spectrum serine hydrolase probe FP-alkyne, and SILAC light- and heavy-labeled samples combined. Next, biotin-azide was conjugated by copper catalyzed azide-alkyne cycloaddition (CuAAC (Rostovtsev et al., 2002)) followed by avidin enrichment, on-bead trypsin digestion, and tandem liquid chromatography-mass spectrometry LC-MS/MS ABPP to measure changes in serine hydrolase activity in BMDM proteomes (Fig 2C). The activity profiles across the 45 detected serine hydrolases in BMDMs were largely unchanged (median SILAC ratio (light/heavy) or SR of ~1) between proteomes from vehicle and KT109-treated cells with the exception of DAGLβ, which showed potent blockade of activity (SR >5, Fig 2C). We also identified PLA2G15 as a moderately-competed target of KT109 (SR >3, Fig 2C), which matched our previous ABPP studies in primary macrophages (Hsu et al., 2012). In contrast, the negative control molecule KT195 was largely inactive against DAGLβ (SR = 1) but showed potent and moderate activity against PLA2G15 and PLA2G7, respectively, in compound-treated BMDMs (Fig 2C). Finally, we could not detect DAGLα activity in our LC-MS studies, which is in agreement with our HT-01-based ABPP studies (Fig S1A and C).
In summary, our ABPP-SILAC studies identified DAGLβ as the active DAGL isoform in BMDMs and confirmed that KT109 and KT195 can be used as complementary probes to discern DAGLβ-specific from non-specific pharmacological effects in our metabolomics studies.
DAGLβ inhibitors recapitulate PUFA-TAG accumulation observed in KO BMDMs
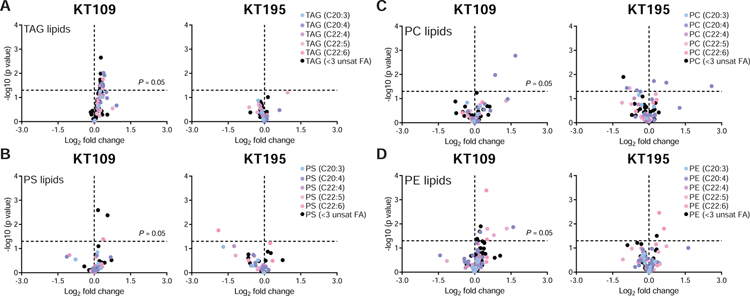
From our ABPP-SILAC studies, we selected compound treatment conditions (200 nM compounds, 4 h, 37 °C) for untargeted metabolomics analyses of BMDMs. We first validated that treatment of BMDMs with KT109 but not KT195 resulted in lipid alterations that support functional blockade of DAGLβ (Fig S2B). Closer inspection of our untargeted metabolomics data revealed a moderate but statistically significant (P < 0.05) PUFA-TAG accumulation in KT109- but not KT195-treated BMDMs (Fig 3A). Akin to Daglb−/− BMDMs, we observed minimal accumulations in saturated-TAG species in KT109-treated cells (Fig 3A). Importantly, the PUFA-TAG changes observed were specific given that acute blockade of DAGLβ did not result in obvious perturbations in phospholipids detected including PS, PC, and PE species of varying fatty acyl compositions (Fig 3B–D).
Figure 3. Pharmacological disruption recapitulates DAGLβ KO BMDM lipidomic profiles.
(A) Untargeted lipidomics profiling revealed statistically significant accumulation of PUFA-TAGs in DAGLβ WT BMDMs treated with KT109 but not KT195 (200 nM compounds, 4 h, 37 °C; n=5 biological samples). (B-D) Analysis of additional lipid species including PS, PC, and PE reveal KT109-mediated lipid changes appear to be specific for PUFA-TAGs (n=5 biological samples). Dashed lines in volcano plots indicate statistical significance cutoff (horizontal; P = 0.05) and direction of fold change in inhibitor/DMSO comparisons (vertical; left quadrant – decreased abundance, right quadrant - increased abundance). Statistical significance was determined using a Holm-Sidak correction following a two-sided binomial test. All data shown are representative of two independent experiments (n=2 biologically independent experiments).
To assess the impact of DAGLβ inactivation on bulk TAG levels in BMDMs, we compared the abundance of PUFA- and saturated-TAGs detected in BMDM lipidomes based on ion intensities of MS1 peaks. Unlike PC, PE, and PS lipids, the abundance of PUFA-TAGs represented a minor fraction (~29%) of total TAGs, supporting that disruption of DAGLβ will not likely result in general TAG accumulation as observed for other TAG lipases (Haemmerle et al., 2006; Inloes et al., 2014) (Fig S3). In summary, our pharmacology supports our genetic findings of an additional metabolic function for DAGLβ in BMDMs.
Biochemical validation that DAGLβ exhibits PUFA-specific TAG lipase activity
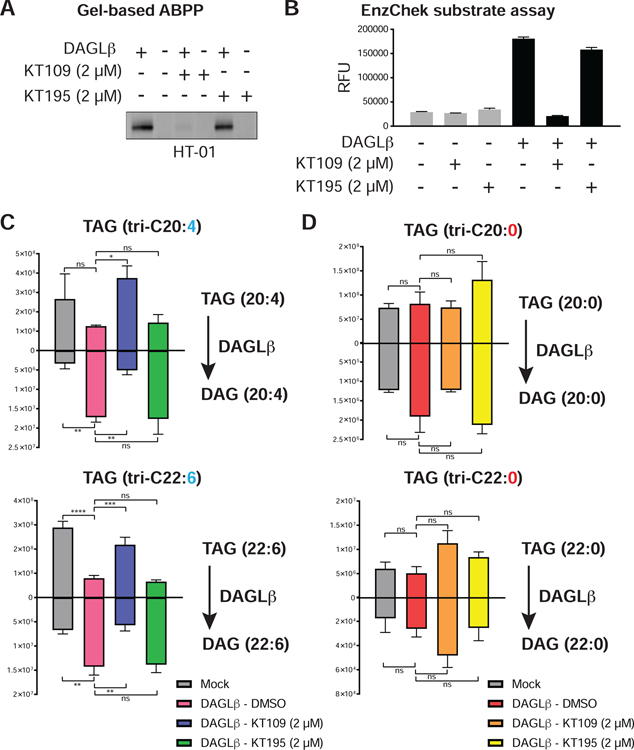
We next examined whether DAGLβ could hydrolyze synthetic TAGs in a biochemical substrate assay. These studies were important for establishing TAGs as authentic substrates of DAGLβ given that changes observed in cellular studies could arise from secondary effects due to DAGLβ blockade. We transiently overexpressed mouse DAGLβ in HEK293T cells and measured recombinant DAGLβ activity in membrane fractions by gel-based ABPP with HT-01. We confirmed that treatment with KT109 but not KT195 blocked HT-01 labeling of recombinant DAGLβ overexpressed-HEK293T membrane proteomes (Fig 4A). We also demonstrated that recombinant DAGLβ showed robust hydrolysis of a fluorogenic lipase substrate compared with non-transfected mock samples and this catalytic activity could be blocked by >80% with KT109 but not KT195 pretreatment (EnzChek™, Fig 4B and Fig S4).
Figure 4. Biochemical validation of PUFA-TAGs as authentic DAGLβ substrates.
(A) Gel-based ABPP analyses confirmed recombinant mouse DAGLβ overexpressed in HEK293T membrane proteomes was active as judged by HT-01 probe labeling (1 μM probe, 30 min, 37 °C) that could be blocked in vitro by KT109 but not KT195 (2 μM inhibitor, 30 min, 37 °C). (B) A fluorogenic lipase substrate assay (EnzChek™) was used to confirm recombinant DAGLβ was catalytically active (n=4 biological samples). A micelle-based substrate assay was used to evaluate PUFA- (tri-C20:4 and -C22:6) and matching saturated-TAGs (tri-C20:0 and -C22:0) as substrates for recombinant DAGLβ (n=3 biological samples). Recombinant DAGLβ-HEK293T membrane proteomes were incubated with TAG substrates and TAG hydrolysis activity was monitored by targeted LC-MS on a Q-Exactive Plus mass spectrometer configured for parallel reaction monitoring (PRM) of lipid substrate and product abundances. Recombinant DAGLβ efficiently hydrolyzed PUFA-TAGs (C) but not saturated TAG counterparts (D). All data shown represent mean ± s.e.m. and are representative of two independent experiments (n=2 biologically independent experiments). See STAR Methods for details of substrate assays and LC-MS studies.
Next, we tested whether recombinant DAGLβ could hydrolyze synthetic TAG substrates and if this lipase showed substrate preference based on fatty acyl composition. We compared saturated and unsaturated TAG pairs to directly test the importance of FA unsaturation on TAG lipase activity of recombinant DAGLβ. Incubation of recombinant DAGLβ membrane proteomes with triarachidonin (C20:4 FA) or tridocosahexaenoin (C22:6 FA) resulted in robust hydrolysis activity that could be blocked with KT109 but not KT195 pretreatment, which supports DAGLβ-specific activity (Fig 4C). The saturated TAG counterparts (C20:0 and C22:0 FA) were not hydrolyzed by DAGLβ to any appreciable degree (Fig 4D). In summary, our findings support DAGLβ as a TAG lipase with specificity for lipid species containing PUFAs.
DISCUSSION
Assigning substrate specificity to lipid enzymes in vivo is challenging but key to understanding cell metabolism and signaling (Saghatelian et al., 2004). Here, we apply chemical proteomics and metabolomics to assign a PUFA-specific triacylglycerol lipase activity for DAGLβ in primary macrophages. Combined with previous reports that DAGLβ can regulate cellular DAG levels (Hsu et al., 2012), our current findings describe DAGLβ as a TAG/DAG lipase that can remodel lipidomes to support the metabolic and signaling demands of macrophages.
We employed untargeted MS-metabolomics and ABPP-SILAC to discover a striking metabolic phenotype in DAGLβ-inactivated primary macrophages that is characterized by accumulation of a minor subset of TAGs that are composed of PUFA chains. Our findings are in agreement with previous studies that showed negligible TAG lipase activity of DAGLβ against mono-unsaturated TAG substrates (Bisogno et al., 2003) (Fig 1B). Instead, we demonstrated using both genetic and pharmacological approaches that DAGLβ blockade results in cellular accumulation of PUFA-TAGs while saturated and low unsaturated counterparts (<3 unsaturation units) remain largely unchanged (Fig 1C and 3A). The integration of pharmacological and genetic disruption models was important to provide evidence in support of direct DAGLβ-specific effects on TAG metabolism given that long-term loss of protein could result in network wide compensation. To validate PUFA-TAGs as authentic substrates, we performed biochemical assays to show DAGLβ hydrolyzed PUFA-TAG but not saturated FA counterparts, which further supports its unique specificity for TAG substrates (Fig 4C and D).
Our analysis of relative lipid abundances showed that the majority of TAG species found in BMDMs were composed of saturated/low unsaturated FAs (<3 unsaturation units) and that PUFA-TAGs represented a low abundance subset. The differences in lipid abundances based on FA composition was not a general phenomenon in BMDMs because PC, PS, and PE showed more equivalent distribution between PUFA- and saturated-lipid species (Fig S3). Thus, DAGLβ disruption is not likely to result in general TAG accumulation as observed for adipose triglyceride lipase (ATGL) (Chandak et al., 2010), which can lead to metabolic disorders (Haemmerle et al., 2011). The ability of DAGLβ to hydrolyze DAGs and TAGs in macrophages suggests a broader role of this enzyme in neutral lipid metabolism and provides a potential connection between endocannabinoid and bioenergetic pathways. Future studies will focus on whether DAGLβ is involved in regulation of lipolysis (Zechner et al., 2012).
In conclusion, our findings illustrate the utility of unbiased global metabolomics to assign unique substrate specificities of enzymes in living systems. DAGLβ has emerged as a promising target for attenuating innate immune activation in animal models of pain and (neuro)inflammation (Shin et al., 2018; Viader et al., 2016; Wilkerson et al., 2016) and our results illuminate additional lipid substrates regulated by this enzyme to mediate the metabolic and signaling functions of immune cells.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to the Lead Contact, Ku-Lung Hsu (kenhsu@virginia.edu). This study did not generate any unique/stable reagents however, all other materials are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice.
All studies were conducted in 6–12-week-old C57BL/6J, DAGLβ, and DAGLα mice. Mice were housed in the MR6 animal facility (UVa), and Gilmer animal facility (UVa). All studies were carried out under a protocol approved by the ACUC/University of Virginia. All animals were allowed free access to a standard chow diet and water. The animals were housed according to the ACUC policy on social housing of animals. For experiments involving BMDMs, both male and female mice were used in performing described work. Each experiment was controlled by using same-sex littermates.
Bone Marrow Derived Macrophages (BMDMs) differentiation.
BMDMs were cultured as previously described (Serbulea et al., 2018). In brief, the bone marrow from the hind legs of both male and female mice was extracted and incubated for 5 min with 0.83% ammonium chloride, to clear erythrocyte progenitors. The bone marrow was cultured with RPMI media supplemented with 10% FBS (Omega Scientific), 5% HEPES (Gibco), 1% antibiotic-antimitotic (penicillin-streptomycin, Gibco), and 10% L929-conditioned media (L929 cells purchased from ATCC). For SILAC BMDMs, the L929-conditioned media was instead supplemented with 10% dialyzed FBS (Omega Scientific) and either “light” L-arginine and L-lysine (100 μg ml−1) or “heavy” [13C6 15N4]L-arginine and [13C6 15N2]L-lysine (100 μg ml−1). The culture continued for 7 days, with media changes every 3–4 days, after which the media was exchanged for one lacking L929-conditioned media. On day 7, the BMDMs were gently separated from petri dishes using 0.25% trypsin (Gibco), followed by centrifugation and media replacement. Finally, BMDMs were plated on non-tissue culture treated petri dishes for various treatments and analyses.
HEK293T Cells.
HEK293T cells were obtained from ATCC and cultured in complete DMEM media (10% FBS (U.S. Source, Omega Scientific) and 1% L-glutamine (Thermo Fisher Scientific)) in 10 cm2 plates. All cells were grown to ~80% confluency in a 37°C incubator with 5% CO2.
L929 Cells.
L929 cells were obtained from ATCC and cultured in complete RPMI media (10% FBS (U.S. Source, Omega Scientific), 2 mM L-glutamine (Thermo Fisher Scientific), 100 μg mL−1 penicillin-streptomycin (Sigma-Aldrich), and 20 mM HEPES (pH 7.4)) in Corning™ CELLSTACK™ Culture Chambers. 678,400 cells were seeded across the two chambers of the cell stack in 423 mL of complete RPMI media in a 37°C incubator with 5% CO 2. Media was harvested from the cell stack after each week for two weeks and replaced with fresh complete RPMI media.
METHOD DETAILS
Reagents.
Unless otherwise specified, reagents used were purchased from Fisher Scientific. Bio-beads SM-2 Adsorbants (Bio-Rad) catalog# 1528920. Polyethyleneimine (Polysciences, Inc.) catalog# 24765, 2-Mercaptoethanol (Sigma-Aldrich) catalog# M6250–250ML, recombinant M-CSF (Peprotech) catalog# 315–02. KT109 (synthesized in house), KT195 (Cayman Chemical Company) catalog# 14818. Anti-FLAG antibody produced in rabbit (Sigma-Aldrich) catalog# F7425, Goat anti-rabbit DyLight 550 (Thermo Fisher Scientific) catalog# 84541. PE/Cy7 anti-mouse/human CD11b antibody (Biolegend) catalog# 101215. Rat anti-mouse F4/80 antibody (Bio-Rad) catalog# MCA497A488T. Arachidonic acid-d8 (Cayman Chemical Company) catalog# 390010, 2-Arachidonyl glycerol-d5 (Cayman Chemical Company) catalog# 362162, Tritetradecanoin (C14:0) (Nu-Chek Prep) catalog# T-140, Trihexadecanoin (C16:0) (Nu-Chek Prep) catalog# T-150, Trioctadecanoin (C18:0) (Nu-Chek Prep) catalog# T-160, Trieicosanoin (C20:0) (Nu-Chek Prep) catalog# T-170, Tridocosanoin (C22:0) (Nu-Chek Prep) catalog# T-180, Triarachidonin (C20:4) (Nu-Chek Prep) catalog# T-295, Trieicosapentaenoin (C20:5) (Nu-Chek Prep) catalog# T-325, Tridocosahexaenoin (C22:6) (Nu-Chek Prep) catalog# T-310. Fetal bovine serum (FBS) and dialyzed FBS were obtained from Omega Scientific.
Overexpression of DAGLβ in HEK293T Cells.
Recombinant mouse DAGLβ protein was produced by transient transfection of HEK293T cells with recombinant DNA. The transient transfection was performed as described previously (Hsu et al., 2012). In brief, HEK293T cells were seeded in 10 cm2 cell culture treated plate at 400,000 cells in complete DMEM (DMEM, 10% FBS, 10 units and 100 μg mL−1 penicillin-streptomycin, 2 mM L-Glutamine). After cells were allowed to grow to 40–60% confluency, 20 μL of sterilized polyethyleneimine (PEI) solution (1 mg mL−1, pH 7.4) and 2.6 μg of DNA were added to 600 μL of DMEM and incubated at 25°C for 30 min. The combined solution was added to 10 cm plate dropwise and mixed gently. Cells were harvested after two days of growth and snap frozen in liquid nitrogen and stored at −80°C freezer until further analysis.
Western Blot Analysis.
Cell lysates were separated via centrifugation at 100,000 x g for 45 min at 4°C. Proteins separated by SDS-PAGE (7.5% po lyacrylamide, TGX Stain-Free Mini Gel) at 150 V for 55 min. Gel transfers were performed using the Bio-Rad Trans-Blot Turbo RTA Midi Nitrocellulose Transfer Kit with a Bio-Rad Trans-Blot Turbo Transfer System (25V, 10 min). The nitrocellulose blot was then incubated in blocking solution (30 mL, 3% BSA in TBS-T (1.5 M NaCl, 0.25 M Tris pH 7.4 in ddH2O)) for 1 h at 25°C with gentle shaking. The blot was then transferred immediately to primary antibody solution (1:1,000 anti-FLAG or 1:10,000 anti-HA in TBS-T) and incubated overnight at 4°C with gentle s haking. The blot was then rinsed 5 times for 5 min in TBS-T, transferred immediately into secondary antibody solution (1:10,000 anti-species DyLight 550 in TBS-T), and incubated for 1 h at 25°C with gentle shaking. The blot was then rinsed 5 times for 5 min in TBS-T, transferred into ddH2O, and imaged by in-blot fluorescence scanning on a ChemiDoc MP Imaging System.
Lipid Extraction.
BMDM and HEK293T cell pellets (3 million cells) were extracted using the Folch extraction method (2:1:1 chloroform:methanol:water) as previously described (Folch et al., 1957). BHT (50 μg mL−1) was added to organic solvents used for extraction. Chloroform/methanol solution (3 mL) was added with 1 mL of resuspended cells in water. Samples were briefly vortexed and centrifuged at 2,000 x g for 5 min. Organic layer was transferred and aqueous layer was added with 1.5 mL of additional 2:1 chloroform:methanol solution for extraction followed by centrifugation at 2,000 x g for 5 min. Organic layers were combined and dried under stream of nitrogen gas. Samples were resolubilized in 120 μL of 1:1 methanol:isopropanol solution and stored at −80°C u ntil further analysis.
LC-MS/MS Lipidomics.
A Dionex Ultimate 3000 RS UHPLC system was used with the analytical column (Kinetex® 1.7 μm C18 100 Å, LC column 100 X 2.1 mm) and reverse phase LC solvents (A: 0.1% formic acid, 10 mM ammonium formate, 50% water, 50% acetonitrile, B: 0.1% formic acid, 10 mM ammonium formate, 10% acetonitrile, 88% isopropanol, 2% water) and the following gradient: 0–4 min, 35% to 60% B; 4–12 min, 60% to 85% B; 12–24 min, 85% to 100% B, flow rate at 250 μL/min. The eluted lipids were electrosprayed using a HESI-II probe into an Orbitrap Q-Exactive Plus mass spectrometer (Thermo Scientific), which was operated under one of two methods: 1) A top 10 data-dependent (ddMS2) acquisition method that consisted of one full MS1 scan (250–1,200 m/z) followed by 10 MS2 scans of the most abundant ions recorded in the MS1 scan. 2) A targeted parallel reaction monitoring (PRM) acquisition method that performs multiple MS2 scans from a pre-specified MS1 precursor scan of a known ion m/z and retention window. Isolation window was set at 1.2 m/z. Data acquisitions were performed using both ddMS2 global analysis and PRM targeted methods, in both positive and negative ionization modes to enhance the confidence of lipid identification. PRM targeting of diacylglycerol and triacylglycerol lipid species was accomplished by filtering for the [M+NH4]+ adduct ions and subsequent MS2 detection of expected diagnostic fragment ions (neutral loss of fatty acyl chains in positive ion mode). Intensities of the lipid species were measured using TraceFinder™ software, which identified fragment ions across multiple samples based on the MS2 filter defined in a targeted list of lipids and aligned them according to the intensities of the ion species found in each raw file. Raw intensities were normalized to base peak ion intensities for each sample acquisition to account for instrument injection variation. Lipid identifications and peak alignments were performed using LipidSearch™ software while quantitative analysis of the aligned intensities was exported and analyzed using GraphPad Prism version 7.03.
Lipid Identification by LC-MS/MS.
Data analysis was performed using the LipidSearch™ software package, which used MS1 monoisotopic precursor ions for the in-silico database search followed by the product ions search from MS2 spectra (search parameters: Target database: Q Exactive, SearchType: product, ExpType: LC-MS, Precursor tolerance 5.0 ppm, product tolerance 5.0 ppm, intensity threshold: product ion 1.0%, m-score threshold 2.0). Results from positive and negative ion mode analyses were merged to generate a matched lipid dataset. The searched data were aligned with selected parameters (ExpType: LC-MS, Alignment Method: Median, Toprank Filter: on, Main node Filter: Main isomer peak, m-Score Threshold: 5.0, ID quality filter: A, B, C, D) and curated with species with high-confidence grade scores “A” and “B” for further analysis.
Sample preparation for LC-MS/MS Substrate Assay.
Cell pellets were dounce homogenized in lysis buffer (0.25 M sucrose, 20 mM HEPES at pH 7.4, 2 mM DTT) and ultracentrifuged at 100,000 x g for 45 min at 4°C. Pellets were resuspended by sonication in activity buffer (50 mM HEPES at pH 7.4, 100 mM NaCl, 5 mM CaCl2, 0.1% Triton X-100 v/v, 10% DMSO and 0.5 mM DTT). Protein concentration was standardized to 0.37 mg mL−1 (membrane fraction) and treated with respective inhibitors for 30 min (KT109 2 μM, KT195 2 μM). Lipid substrate was prepared as follows: 250 pmol of respective lipid substrate, dissolved in chloroform, was placed in a dram vial, dried under a stream of nitrogen gas, brought up in 460 μL of activity buffer, and briefly sonicated to create a turbid solution. 40 μL of the pre-treated lysates were then added to the lipid substrate mixture at a final volume of 500 μL in activity buffer. The reaction was vortexed and left to continue at room temperature for 4 h. The reaction was quenched via organic extraction. The extraction was carried out by adding 1.5 mL of hexane (3:1 hexane: H2O) with 80 mg of bio-beads (Bio-Rad). Reaction mixture was vortexed and centrifuged at 2,000 x g for 5 min. Additional wash with 1.5 mL of hexane was performed and hexane layers were combined. Hexane layer was transferred to a new dram vial and added with 1 mL of water, and 80 mg of bio-beads. After vortex/centrifugation, hexane layer was transferred to a clean dram vial and dried down under nitrogen. Samples were resuspended in 1:1 isopropanol:methanol and stored at −80°C until further analysis.
LC-MS/MS Substrate Assay.
LC-MS/MS conditions used for the substrate assay lipid identification are identical to the conditions described in “LC-MS/MS Analysis of Lipid Extracts” with slight changes in LC configuration. External hardware divert valves were used to discard eluents from the initial 6 min of the analysis in order to prevent residual triton X-100 from being introduced to mass spectrometer.
Sample Preparation for Gel-based Competitive Activity Based Protein Profiling (ABPP).
Cell pellets were processed using dounce homogenizer. Supernatant was isolated and centrifuged at 100,000 x g for 45 min. Resulting supernatant fraction and resuspended pellet solution were referred as soluble and membrane fraction, respectively. Proteomes (1 mg mL−1) were treated with HT-01 probe at 1 μM final concentration for 30 min at 37°C. The reaction was quenched using SDS-PAGE loading buffer. After separation by SDS-PAGE (10% acrylamide), samples were visualized by in-gel fluorescence scanning using ChemiDoc MP imaging system (Bio-Rad).
ABPP-SILAC Analysis.
SILAC proteomes were diluted to 1 mg mL−1 in PBS and sample aliquots (432 μl) were treated with fluorophosphonate (FP) alkyne probe at 100 μM (5 μl, 100X stock in DMSO; final concentration of 1 μM), mixed gently and incubated for 2 h at room temperature. Light and heavy probe-modified proteomes were mixed 1:1 and subjected to CuAAC conjugation to desthiobiotin-PEG3-azide (10 μl of 10 mM stock in DMSO; final concentration of 200 μM) using TCEP (10 μl of fresh 50 mM stock in water, 1 mM final concentration), TBTA ligand (33 μl of a 1.7 mM 4:1 t-butanol/DMSO stock, 100 μM final concentration) and CuSO4 (10 μl of 50 mM stock, 1 mM final concentration). Samples were vortexed and then incubated for 1 h at room temperature. Excess reagents were removed by chloroform-methanol extraction as previously described (Franks et al., 2017). Protein pellets were resuspended in 500 μL of 6 M urea:25 mM ammonium bicarbonate followed by dithiothreitol reduction and iodoacetamide alkylation as previously described (Franks et al., 2017). Excess reagents were removed by chloroform-methanol extraction as described above, and the protein pellet was resuspended in 500 μL of 25 mM ammonium bicarbonate and then digested to peptides as trypsin/Lys-C (7.5 μg in 15 μL of ammonium bicarbonate, Promega catalog # V5073) was added to the mixture and incubated for 3 h at 37°C. Probe-modified peptides were enriched by avidin affinity chromatography, eluted and prepared for LC-MS analysis as previously described (Franks et al., 2017). Nano-electrospray ionization–LC–MS/MS analyses were performed using an Ultimate 3000 RSLC nanoSystem-Orbitrap Q Exactive Plus mass spectrometer (Thermo Scientific) as previously described (Hahm et al., 2019). The eluting peptides were electrosprayed into an Orbitrap Q Exactive Plus mass spectrometer (Thermo Scientific), which was operated with a top 10 data-dependent acquisition method that consisted of one full MS1 scan (375 – 1,500 m/z) followed by 10 MS2 scans of the most abundant ions recorded in the MS1 scan. Data analysis was accomplished using the IP2 (Integrated Proteomics Applications) software package, in which RawConverter was used to generate searchable MS1 and MS2 data from the .raw file followed by using the ProLuCID algorithm to search the data against a mouse protein database (UniProt mouse protein database, angiotensin I and vasoactive intestinal peptide standards, 55,513 proteins) with the following parameters: static carbamidomethyl modification (+57.02146 Da), differential modification of oxidized methionine (+15.9949 Da), added masses of the SILAC “heavy”-labeled amino acids (+10.0083 Da for R, +8.0142 Da for K), and trypsin enzyme specificity with 1 missed cleavage. The resulting MS2 spectra matches used to generate SILAC ratios (SR) of light/heavy (vehicle/compound treated) peptides was done as described previously (Franks et al., 2017).
Flow Cytometry Cell Sample Preparation and Analysis.
After BMDMs were differentiated, suspension cells were seeded in a 96-well plate in 50 mL volume at 100,000 cells per well in complete BMDM differentiation media. Supernatant was collected in V-bottom 96 well plate and centrifuged at 1,400 x g for 5 min to pellet residual BMDCs. Supernatant was transferred to a separate 96 well plate and stored at −80°C until fu rther analysis. Cytokine profiling was performed at the UVa flow cytometry core. In brief, antibody-immobilized beads were prepared by sonication/vortexing for 1 min with 60 mL of each individual antibody-bead: PE/Cy7 anti-mouse/human CD11b [M1/70] (Catalog# 101215) and Rat anti Mouse F4/80 (Catalog# MCA497A488T) and combining them in a total volume of 3.0 mL of assay buffer (Millipore corporation, catalog# L-AB). Wells were washed, detected, and analyzed using the Luminex MAGPIX system following manufacturer’s recommended protocols. Samples were analyzed using median fluorescent intensity (MFI) data using a 5-parameter logistic for calculating the cytokine concentrations in samples. Exported data were analyzed using FloJo (version 10.6.1).
Quantification and Statistical Analysis.
For the lipid analyses, the intensities of identified lipid species were used for the semi-quantitative determination of lipid concentrations within a sample. Intensities of lipid species were divided by the base peak intensities of each injected sample. The calculated lipid amounts were normalized based on the number of cells for each respective sample. Statistical analyses were performed in GraphPad Prism version 7.03. In order to determine statistically significant differences between groups analyzing multiple lipid species, a Holm-Sidak multiple comparison test (do not assume consistent standard deviation, confidence interval of 95%) was applied. Otherwise an unpaired t-test was implemented to demonstrate significant differences between two groups with respect to individual lipid changes while one-way ANOVA was used for comparisons across three or more conditions. Statistical significance was set at P < 0.05. The number of experimental biological replicates of cell populations in each sample group are indicated by n and can be found in respective figure legends as well as the number of times individual experiments were repeated with similar results. Data are shown as mean ± standard error of mean (s.e.m.).
Data and Software Availability.
All data produced or analyzed for this study are included in the published article (and its supplemental information files) or are available from the Lead Contact on reasonable request.
Supplementary Material
Lipids detected by LC-MS analyses configured for data-dependent acquisition (ddMS2) and identified using the LipidSearch™ post-acquisition analysis workflow. Data listed in this dataset were used to generate Figures 1, 3, and S3 (n=5 biological samples). Statistical significance was determined using a Holm-Sidak correction following a two-sided binomial test.
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti-FLAG antibody produced in rabbit | Sigma-Aldrich | Clone: polyclonal, catalog# F7425, RRID: AB_439687 |
| Goat anti-rabbit DyLight 550 | Thermo Fisher Scientific | Clone: polyclonal, catalog# 84541, RRID: AB_10942173 |
| PE/Cy7 anti-mouse/human CD11b [M1/70] | Biolegend | Clone: monoclonal, catalog# 101215, RRID: AB_312798 |
| Rat anti Mouse F4/80 | Bio-Rad | Clone: monoclonal, catalog# MCA497A488T, RRID: AB_1102554 |
| Chemicals, Peptide, Recombinant proteins | ||
| Polyethyleneimine | Polysciences, Inc. | catalog# 24765 |
| Recombinant M-CSF | Peprotech | catalog# 315–02 |
| 2-Mercaptoethanol | Sigma-Aldrich | catalog# M6250–250ML |
| Bio-beads SM-2 Adsorbants | Bio-Rad | catalog# 1528920 |
| HT-01 probe | Synthesized in house | N/A |
| FP-alkyne probe | Synthesized in house | N/A |
| Arachidonic acid-d8 | Cayman Chemical | catalog# 390010 |
| 2-Arachidonyl glycerol-d5 | Cayman Chemical | catalog# 362162 |
| Tritetradecanoin (C14:0) | Nu-Chek Prep | catalog# T-140 |
| Trihexadecanoin (C16:0) | Nu-Chek Prep | catalog# T-150 |
| Trioctadecanoin (C18:0) | Nu-Chek Prep | catalog# T-160 |
| Trieicosanoin (C20:0) | Nu-Chek Prep | catalog# T-170 |
| Tridocosanoin (C22:0) | Nu-Chek Prep | catalog# T-180 |
| Triarachidonin (C20:4) | Nu-Chek Prep | catalog# T-295 |
| Trieicosapentaenoin (C20:5) | Nu-Chek Prep | catalog# T-325 |
| Tridocosahexaenoin (C22:6) | Nu-Chek Prep | catalog# T-310 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mouse: DAGLβ KO | Taconic | N/A |
| Mouse: DAGLα KO | TSRI | N/A |
| HEK293T cells | ATCC | CVCL_0063 |
| L929 cells | ATCC | CVCL_0462 |
| Software and Algorithms | ||
| GraphPad Prism 7.03 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| TraceFinder 4.0 | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/OPTON-30491 |
| Xcalibur 3.0 | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/OPTON-30487 |
| LipidSearch 4.0 | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/IQLAAEGABSFAPCMBFK?SID=srch-srp-IQLAAEGABSFAPCMBFK |
| ImageLab software 5.2 | Bio-Rad | https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| FlowJo 10.6.1 | BD Life Sciences | https://www.flowjo.com/solutions/flowjo |
| RawConverter 1.1.0.22 | Scripps | http://fields.scripps.edu/rawconv/ |
| ProLuCID 4.0 | Integrated Proteomics Applications | http://www.integratedproteomics.com/index.html |
| Skyline-daily 19.1 | MacCoss Lab Software | https://skyline.ms/project/home/software/Skyline/begin.view |
| Data and Software Availability | ||
| Lipidomics data (mass spec) | Available from the Lead Contact on reasonable request. | |
| Proteomics data (mass spec) | Available from the Lead Contact on reasonable request. | |
| Flow Cytometry data | Available from the Lead Contact on reasonable request. | |
SIGNIFICANCE.
Assigning substrate specificity to enzymes in living systems is critical for understanding specificity of cell metabolism. Here, we apply quantitative chemoproteomics and discovery metabolomics to assign a polyunsaturated fatty acid (PUFA)-specific triacylglycerol (TAG) lipase activity for diacylglycerol lipase-beta (DAGLβ) in macrophages. We provide genetic and pharmacological evidence that disruption of DAGLβ results in cellular accumulation of TAGs containing polyunsaturated fatty acids (PUFAs) in primary bone marrow-derived macrophages (BMDMs). Interestingly, bulk TAG levels were unchanged in DAGLβ-disrupted BMDMs, supporting DAGLβ as a PUFA-specific TAG lipase that was validated by biochemical substrate assays. Our findings support DAGLβ as a lipase capable of remodeling the neutral lipidome via fatty acyl-specific DAG and TAG metabolism. The significance of our findings is providing a connection between endocannabinoid and bioenergetic pathways and provides future opportunities to investigate potential cross-talk between DAGLβ and lipolytic pathways.
Highlights.
Differentiation of SILAC BMDMs for DAGLβ chemoproteomics in primary macrophages
Discovery metabolomics to identify PUFA-TAGs as DAGLβ substrates in live BMDMs
Biochemical validation that DAGLβ exhibits PUFA-specific TAG lipase activity
ACKNOWLEDGMENTS
We thank Mark Ross and all members of the Hsu Lab for helpful discussions. This work was supported by the University of Virginia (start-up funds to K.-L.H.) and National Institutes of Health (DA035864 and DA043571 to K.-L.H.; GM801868 to T.B.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References:
- Baggelaar MP, Chameau PJ, Kantae V, Hummel J, Hsu KL, Janssen F, van der Wel T, Soethoudt M, Deng H, den Dulk H, et al. (2015). Highly Selective, Reversible Inhibitor Identified by Comparative Chemoproteomics Modulates Diacylglycerol Lipase Activity in Neurons. J Am Chem Soc 137, 8851–8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisogno T, Howell F, Williams G, Minassi A, Cascio MG, Ligresti A, Matias I, Schiano-Moriello A, Paul P, Williams EJ, et al. (2003). Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol 163, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandak PG, Radovic B, Aflaki E, Kolb D, Buchebner M, Frohlich E, Magnes C, Sinner F, Haemmerle G, Zechner R, et al. (2010). Efficient phagocytosis requires triacylglycerol hydrolysis by adipose triglyceride lipase. J Biol Chem 285, 20192–20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, and Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226, 497–509. [PubMed] [Google Scholar]
- Gao Y, Vasilyev DV, Goncalves MB, Howell FV, Hobbs C, Reisenberg M, Shen R, Zhang MY, Strassle BW, Lu P, et al. (2010). Loss of Retrograde Endocannabinoid Signaling and Reduced Adult Neurogenesis in Diacylglycerol Lipase Knock-out Mice. J Neurosci 30, 2017–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, et al. (2006). Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737. [DOI] [PubMed] [Google Scholar]
- Haemmerle G, Moustafa T, Woelkart G, Buttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, et al. (2011). ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat Med 17, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm HS, Toroitich EK, Borne AL, Brulet JW, Libby AH, Yuan K, Ware TB, McCloud RL, Ciancone AM, and Hsu KL (2019). Global targeting of functional tyrosines using sulfur-triazole exchange chemistry. Nat Chem Biol [DOI] [PMC free article] [PubMed]
- Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, and Cravatt BF (2012). DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat Chem Biol 8, 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inloes JM, Hsu KL, Dix MM, Viader A, Masuda K, Takei T, Wood MR, and Cravatt BF (2014). The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc Natl Acad Sci U S A 111, 14924–14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, and Yokomizo T (2018). Applications of mass spectrometry-based targeted and non-targeted lipidomics. Biochem Biophys Res Commun 504, 576–581. [DOI] [PubMed] [Google Scholar]
- Long JZ, and Cravatt BF (2011). The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev 111, 6022–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M (2006). Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol 7, 952–958. [DOI] [PubMed] [Google Scholar]
- Niphakis MJ, and Cravatt BF (2014). Enzyme Inhibitor Discovery by Activity-Based Protein Profiling. Annual Review of Biochemistry 83, 341–377. [DOI] [PubMed] [Google Scholar]
- Ogasawara D, Deng H, Viader A, Baggelaar MP, Breman A, den Dulk H, van den Nieuwendijk AM, Soethoudt M, van der Wel T, Zhou J, et al. (2016). Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition. Proc Natl Acad Sci U S A 113, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, and Sharpless KB (2002). A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angewandte Chemie International Edition 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, and Cravatt BF (2004). Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry 43, 14332–14339. [DOI] [PubMed] [Google Scholar]
- Serbulea V, Upchurch CM, Schappe MS, Voigt P, DeWeese DE, Desai BN, Meher AK, and Leitinger N (2018). Macrophage phenotype and bioenergetics are controlled by oxidized phospholipids identified in lean and obese adipose tissue. Proc Natl Acad Sci U S A 115, E6254–E6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Buckner A, Prince J, Bullock TNJ, and Hsu KL (2019). Diacylglycerol Lipase-beta Is Required for TNF-alpha Response but Not CD8(+) T Cell Priming Capacity of Dendritic Cells. Cell Chem Biol 26, 1036–1041 e1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Snyder HW, Donvito G, Schurman LD, Fox TE, Lichtman AH, Kester M, and Hsu KL (2018). Liposomal Delivery of Diacylglycerol Lipase-Beta Inhibitors to Macrophages Dramatically Enhances Selectivity and Efficacy in Vivo. Mol Pharm 15, 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi R, Nishijima M, and Shimizu T (2007). Basic Analytical Systems for Lipidomics by Mass Spectrometry in Japan In Methods in Enzymology (Academic Press; ), pp. 185–211. [DOI] [PubMed] [Google Scholar]
- Tanimura A, Yamazaki M, Hashimotodani Y, Uchigashima M, Kawata S, Abe M, Kita Y, Hashimoto K, Shimizu T, Watanabe M, et al. (2010). The endocannabinoid 2-arachidonoylglycerol produced by diacylglycerol lipase alpha mediates retrograde suppression of synaptic transmission. Neuron 65, 320–327. [DOI] [PubMed] [Google Scholar]
- Viader A, Ogasawara D, Joslyn CM, Sanchez-Alavez M, Mori S, Nguyen W, Conti B, and Cravatt BF (2016). A chemical proteomic atlas of brain serine hydrolases identifies cell type-specific pathways regulating neuroinflammation. eLife 5, e12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson JL, Ghosh S, Bagdas D, Mason BL, Crowe MS, Hsu KL, Wise LE, Kinsey SG, Damaj MI, Cravatt BF, et al. (2016). Diacylglycerol lipase beta inhibition reverses nociceptive behaviour in mouse models of inflammatory and neuropathic pain. Br J Pharmacol 173, 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, and Madeo F (2012). FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell metabolism 15, 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lipids detected by LC-MS analyses configured for data-dependent acquisition (ddMS2) and identified using the LipidSearch™ post-acquisition analysis workflow. Data listed in this dataset were used to generate Figures 1, 3, and S3 (n=5 biological samples). Statistical significance was determined using a Holm-Sidak correction following a two-sided binomial test.