Abstract
Microalgae, a diverse group of single-celled organisms exhibiting versatile traits, find broad applications in industry. However, high production costs require further efforts to optimize their production and to enhance biomass yields. In the present study, co-occurrence of algae and methylobacteria was observed when naturally occurring microalgae biofilms were subjected to 16S rRNA gene fragment amplicon sequencing. This bacterial group is so far less explored than other microalgae-associated bacteria in terms of mutualistic relationships that might be exploitable for biotechnological applications. In order to assess the potential of four plant growth-promoting strains from the genus Methylobacterium for increased algae biomass production, co-cultivation experiments were conducted with three industrially relevant microalgae (Chlorella vulgaris, Scenedesmus vacuolatus, and Haematococcus lacustris). For S. vacuolatus and H. lacustris, a significant increase in algal biomass formation of 1.3-fold to up to 14-fold was observed after 7 days of co-incubation. Visualization of mixed cultures using confocal laser scanning microscopy revealed a high abundance of methylobacteria in the phycosphere of H. lacustris and S. vacuolatus, visually attached to the algae’s surface forming a biofilm-like assemblage. Genome analyses revealed that features attributable to enhanced algal growth include genes involved in the synthesis of vitamins, siderophores and plant hormones. Our results provide evidence for the constructability of novel symbiotic algae-bacteria relationships with inter-kingdom supportive capacities, underlining the potential of microbial consortia as promising tool for sustainable biotechnology and agriculture.
Keywords: microalgae, microbiota, growth-promotion, methylobacteria, symbiosis, algae–bacteria interactions
Introduction
In the recent past, the potential of microalgae for industrial purposes increasingly gained importance at a global scale. Due to their versatile characteristics including high lipid content and their ability to accumulate high-value compounds, they can be used as supplements for food and animal feed, as pharmaceuticals, as feedstock for biofuel production, or as sustainable alternative to synthetic fertilizers (Spolaore et al., 2006; Brennan and Owende, 2010; Renuka et al., 2018). In order to meet industrial demands, microalgae are cultivated either in open pond systems for bulk material production or in closed photobioreactors for the production of high-value compounds. However, both cultivation methods often fail to reach economic competitiveness in terms of biomass yields, contamination prevention and the involved costs (Benemann and Oswald, 1996; Wang et al., 2013). Therefore, further efforts need to be made in order to reduce costs and enhance biomass yields by optimizing cultivation conditions for efficient microalgae production that can compete with non-sustainable alternatives. One means of increasing microalgae populations and thus enhancing yields might be the co-inoculation with beneficial microorganisms as already successfully applied in agriculture for increasing yields of vascular land plants (Berg, 2009; Lugtenberg and Kamilova, 2009; Berg et al., 2013). This strategy is primarily applicable for large-scale production where cultivation conditions are not optimal and contaminations with other microorganism are unavoidable. Algae were shown to benefit from micro- and macronutrients provided by bacteria, and in return, release dissolved organic carbon into their surroundings. Efforts have been made in identifying bacteria that positively contribute to the algal fitness; especially nitrogen-fixing bacteria of the genera Azospirillum, Rhizobium, and Bacillus, known for their beneficial effect on plants, are promising candidates (Gonzalez and Bashan, 2000; de-Bashan et al., 2008; Hernandez et al., 2009; de-Bashan and Bashan, 2010; Kim et al., 2014; Amavizca et al., 2017). While binary algae-bacteria interactions were explored in the past, little is known about the algae-associated microbiota as well as potentially beneficial algae-microbe interactions with distinct members of the plant microbiota.
The species-specific plant microbiota forms complex networks and contributes multiple aspects to the functioning of the plant holobiont, such as (i) seed germination and growth, (ii) nutrient supply, (iii) resistance against biotic and abiotic stress factors, and (iv) production of bioactive metabolites (Berg et al., 2017). Mutualistic inter-kingdom symbioses are not only limited to land plants, but also extend to green algae (Xie et al., 2013; Hom et al., 2015). Following the same principles of plant growth-promoting rhizobacteria, algae-associated bacteria harbor potential to stimulate growth and morphogenesis of algae by releasing essential minerals, vitamins, auxins and quorum sensing signaling molecules (Joint et al., 2002; Croft et al., 2006; Goecke et al., 2010; Amavizca et al., 2017). The equivalent to the rhizosphere – the immediate surrounding of roots with increased biological and chemical activity in soil – is the phycoscphere, which is the area surrounding a phytoplankton cell, rich in dissolved organic matter and metabolites exuded by the cell in the surrounding water and stimulating the growth of heterotrophic bacteria (Seymour et al., 2017). Compared to the plant rhizosphere, less is known about the structure and function of the phycosphere microbiota, although studies show negative (Hom et al., 2015) and positive impacts of bacteria on algal growth (Croft et al., 2005; Amin et al., 2009; Hernandez et al., 2009; Kim et al., 2014; Cooper and Smith, 2015).
In order to deepen our understanding of the phycosphere microbiota, we analyzed the naturally occurring microbiome of algae in order to detect potential algae-bacteria associations. Based on the preliminary findings we decided to focus on a bacterial group known for its symbiotic interactions with agricultural plants and three eukaryotic model organisms representing industrially relevant microalgae. The microalgae Chlorella vulgaris, Scenedesmus vacuolatus and Haematococcus lacustris are used as feedstock for biofuel production due to their high amount of carbohydrates and as source material for the production of astaxanthin, a strong antioxidant used in food and pharmaceutical industry (Sharma and Sharma, 2017; Khan et al., 2018). In order to explore the transferability of beneficial, host-associated microorganisms, methylobacteria that are already known for their plant-beneficial traits, were selected from our in-house strain collection. The selected isolates belong to three different species, Methylobacterium extorquens, M. mesophilicum, and M. goesingense, and were originally isolated from different plant species (Verginer et al., 2010). By implementing co-cultivation experiments, the potential of methylobacteria to induce and promote the growth of microalgae was evaluated. In order to provide deeper insights into the mechanisms and bacterial features associated with growth-promoting effects, the whole genomes of M. extorquens Rab1 and M. mesophilicum Sab1 were sequenced and analyzed. The obtained results provide the background for novel co-cultivation approaches that make use of inter-kingdom supporting capacities of beneficial, plant-associated bacteria.
Materials and Methods
Sampling Procedure
Due to the generally higher prevalence beneficial interactions in nutrient-poor systems, naturally occurring algae communities from artificial surfaces were selected for phycobiome analyses. Samples were collected on November 1st 2016 by removing a natural biofilm on the surface of outdoor furniture in Graz (Austria; 47° 4′′ N, 15° 26′′ O) at an altitude of 353 m above sea level, in three replicates, where macroscopic observations and coloration of biofilm indicated the occurrence of microalgae (Supplementary Figure S1A). Upon arrival in the laboratory, samples were resuspended in 3 mL sterile NaCl (0.85%) and visualized microscopically using a light microscope (Leitz, Wetzlar, Germany) at 200 × magnification in order to confirm the occurrence of microalgae (Supplementary Figure S1B).
Isolation and Identification of Microalgae
In order to isolate microalgae, the aforementioned samples were plated in dilution series on modified Bolds Basal Medium agar plates (BBM) containing 250 mg/L NaNO3, 175 mg/L KH2PO4, 75 mg/L K2HPO4, 75 mg/L MgSO4 × 7 H2O, 25 mg/L CaCl2, 25 mg/L NaCl, 2.6 mg/L H3BO3, 5 mg/L FeSO4 × 7 H2O, 8.8 mg/L ZnSO4 × 7 H2O, 1.4 mg/L MnCl2 × 4 H2O, 1.4 mg/L MoO3, 1.6 mg/L CuSO4 × 5 H2O, 0.5 mg/L Co(NO3)3 × 6 H2O, 0.5 mg/L EDTA, 0.3 mg/L KOH, 0.017 mg/L vitamin B12, 0.013 mg/L 4-aminobenzoate, 0.003 mg/L biotin, 0.013 mg/L nicotinic acid, 0.017 mg/L hemicalcium-pentathenate, 0.05 mg/L pyridoxamine-HCl, 0.033 mg/L thiaminiumdichlorid, 0.0091 mg/L thioctic-acid, 0.01 mg/L riboflavin, 0.0049 mg/L folic acid and 18 g/L agar-agar. Vitamins and heat sensitive components were added after autoclaving by sterile filtration (0.20 μm pore size). In order to obtain pure cultures single algae colonies were repeatedly subcultured on modified BBM-agar and incubated at 23°C at a light dark cycle (L:16/D:8). The lighting was supplied by cool-white fluorescent lamps TL-D 36W/840 REFLEX Eco (Philips, Amsterdam, Netherlands) with a photosynthetic photon flux of 115 μmol s–1.
In order to identify isolated microalgae species, cells were resuspended in 300 μL NaCl (0,85%) and transferred in sterile Eppendorf tubes filled with glass beads. After mechanical disruption using a FastPrep FP120 instrument (MP Biomedicals, Germany) suspensions were centrifuged at 3,000 rpm for 5 min. Supernatant served as template for the following PCR reaction. Partial 18S rRNA gene fragments were amplified using primer pair NS1 (5′-GTA GTC ARA RGC CTT GTC TC-3′) and NS8 (5′-TCC GCA GGT TCA CCT ACG GA-3′) in a reaction mix (30 μL) containing 16.2 μL ultrapure H2O, 6 μL Taq&Go [5×], 1.2 μL of each primer [10 μM], 2.4 μL MgCl2 [25 mM] and 3 μL DNA template. The cycling program was adjusted to the following settings: 95°C, 10 min; 40 PCR products were then purified using the Wizard SV Gel and PCR-Clean-Up System (Promega Corporation, Madison, WI, United States) according to manufacturer’s protocol. The amplified 18S rRNA gene fragments were sequenced by Sanger sequencing (LGC genomics; Berlin, Germany) and subsequently aligned against the NCBI nucleotide collection database for taxonomic assignments. Microscopic visualization of the pure cultures was used to verify the absence of other microorganisms in the pure cultures of the isolated microalgae C. vulgaris G1-G, S. vacuolatus G1-O and H. lacustris G1-R (Supplementary Figure S2).
Algae-Associated Microbiome Analyses Using 16S rRNA Gene Fragment Amplicon Libraries
Total Community DNA Extraction and Barcoding
Total community DNA of three biological replicates was extracted using the FastDNA Kit for Soil (MP Biomedicals, Heidelberg, Germany) according to the manufacturer’s protocol. The 16S rRNA gene fragments were amplified in three technical replicates covering the hypervariable region 4 using the Unibac II 515f (5′-GTG YCA GCM GCC GCG GTA A-3′) and 806r (5′-GGA CTA CHV GGG TWT CTA AT-3′) primer pair (Caporaso et al., 2011), which included sample-specific barcodes and Illumina sequencing adaptors. Peptide nucleic acid (PNA) was added to the PCR mix to prevent the amplification of mitochondrial (mPNA) and plastidial (pPNA) DNA from eukaryotes (Lundberg et al., 2013). The PCR was performed by using a total volume of 30 μL containing 20.15 μL ultrapure water, 6 μL Taq&Go [5×], 1.2 μL of each primer (5 μM), 0.225 μL pPNA [100 μM], 0.225 μL mPNA [100 μM] and 1 μL DNA template. The cycling program was adjusted to an initial denaturation temperature at 96°C for 5 min, followed by 30 cycles of 96°C for 1 min, 78°C for 5 s, 54°C for 1 min, and 74°C for 1 min. The final extension was done at 74°C for 10 min. The PCR products of all samples were quality checked by gel electrophoresis. Subsequently, the PCR products were purified using the Wizard SV Gel and PCR-Clean-Up System according to manufacturer’s protocol. Equimolar DNA concentrations of each barcoded amplicon were sent for paired end Illumina HiSeq sequencing (read length: 2 × 300 bp) to GATC Biotech AG (Konstanz, Germany).
Bioinformatic Analyses of 16S rRNA Gene Fragments Amplicons
Initial data processing, including joining of forward and reverse read pairs was done using software package QIIME 1.9.1 (Caporaso et al., 2010). After removing barcodes, primer and adapter sequences reads as well as metadata were imported into QIIME 2 (2018.11 release). Further analyses of sequencing data were performed using the QIIME 2 pipeline according to tutorials provided by the QIIME developers (Caporaso et al., 2010). The DADA2 algorithm (Callahan et al., 2016) was used to demultiplex and denoise truncated reads and remove chimeras. Taxonomic analyses are based on a Naïve-Bayes classifier trained on the SILVA 128 release database (Quast et al., 2013) clustering at 99% similarity. After removing mitochondrial, chimeric and plastid sequences, the 16S rRNA dataset was normalized to 264,542 reads. A normalized feature table served as input for the OTU table (make_out_network.py) using QIIME 1.9.1. OTU-network was generated and rendered using Cytoscape version 3.7.0 (Shannon et al., 2003). Features assigned to the genus Methylobacterium were manually aligned against the NCBI nucleotide collection using the BLAST algorithm (Altschul et al., 1990).
Bacterial Strains and Initial Culture Conditions
For growth assays M. extorquens Rab1 (deposited at DSMZ; DSM 21961), M. mesophilicum Sab1 (deposited at DSMZ; DSM 21962), M. goesingense Vab1 and M. goesingense Vab2 were selected from the in-house strain collection SCAM (Strain Collection of Antagonistic Microorganisms; Institute of Environmental Biotechnology, Graz University of Technology). They were originally isolated from different plant species (Verginer et al., 2010). Bacteria were cultured on MIS (methanol, inorganic salt) agar plates containing 1.8 g/L (NH4)2SO4, 0.2 g/L MgSO4 × 7 H2O, 1.4 g/L NaH2PO4 × 2 H2O, 1.9 g/L K2HPO4 and 18 g/L agar-agar. After autoclaving 5 mL/L methanol and 1 mL/L sterile filtered (0.20 μm pore size) trace element solution containing 500.0 mg/L EDTA, 200.0 mg/L FeSO4 × 7 H2O, 10.0 mg/L ZnSO4 × 7 H2O, 3.0 mg/L MnCl2 × 4 H2O, 30.0 mg/L H3BO3, 20.0 mg/L CoCl2 × 6H2O, 1.0 mg/L CuCl2 × 6 H2O, 2.0 mg/L NiCl2 × 6 H2O, and 3.0 mg/L Na2MoO4 × 2 H2O were added.
Evaluation of Microalgae Growth-Promoting Effects of Methylobacterium spp. Through Co-cultivation Experiments
The effect of four different methylobacteria strains on the growth of three different microalgal genera, including Chlorella, Scenedesmus and Haematococcus was evaluated through co-cultivation experiments. In order to explore the effect of the bacterial load on the performance of the microalgae, bacteria were added in two different cell concentrations (OD600 0.2 and OD600 0.5) at the beginning of the experiments. For the preparation of bacterial pre-cultures, 100 μL of the respective bacterial suspension were plated on 15–20 MIS agar plates. After 7 days of incubation at 30°C bacterial lawn was harvested using sterile object slide, resuspended in NaCl (0.85%) and used to reach the targeted absorbance OD600 of 0.5 and 0.2 in the main cultures. For C. vulgaris G1-G and S. vacuolatus G1-O, co-culture experiments were performed in 12 replicates in Parafilm® -sealed 12-well plates containing a final volume of 3 mL vitamin-depleted BBM medium (dBBM) containing 250 mg/L NaNO3, 175 mg/L KH2PO4, 75 mg/L K2HPO4, 75 mg/L MgSO4 × 7 H2O, 25 mg/L CaCl2, 25 mg/L NaCl, 2.6 mg/L H3BO3, 5 mg/L FeSO4 × 7 H2O, 8.8 mg/L ZnSO4 × 7 H2O, 1.4 mg/L MnCl2 × 4 H2O, 1.4 mg/L MoO3, 1.6 mg/L CuSO4 × 5 H2O, 0.5 mg/L Co(NO3)3 × 6 H2O, 0.5 mg/L EDTA, 0.3 mg/L KOH. Algal pre-cultures were obtained by inoculating 50 mL BBM with a single colony of the respective microalgae. After a sufficient cell density was reached, these pre-cultures served as inoculum for co-cultivation experiments to reach a FI of 154 ± 29, which corresponds to 5.08 ± 0.94 × 104 CFU/mL for C. vulgaris G1-G and to a FI of 152 ± 21, corresponding to 3.51 ± 0.47 × 104 CFU/mL for S. vacuolatus G1-O. For H. lacustris G1-R, the co-culture experiments were performed in 12 replicates in sterile 100-mL flasks sealed with aluminum foil containing a total volume of 10 mL dBBM. Algal pre-cultures were obtained by rinsing two BBM agar plates containing H. lacustris cultures with dBBM. The FI was adjusted to 33 ± 8 at the beginning of the experiments, corresponding to 7.04 ± 1.65 × 103 algal CFU/mL.
All co-cultures were incubated at 23°C at a light dark cycle (L:16/D:8). The lighting was supplied by cool-white fluorescent lamps TL-D 36W/840 REFLEX Eco (Philips, Amsterdam, Netherlands) with a photosynthetic photon flux of 115 μmol s–1. Biomass formation was determined by measuring the fluorescence intensity at 685 nm when excited at 450 nm using an infinite M200 spectrofluorimeter (TECAN; Switzerland) after 7 days for all included algae and additionally after 14 days for C. vulgaris and S. vacuolatus. The second measurement (14 days) was not conducted for H. lacustris G1-R due to the onset of encapsulation. In order to correlate the FI with algal cell counts, fluorescence measurements and subsequent plating of the respective dilutions on BBM agar were conducted. Linear regression allowed the correlation between FI and microalgae cell counts (C. vulgaris G1-G: R2 = 0.95, Supplementary Figure S3A; S. vacuolatus G1-O: R2 = 0.98, Supplementary Figure S3B; H. lacustris G1-R: R2 = 0.87; Supplementary Figure S3C).
Visualization of Co-cultures
All microscopic visualizations were done using a Leica TCS SPE confocal laser scanning microscope (Leica Microsystems GmbH, Mannheim, Germany) and the oil immersion objectives Leica ACS APO 40.0 × 1.15 (183.33 μm × 183.33 μm) and ACS APO 63 × 1.30 (116.40 μm × 116.40 μm). Solid-state lasers were used with 405, 488, 532, 635 nm excitation. The micrograph included in the OTU network was obtained with the natural community within the sampled biofilm following staining with the LIVE/DEAD bacterial viability kit (Thermo Fisher Scientific, MA, United States) in combination with calcofluor white (0.15%; Sigma-Aldrich, Missouri, United States). Micrographs of co-cultures were obtained with M. goesingense Vab1 and C. vulgaris, S. vacuolatus and H. lacustris, respectively, with an initial bacterial optical density of 0.5 after 7 days of incubation. For visualization of mixed cultures, 1 mL of the respective cultures were transferred into 1.5 mL reaction tubes and let stand for 2–3 h to allow sedimentation of the microorganisms. The supernatant was discarded and the remaining cultures were stained using the LIVE/DEAD bacterial viability kit in combination with calcofluor white at a concentration of 0.15%.
Genomic DNA Extraction
Methylobacterium extorquens Rab1 and M. mesophilicum Sab1 were grown on solid MIS medium and incubated for 7 days at 30°C. Cells were then collected from the plates using a sterile pipet tip and resuspended in 500 μL sterile NaCl (0.85%). Genomic DNA was extracted using the MasterPure DNA purification kit (Epicentre, WI, United States). DNA quality and quantity were checked by agarose gel electrophoresis, fluorometry (Qubit 4, Thermo Fisher Scientific, MA, United States) and spectrophotometry using a UV-Vis spectrophotometer (NanoDrop 2000c, Thermo Fisher Scientific, MA, United States). Subsequently, genomic DNA of M. extorquens Rab1 (5.05 μg; 0.19 μg/mL) and M. mesophilicum Sab1 (8.02 μg; 0.30 μg/mL) was sent for Illumina NextSeq 500/550 V2 (150 bp paired-end sequencing; LGC Genomics GmbH, Berlin, Germany).
Genome Assembly and Annotation
The genomes of M. extorquens Rab1 and M. mesophilicum Sab1 were assembled using SPAdes (Bankevich et al., 2012) and arranged into scaffolds. The scaffolds were then reassembled by using reference genomes (M. extorquens Rab1: NZ_CP021054.1; M. mesophilicum Sab1: NC_010505.1) and AlignGraph (Bao et al., 2014) in order to extend and join scaffolds. The annotation was done by using the Rapid Annotation using Subsystem Technology (RAST; Aziz et al., 2008).
Statistical Analyses
Each co-cultivation experiment was conducted with 12 replicates that were separately assessed. The resulting data was tested for normal distribution in order to select appropriate statistical tests. Significances were then determined using ANOVA for normally distributed values and the Kruskal-Wallis test for non-parametric analyses including Bonferroni multiple test correction.
Results
Microalgae and Methylobacteria Naturally Co-occur in Mixed-Community Biofilms
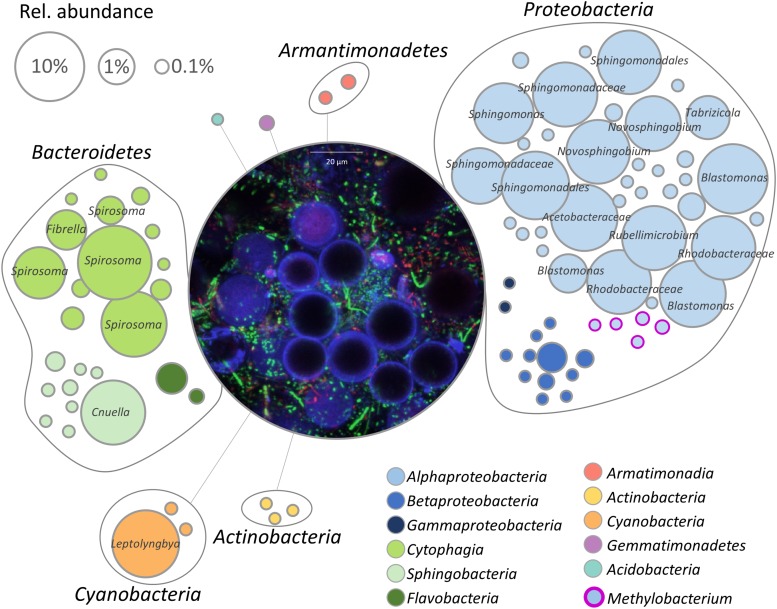
The bacterial fraction occurring in the microalgae-dominated biofilm was investigated by 16S rRNA gene fragment amplicon sequencing. After removal of chimeric, mitochondrial and chloroplast sequences, 264,542 reads remained in the dataset, resulting in 92 features. The bacterial microbiome consisted of seven phyla with Proteobacteria (61%) being the most dominant followed by Bacteroidetes (32%) and Cyanobacteria (7%). Acidobacteria, Actinobacteria, Armatimonadetes, and Gemmatimonadetes occurred in relative abundance less than 1%. On class level, Alphaproteobacteria (58%) were the dominating group followed by Cytophagia (27%) and Cyanobacteria (7%); Sphingobacteriia (4%), Betaproteobacteria (2%), Flavobacteria (1%), Acidobacteria (<1%), Actinobacteria (<1%), Armatimonadia (<1%), and Gammaproteobacteria (<1%) occurred in a substantially lower relative abundance. Members of the order Sphingomonadales (41%) were most frequently detected, followed by Cytophagales (27%), Rhodobacteriales (9%), and Rhodospirillales (8%). In total, 49 bacterial genera were identified with Spirosoma (24%) being the dominating fraction, followed by Porphyrobacter (11%) and not further classified Sphingomonadales (11%). Despite Methylobacterium spp. accounted only for a low proportion of the total bacterial community, their occurence in the core microbiome indicates that they are potentially embedded in a mutualistic interaction with microalgae (Figure 1). Five distinct features in the analyzed sample represented the genus and were also manually aligned against the NCBI nt collection; Supplementary Table S1 lists respective feature ID and closest hit after BLAST search. Visualization of the natural biofilm using a confocal laser scanning microscope revealed H. lacustris as the dominating algal taxon (Figure 1 and Supplementary Figure S1).
FIGURE 1.
Network visualization of the bacterial community within the biofilm. Each node represents a feature (99% cut-off level), whereas the node size corresponds to their relative abundance. Taxonomy of highly abundant features (>1%) is included. The central micrograph shows the natural microbial community within the biofilm after staining with the LIVE/DEAD bacterial viability kit in combination with calcofluor white (0.15%). Blue: algal cell wall (cellulose, chitin), green: viable bacterial cells, red: dead bacteria.
Co-cultivation of Distinct Microalgae and Methylobacteria Leads to Significantly Increased Biomass Formation
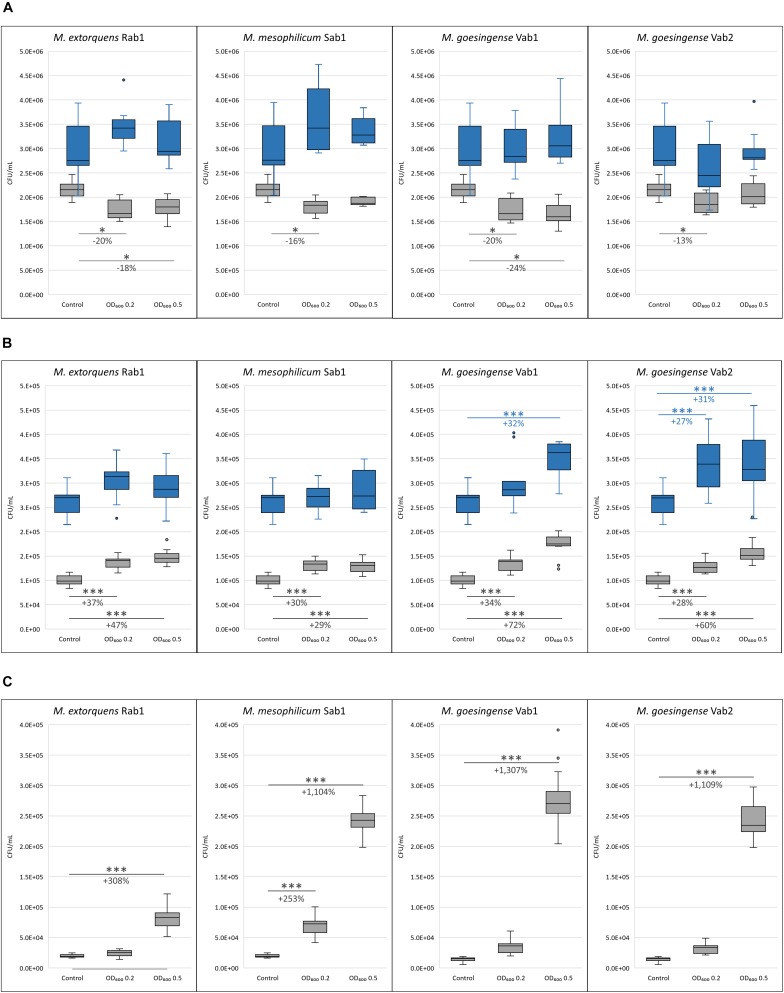
Co-cultivation experiments revealed varying effects of methylobacteria on the growth of different microalgal genera. While co-cultivation with methylobacteria led to a significant decrease in algal cell count for C. vulgaris G1-G, contrary effects were revealed for S. vacuolatus G1-O and H. lacustris G1-R. Both, growth-promoting as well as growth-inhibiting effects of methylobacteria were stronger after 7 days of co-cultivation but decreased after 14 days of incubation. In general, the growth-promotion by methylobacteria was more explicit, when the bacterial population density was higher at the beginning of the experiments. In detail, a significant decrease in microalgal biomass was observed for C. vulgaris G1-G when co-cultured with all tested methylobacteria with an initial optical density of 0.2 after 7 days of incubation compared to the axenic control. For M. extorquens Rab1 and M. goesingense Vab1 a significant decrease in microalgal biomass formation was also observed when the initial bacterial cell density was adjusted to an OD600 of 0.5. After 14 days of incubation, no significant differences in C. vulgaris G1-G cell count were measured between the control samples and co-cultures (Figure 2A). Co-cultivation of S. vacuolatus G1-O and all tested methylobacteria led to a significant increase in algal cell count after 7 days of incubation, independent from the initial bacterial cell load; however, a higher initial cell density of both M. goesingense strains resulted in even better performance of the microalga after 7 days of co-culturing. After 14 days of incubation, the effect of algal-growth promotion diminished; significant increase in algal cell count was only observable for co-cultures with M. goesingense Vab1 with an initial bacterial OD600 of 0.5 and for co-cultures with M. goesingense Vab2 (Figure 2B). The effect of microalgae growth-promotion was greatest for H. lacustris; co-culturing with an initial bacterial optical density of 0.5 led to a 4-fold, 12-fold, 14-fold, and 12-fold amount of biomass for M. extorquens Rab1, M. mesophilicum Sab1, M. goesingense Vab1 and M. goesingense Vab2 respectively when compared to the control group. Co-inoculation with a lower load of bacterial cells (OD600 0.2) led only in the case of M. mesophilicum Sab1 to a significant increase in H. lacustris biomass (3.5-fold) after 7 days of incubation (Figure 2C). The growth rate of H. lacustris increased by 51% when co-cultivated with M. goesingense Vab1; this was the highest observed increase for all tested combinations. Algal CFUs for each treatment are provided in detail in Supplementary Tables S2A, S3A, and S4A together with the corresponding fluorescence intensities in Supplementary Tables S2B, S3B, and S4B.
FIGURE 2.
Microalgal cell counts after seven (grsy boxes) and 14 (blue boxes) days of incubation. C. vulgaris G1-G (A), S. vacuolatus G1-O (B), and H. lacustris G1-R (C) were co-inoculated with four different Methylobacterium strains in two differing cell concentrations (initial cell densities: OD600 0.2 and 0.5). Algal CFU was determined by measuring the fluorescence intensity (FI). For each box, the central line indicates the median, while the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers. Asterisks indicate significant differences in algal cell count compared to the control (∗p-value ≤ 0.05; ∗∗p-value ≤ 0.01; ∗∗∗p-value ≤ 0.001). The percentage of increased/reduced microalgal cell counts compared to the control is included.
Visualization of Mixed Cultures Using Confocal Laser Scanning Microscopy
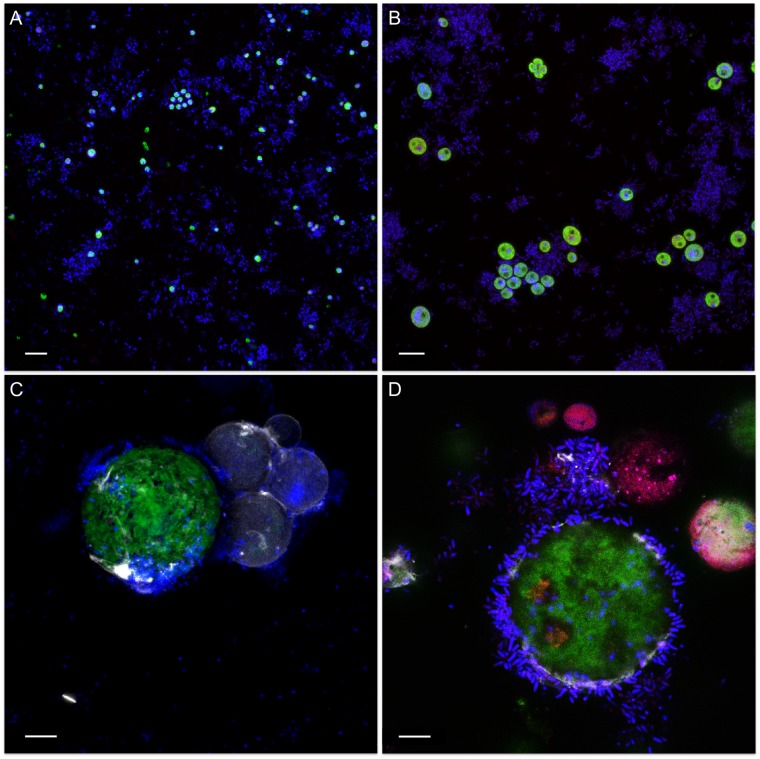
Visualization of mixed cultures revealed very loose associations between methylobacteria and C. vulgaris G1-G. Here, methylobacteria and algae were randomly and evenly distributed in the media and no distinct clustering was observed (Figure 3A). On the contrary, methylobacteria seem to embed S. vacuolatus cells and form cell aggregates (Figure 3B) and were found closely attached to the surface of viable H. lacustris cells (Figures 3C,D). Methylobacteria were also found in close proximity to disrupted, leaking H. pluvialis cells, seemingly feeding on microalgal cell debris (Figure 3D).
FIGURE 3.
Confocal laser scanning micrographs of algae-bacteria cultures. The micrographs show different algae (A: C. vulgaris G1-G; B: S. vacuolatus G1-O; C,D: H. lacustris G1-R) that were inoculated with M. goesingense Vab1 with an initial optical density of 0.5 and visualized after 7 days of incubation. The microbial communities were stained with the LIVE/DEAD bacterial viability kit (Thermo Fisher Scientific, MA, United States) that allows differentiating between living and dead cells in combination with calcofluor white (0.15%) to enhance fluorescence of the algae. Green: autofluorescence of algae; blue: living cells; red: dead cells. Scale bar: 10 μm.
Whole Genome Sequencing of Methylobacteria Provides Indication for Various Beneficial Traits
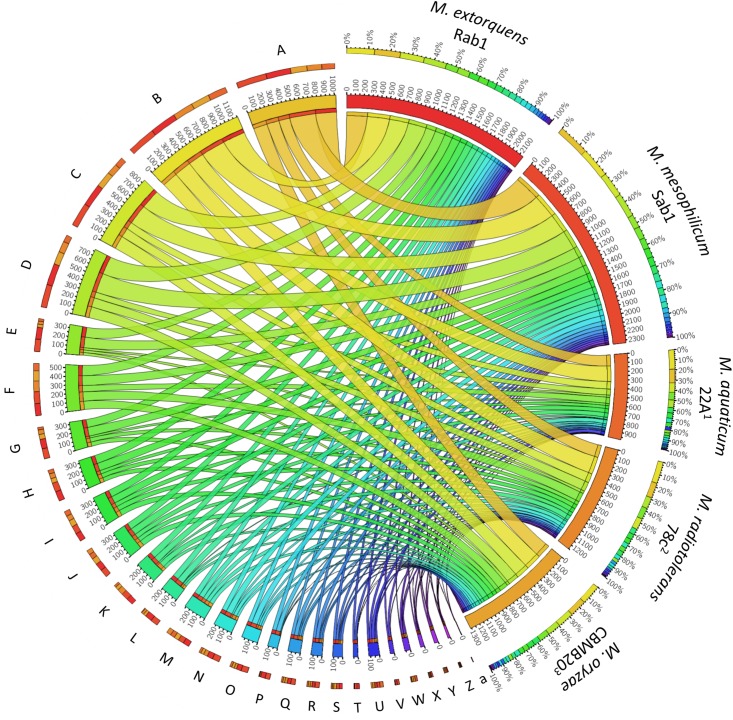
The total genome sizes were determined to be 5,747,629 bp, featuring a GC content of 68.2% (M. extorquens Rab1) and 6,367,318 bp, with a GC content of 68.6% (M. mesophilicum Sab1). The genome analyses revealed 5,952 and 7,795 coding sequences for M. extorquens Rab1 and M. mesophilicum Sab1, respectively. Detailed statistics are provided in Table 1 including reference genomes of plant-associated methylobacteria. Identified features were assigned to subcategories with the SEED database (Overbeek et al., 2005). The analyzed genomes harbored between 221 (M. extorquens Rab1) and 234 (M. mesophilicum Sab1) genes associated with the synthesis of cofactors, vitamins, prosthetic groups and pigments, including genes responsible for the synthesis of biotin, riboflavin, tetrapyrroles as well as folate. Between 18 and 23 genes were related to iron acquisition and metabolism, whereas M. extorquens Rab1 harbored eleven and M. mesophilicum Sab1 16 genes associated with the production of siderophores. Both investigated methylobacteria harbored four genes associated with the synthesis of auxins. Details related to the distribution of features within both analyzed genomes as well as reference genomes of other plant-associated methylobacteria are displayed in Figure 4 and Table 2, including detailed feature counts per organism per subcategory.
TABLE 1.
Detailed genome statistics for the analyzed Methylobacterium isolates.
| M. extorquens Rab1 | M. mesophilicum Sab1 | M. oryzae CBMB201 | M. aquaticum 22A2 | M. radiotolerans 78c3 | |
| Total genome size [bp] | 5,747,629 | 6,367,318 | 6,286,629 | 5,348,274 | 6,788,652 |
| GC content [%] | 68.2 | 68.6 | 69.8 | 71.1 | 71.2 |
| N50 | 252,941 | 132,863 | − | − | 57,566 |
| L50 | 8 | 13 | 1 | 1 | 38 |
| Number of scaffolds | 343 | 1660 | 1 | 1 | 271 |
| Number of features | 6,004 | 7,852 | 6,308 | 5,315 | 7,790 |
| Number of coding sequences | 5,952 | 7,795 | 6,243 | 5,212 | 7,740 |
| Number of RNAs | 52 | 57 | 65 | 103 | 50 |
The genomes were sequenced using Illumina NextSeq paired-end sequencing. Information about reference genomes of plant-associated methylobacteria is included. Strain references:1Kwak et al., 2014, 2Tani et al., 2015, 3Eevers et al., 2015.
FIGURE 4.
Feature distribution within the analyzed Methylobacterium genomes and reference genomes of plant-associated methylobacteria. The two algae-growth-promoting isolates M. extorquens Rab1 and M. mesophilicum Sab1 were compared with other isolates in order to identify distinct genetic features. Detected genes were grouped according to hierarchical assignment to SEED functions and visualized with Circos (http://circos.ca) to highlight the distribution of functional categories in each genome. A detailed feature count per organism per subcategory is included in Table 2 according to the SEED database after annotation using RAST (Aziz et al., 2008). Strain references: 1Kwak et al., 2014, 2Tani et al., 2015, 3Eevers et al., 2015.
TABLE 2.
Detailed feature count for each microorganism per subcategory according to the SEED database after annotation using RAST (Aziz et al., 2008).
| Subcategory | M. extorquens Rab1 | M. mesophilicum Sab1 | M. aquaticum 22A1 | M. radiotolerans 78c2 | M. oryzae CBMB203 | |
| A | Carbohydrates | 253 | 314 | 133 | 150 | 169 |
| B | Amino Acids and Derivatives | 272 | 294 | 189 | 210 | 192 |
| C | Protein Metabolism | 235 | 242 | 91 | 111 | 161 |
| D | Cofactors, Vitamins, Prosthetic Groups, Pigments | 221 | 234 | 73 | 113 | 131 |
| E | RNA Metabolism | 120 | 128 | 26 | 32 | 32 |
| F | Respiration | 124 | 119 | 76 | 111 | 103 |
| G | Stress Response | 98 | 105 | 25 | 47 | 51 |
| H | Fatty Acids, Lipids and Isoprenoids | 99 | 93 | 39 | 49 | 49 |
| I | Nucleosides and Nucleotides | 85 | 83 | 50 | 55 | 65 |
| J | DNA Metabolism | 78 | 82 | 53 | 46 | 66 |
| K | Membrane Transport | 74 | 70 | 24 | 60 | 36 |
| L | Cell Wall and Capsule | 71 | 77 | 8 | 23 | 29 |
| M | Motility and Chemotaxis | 63 | 60 | 47 | 47 | 47 |
| N | Virulence, Disease and Defense | 59 | 71 | 21 | 42 | 29 |
| O | Regulation and Cell signaling | 49 | 68 | 22 | 27 | 24 |
| P | Sulfur Metabolism | 48 | 49 | 10 | 3 | 4 |
| Q | Miscellaneous | 35 | 46 | 16 | 27 | 18 |
| R | Phosphorus Metabolism | 31 | 43 | 13 | 20 | 19 |
| S | Nitrogen Metabolism | 30 | 28 | 12 | 16 | 27 |
| T | Cell Division and Cell Cycle | 25 | 24 | 2 | 2 | 2 |
| U | Metabolism of Aromatic Compounds | 25 | 27 | 34 | 16 | 20 |
| V | Iron acquisition and metabolism | 18 | 23 | 0 | 5 | 0 |
| W | Phages, Prophages, Transposable elements Plasmids | 7 | 21 | 6 | 17 | 15 |
| X | Potassium metabolism | 11 | 12 | 5 | 7 | 7 |
| Y | Photosynthesis | 11 | 11 | 0 | 10 | 10 |
| Z | Secondary Metabolism | 5 | 6 | 5 | 5 | 6 |
| a | Dormancy and Sporulation | 1 | 3 | 0 | 1 | 1 |
Strain references: 1Kwak et al., 2014, 2Tani et al., 2015, 3Eevers et al., 2015.
Discussion
Our study revealed a co-occurrence of microalgae and methylobacteria in a natural biofilm when the whole bacterial community was assessed with high-throughput sequencing. Although the identified bacterial features assigned to the genus Methylobacteria occurred in rather low abundances, when compared to other bacterial constituents, we hypothesized that a synergistic relationship might occur between microalgae and distinct members of this genus. This is mainly due to their known association with land plants and the K-strategic lifestyle of methylobacteria based on their capability to grow on C1-compounds as sole carbon and energy source possibly exuded by algae. The synergistic relationship was confirmed by implementing different model organisms in co-cultivation experiments. Plant-associated methylobacteria were shown to significantly increase biomass formation at laboratory scale of two industrially relevant green algae in co-cultivation experiments. Genome analyses of the employed M. extorquens and M. mesophilicum strains revealed a number of features attributable to the observed algae growth-promotion, including genes involved in the production of vitamins, siderophores and auxins. The findings are supported by previous studies, where methylobacteria were identified in the phycosphere of Chlorella, Scenedesmus, Micrasterias, and Chlamydomonas (Levy et al., 2009; Krohn-Molt et al., 2017; Calatrava et al., 2018).
When different combinations were assessed, our co-cultivation experiments provided evidence that methylobacteria can stimulate the growth of certain microalgae through a species-specific relationship. In addition, also negative effects were observed for distinct algae-bacteria combinations and co-incubation periods. After 7 days of incubation, a significant decrease in biomass formation was observed for C. vulgaris irrespective of the applied methylobacteria. We assume that C. vulgaris and methylobacteria might compete for nutrients during the initial growth phase, resulting in a growth inhibition of the algae. Another possible explanation for the lower algal biomass when co-cultured with methylobacteria might be increased shading which attenuates light penetration (Kazamia et al., 2012). Carotenoids are known to have not only photoprotective capacities, but are also involved in the light-harvesting process during photosynthesis by expanding the light absorption spectrum. Detailed analyses of the methylobacteria genomes revealed that both investigated strains harbored genes related to the synthesis of bacterial light-harvesting proteins, as well as the photosynthetic reaction center of the photosystem type-II. Therefore, the beta-carotenoid-containing methylobacteria might also hamper photosynthetic activity of Chlorella through the competition for light (Van Dien et al., 2004). Although methylobacteria are able to produce vitamin B12 – an essential compound for many algae – Croft and colleagues could show that many microalgal species belonging to the genus Chlorella do not necessarily require cobalamin for proliferation (Croft et al., 2006). Moreover, the obtained visualizations of co-cultivated microorganisms revealed very loose associations between algae and methylobacteria in the case of C. vulgaris. The fact, that methylobacteria are not accumulated in the phycosphere of C. vulgaris, and thus impede a direct metabolite exchange, might also hamper a successful symbiotic relationship. Taken all these considerations into account, we suppose that the negative effects due to competition for nutrients, light and space outweigh the possible beneficial impact of the bacteria through the production of vitamins, auxins and siderophores for this bacteria-microalgae combination.
More distinctive effects were observed for S. vacuolatus; a significant increase in biomass formation was observed when it was co-cultured with methylobacteria after 7 days of incubation. The highest increase in biomass formation was achieved with both M. goesingense strains when they were added with an initial bacterial optical density of 0.5. While after 14 days the growth promoting effect of M. extorquens and M. mesophilicum was lost, significantly more algal biomass formation was still monitored in co-cultures with both M. goesingense strains, underlining the specificity of bacteria-microalgae symbioses. M. goesingense might metabolize the available nutrients more efficiently or harbors features which allows it to reduce the dissolved organic carbon produced by S. vacuolatus and thus the growth promoting effect was observed for prolonged time periods. Confocal laser scanning microscopy revealed algae cells that appeared to be embedded in bacterial aggregates. Similar colonization patterns were previously observed with methylobacteria and Chlamydomonas, where bacteria enable the growth of algae in a nitrogen depleted medium by mineralizing certain amino acids and peptides and thereby produce ammonium which consequently can be assimilated by the algae (Calatrava et al., 2018).
The algae growth-promoting effect of all investigated methylobacteria was strongest for H. lacustris, where up to 14-fold more algal biomass was formed after 7 days of incubation compared to the control. As reported by Croft et al. (2006), many Haematococcus species are dependent on an external supply of vitamins. In a vitamin-depleted medium, those essential micro-nutrients are provided by methylobacteria, allowing the microalgae to thrive. Visualization of H. lacustris co-cultures revealed that symbiotic bacteria appeared in close proximity of the microalgae, seemingly attached to the algal surface, allowing direct metabolite exchange. Similar colonization patterns of methylobacteria were observed on higher land plants, where bacteria are found attached to surface areas on leafs, attracted by the emitted methanol toward the stomata. The methanol produced by plants is a result of pectin-methylesterases de-esterifying the pectin within the cell wall during growth (Fall and Benson, 1996; Kutschera, 2007). During its life cycle, H. lacustris undergoes different cell stages; vegetative, motile cells become spherical, non-motile cyst cells under unfavorable conditions (Wayama et al., 2013). After 7 days of incubation, only a few flagellated cells were observable, while mostly green coccoid cells (palmelloid) and transitioning cells were found. In these stages, the microalgae change their extracellular matrix during the formation of a primary cell wall, which results in positive calcofluor-white staining indicating the presence of β-1,4-glycosidic linkages (Hagen et al., 2002). This was also evident during microscopic observations of the present study. Since the cell wall composition of Haematococcus is known to share similarities to those of plants, as they form primary and secondary cell walls consisting of cellulose and pectin along with other polysaccharides, a similar mechanism might be responsible for the attraction of methylobacteria in their surroundings (Wang et al., 2004). We found the highest growth-promoting effects for H. lacustris indicating that physical proximity is important for successful metabolite exchange and thus growth enhancement, as already suggested in previous studies (Gonzalez and Bashan, 2000; Hernandez et al., 2009). It is noteworthy to mention that the co-cultivation of microalgae and bacteria was not conducted under optimal conditions for any of the involved microorganisms. Nevertheless, this reflects conditions that prevail during large-scale cultivations where optimal light exposure and aeration are economically limited.
Microscopic observation of the analyzed biofilm revealed H. lacustris as the predominant algal species, which has likely an effect on the prevalent bacterial community. Natural co-occurrences of H. lacustris and methylobacteria, as revealed through 16S rRNA gene fragment amplicon sequencing, support the hypothesis of specific, evolutionary evolved co-occurrences of certain microalgae and bacteria in natural habitats. Complementary analysis of the two methylobacteria genomes revealed that both strains harbor several features which have already been described to positively contribute to algae growth including genes involved in the production of a variety of vitamins, such as cobalamin, biotin, thiamin and riboflavin, indicating the potential of methylobacteria to support algae growth. Reference data from other strains revealed that those genes are frequently found in different Methylobacterium spp., underlining the potential of methylobacteria as promising growth promoters of algae. In addition to genes involved in the synthesis of vitamins, genes attributable to the production of plant hormones were found in the genomes of the utilized model strains. Several studies have demonstrated that phytohormones can stimulate the growth and lipid production of microalgae (de-Bashan et al., 2008; Amin et al., 2015; Liu et al., 2016; Yu et al., 2017). Additionally, both genomes contained a substantial higher number of genes involved in iron acquisition and metabolism than genomes of other plant-associated methylobacteria that were implemented as a reference. It was previously shown that microalgae benefit from bacterial siderophores as the bioavailability of chelated iron is increased resulting in a “carbon for iron mutualism” where algae assimilate iron complexed in bacterial siderophores and in return provide the for the bacteria essential dissolved organic matter (Amin et al., 2009, 2012, 2015).
Our results provide new insights into the potential of plant-associated methylobacteria to promote the growth of two different, industrially relevant microalgal genera. Moreover, the observations lead us to the conclusion that symbiotic relationships are interchangeable between plant and microalgae hosts, but selective in terms of species specificity. In addition, our results provide evidence that a balanced ratio between symbiotic bacteria and microalgae is essential for algae growth-promotion, which facilitates a mutualistic relationship without interferences through the competition for nutrients, light and space. Deepening exploration of natural algae-bacteria relationships will further facilitate the design of synthetic microbial communities, which are a promising tool for biotechnology and might lead to improved cultivation procedures in the future.
Data Availability Statement
The amplicon datasets used and/or analyzed during the current study are available in the ENA repository (https://www.ebi.ac.uk/ena) under the accession number PRJEB35130; both genome assemblies are available in the ENA repository under the accession number PRJEB35220; and the sequences of the implemented algae strains were deposited in the ENA repository under accession number PRJEB36220.
Author Contributions
GB developed the study design. LK conducted the environmental sampling, performed the laboratory experiments and the bioinformatics analyses. HM processed the genomes. CD and CM discussed results. GB, LK, and TC interpreted the data and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Conflict of Interest
LK was employed by the company acib GmbH. CM and CD were employed by company bio-ferm GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We want to thank Birgit Wassermann (Graz) for her valuable help with microscopy.
Footnotes
Funding. This work has been supported by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol, the Government of Lower Austria and ZIT - Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00427/full#supplementary-material
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Amavizca E., Bashan Y., Ryu C.-M., Farag M. A., Bebout B. M., de-Bashan L. E. (2017). Enhanced performance of the microalga Chlorella sorokiniana remotely induced by the plant growth-promoting bacteria Azospirillum brasilense and Bacillus pumilus. Sci. Rep. 7:41310. 10.1038/srep41310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. A., Green D. H., Hart M. C., Küpper F. C., Sunda W. G., Carrano C. J. (2009). Photolysis of iron–siderophore chelates promotes bacterial–algal mutualism. Proc. Natl. Acad. Sci. U.S.A. 106 17071–17076. 10.1073/pnas.0905512106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. A., Hmelo L. R., van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522 98–101. 10.1038/nature14488 [DOI] [PubMed] [Google Scholar]
- Amin S. A., Parker M. S., Armbrust E. V. (2012). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76 667–684. 10.1128/MMBR.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao E., Jiang T., Girke T. (2014). AlignGraph: algorithm for secondary de novo genome assembly guided by closely related references. Bioinforma. Oxf. Engl. 30 i319–i328. 10.1093/bioinformatics/btu291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benemann J. R., Oswald W. J. (1996). Systems and Economic Analysis of Microalgae Ponds for Conversion of CO2 to Biomass. Final report. Berkeley, CA: California University. [Google Scholar]
- Berg G. (2009). Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84 11–18. 10.1007/s00253-009-2092-7 [DOI] [PubMed] [Google Scholar]
- Berg G., Köberl M., Rybakova D., Müller H., Grosch R., Smalla K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050. 10.1093/femsec/fix050 [DOI] [PubMed] [Google Scholar]
- Berg G., Zachow C., Müller H., Philipps J., Tilcher R. (2013). Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy 3 648–656. 10.3390/agronomy3040648 [DOI] [Google Scholar]
- Brennan L., Owende P. (2010). Biofuels from microalgae—A review of technologies for production, processing, and extractions of biofuels and co-products. Renew. Sustain. Energy Rev. 14 557–577. 10.1016/j.rser.2009.10.009 [DOI] [Google Scholar]
- Calatrava V., Hom E. F. Y., Llamas Á, Fernández E., Galván A. (2018). OK, thanks! A new mutualism between Chlamydomonas and methylobacteria facilitates growth on amino acids and peptides. FEMS Microbiol. Lett. 365:fny021. 10.1093/femsle/fny021 [DOI] [PubMed] [Google Scholar]
- Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M. B., Smith A. G. (2015). Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr. Opin. Plant Biol. 26 147–153. 10.1016/j.pbi.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438 90–93. 10.1038/nature04056 [DOI] [PubMed] [Google Scholar]
- Croft M. T., Warren M. J., Smith A. G. (2006). Algae need their vitamins. Eukaryot. Cell 5 1175–1183. 10.1128/ec.00097-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de-Bashan L., Antoun H., Bashan Y. (2008). Involvement of indole-3-acetic acid produced by the growth-promoting bacterium Azospirillum spp. promoting growth of Chlorella vulgaris. J. Phycol. 44 938–947. 10.1111/j.1529-8817.2008.00533.x [DOI] [PubMed] [Google Scholar]
- de-Bashan L. E., Bashan Y. (2010). Immobilized microalgae for removing pollutants: review of practical aspects. Bioresour. Technol. 101 1611–1627. 10.1016/j.biortech.2009.09.043 [DOI] [PubMed] [Google Scholar]
- Eevers N., Van Hamme J. D., Bottos E. M., Weyens N., Vangronsveld J. (2015). Draft genome sequence of Methylobacterium radiotolerans, a DDE-degrading and plant growth-promoting strain isolated from Cucurbita pepo. Genome Announc. 3:00488-15. 10.1128/genomeA.00488-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall R., Benson A. A. (1996). Leaf methanol — the simplest natural product from plants. Trends Plant Sci. 1 296–301. 10.1016/s1360-1385(96)88175-0 [DOI] [Google Scholar]
- Goecke F., Labes A., Wiese J., Imhoff J. F. (2010). Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409 267–299. 10.3354/meps08607 [DOI] [Google Scholar]
- Gonzalez L. E., Bashan Y. (2000). Increased growth of the microalga Chlorella vulgaris when coimmobilized and cocultured in alginate beads with the plant-growth-promoting bacterium Azospirillum brasilense. Appl. Environ. Microbiol. 66 1527–1531. 10.1128/aem.66.4.1527-1531.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen C., Siegmund S., Braune W. (2002). Ultrastructural and chemical changes in the cell wall of Haematococcus pluvialis (Volvocales, Chlorophyta) during aplanospore formation. Eur. J. Phycol. 37 217–226. 10.1017/s0967026202003669 [DOI] [Google Scholar]
- Hernandez J.-P., de-Bashan L. E., Rodriguez D. J., Rodriguez Y., Bashan Y. (2009). Growth promotion of the freshwater microalga Chlorella vulgaris by the nitrogen-fixing, plant growth-promoting bacterium Bacillus pumilus from arid zone soils. Eur. J. Soil Biol. 45 88–93. 10.1016/j.ejsobi.2008.08.004 [DOI] [Google Scholar]
- Hom E. F. Y., Aiyar P., Schaeme D., Mittag M., Sasso S. (2015). A chemical perspective on microalgal–microbial interactions. Trends Plant Sci. 20 689–693. 10.1016/j.tplants.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Joint I., Tait K., Callow M. E., Callow J. A., Milton D., Williams P., et al. (2002). Cell-to-Cell communication across the prokaryote-eukaryote boundary. Science 298 1207–1207. 10.1126/science.1077075 [DOI] [PubMed] [Google Scholar]
- Kazamia E., Aldridge D. C., Smith A. G. (2012). Synthetic ecology – A way forward for sustainable algal biofuel production? J. Biotechnol. 162 163–169. 10.1016/j.jbiotec.2012.03.022 [DOI] [Google Scholar]
- Khan M. I., Shin J. H., Kim J. D. (2018). The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Fact. 17:36. 10.1186/s12934-018-0879-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-H., Ramanan R., Cho D.-H., Oh H.-M., Kim H.-S. (2014). Role of Rhizobium, a plant growth promoting bacterium, in enhancing algal biomass through mutualistic interaction. Biomass Bioenerg. 69 95–105. 10.1016/j.biombioe.2014.07.015 [DOI] [Google Scholar]
- Krohn-Molt I., Alawi M., Förstner K. U., Wiegandt A., Burkhardt L., Indenbirken D., et al. (2017). Insights into microalga and bacteria interactions of selected phycosphere biofilms using metagenomic, transcriptomic, and proteomic approaches. Front. Microbiol. 8:1941. 10.3389/fmicb.2017.01941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U. (2007). Plant-associated methylobacteria as co-evolved phytosymbionts. Plant Signal. Behav. 2 74–78. 10.4161/psb.2.2.4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M.-J., Jeong H., Madhaiyan M., Lee Y., Sa T.-M., Oh T. K., et al. (2014). Genome information of Methylobacterium oryzae, a plant-probiotic methylotroph in the phyllosphere. PLoS One 9:e106704. 10.1371/journal.pone.0106704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J. L., Stauber J. L., Wakelin S. A., Jolley D. F. (2009). The effect of bacteria on the sensitivity of microalgae to copper in laboratory bioassays. Chemosphere 74 1266–1274. 10.1016/j.chemosphere.2008.10.049 [DOI] [PubMed] [Google Scholar]
- Liu J., Qiu W., Song Y. (2016). Stimulatory effect of auxins on the growth and lipid productivity of Chlorella pyrenoidosa and Scenedesmus quadricauda. Algal Res. 18 273–280. 10.1016/j.algal.2016.06.027 [DOI] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63 541–556. 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Lundberg D. S., Yourstone S., Mieczkowski P., Jones C. D., Dangl J. L. (2013). Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10 999–1002. 10.1038/nmeth.2634 [DOI] [PubMed] [Google Scholar]
- Overbeek R., Begley T., Butler R. M., Choudhuri J. V., Chuang H.-Y., Cohoon M., et al. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33 5691–5702. 10.1093/nar/gki866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renuka N., Guldhe A., Prasanna R., Singh P., Bux F. (2018). Microalgae as multi-functional options in modern agriculture: current trends, prospects and challenges. Biotechnol. Adv. 36 1255–1273. 10.1016/j.biotechadv.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Seymour J. R., Amin S. A., Raina J.-B., Stocker R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2:17065. 10.1038/nmicrobiol.2017.65 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Sharma N. (2017). Industrial and biotechnological applications of algae: a review. J. Adv. Plant Biol. 1:01. [Google Scholar]
- Spolaore P., Joannis-Cassan C., Duran E., Isambert A. (2006). Commercial applications of microalgae. J. Biosci. Bioeng. 101 87–96. 10.1263/jbb.101.87 [DOI] [PubMed] [Google Scholar]
- Tani A., Ogura Y., Hayashi T., Kimbara K. (2015). Complete genome sequence of Methylobacterium aquaticum strain 22A, isolated from Racomitrium japonicum Moss. Genome Announc. 3:e00266-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dien S., Marx C., O’Brien B., Lidstrom M. (2004). Genetic characterization of the carotenoid biosynthetic pathway in Methylobacterium extorquens AM1 and isolation of a colorless mutant. Appl. Environ. Microbiol. 69 7563–7566. 10.1128/aem.69.12.7563-7566.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verginer M., Siegmund B., Cardinale M., Müller H., Choi Y., Míguez C. B., et al. (2010). Monitoring the plant epiphyte Methylobacterium extorquens DSM 21961 by real-time PCR and its influence on the strawberry flavor. FEMS Microbiol. Ecol. 74 136–145. 10.1111/j.1574-6941.2010.00942.x [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang W., Chen L., Wang J., Liu T. (2013). The contamination and control of biological pollutants in mass cultivation of microalgae. Bioresour. Technol. 128 745–750. 10.1016/j.biortech.2012.10.158 [DOI] [PubMed] [Google Scholar]
- Wang S.-B., Hu Q., Sommerfeld M., Chen F. (2004). Cell wall proteomics of the green alga Haematococcus pluvialis (Chlorophyceae). Proteomics 4 692–708. 10.1002/pmic.200300634 [DOI] [PubMed] [Google Scholar]
- Wayama M., Ota S., Matsuura H., Nango N., Hirata A., Kawano S. (2013). Three-dimensional ultrastructural study of oil and astaxanthin accumulation during encystment in the green alga Haematococcus pluvialis. PLoS One 8:e53618. 10.1371/journal.pone.0053618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B., Bishop S., Stessman D., Wright D., Spalding M. H., Halverson L. J. (2013). Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B12-producing bacteria. ISME J. 7 1544–1555. 10.1038/ismej.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Song M., Pei H., Jiang L., Hou Q., Nie C., et al. (2017). The effects of combined agricultural phytohormones on the growth, carbon partitioning and cell morphology of two screened algae. Bioresour. Technol. 239 87–96. 10.1016/j.biortech.2017.04.120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The amplicon datasets used and/or analyzed during the current study are available in the ENA repository (https://www.ebi.ac.uk/ena) under the accession number PRJEB35130; both genome assemblies are available in the ENA repository under the accession number PRJEB35220; and the sequences of the implemented algae strains were deposited in the ENA repository under accession number PRJEB36220.