Dear Editor,
Signet-ring cell carcinoma (SRCC) is a rare subtype of colorectal cancer (CRC) characterized histologically by the accumulation of mucins in the cytoplasm and displacement of nuclei to the cellular periphery, accounting for about 1% CRC (Fig. S1A) (Borger et al., 2007). Compare to common subtypes of CRC, such as adenocarcinoma (AC) and mucinous adenocarcinoma (MAC), SRCC is associated with aggressive behaviors and younger age at presentation (Kang et al., 2005; Sung et al., 2008; Nitsche et al., 2013; Hugen et al., 2014; Inamura et al., 2015). A retrospective analysis of CRC patient’s data at Fudan University Shanghai Cancer Center (FUSCC) also indicated a worse overall and disease-free survival of SRCC patients (Fig. S1B and S1C, Table S1). Due to low incidence and occasionally mixed presence of SRCC with AC or MAC, genome-wide characterization of SRCC at a large scale is challenging. Limited whole-exome sequencing (n = 5) and gene panel sequencing (n = 35) results indicate that, most driver mutations associated with CRC, such as APC, KRAS, and PIK3CA, are mutated at lower rates in SRCC (Nam et al., 2018; Korphaisarn et al., 2019). Mutations and signaling pathways responsible for the tumorigenesis of SRCC remains to be uncovered.
Following microscopic analysis of 4,000 CRC specimens, we identified 29 SRCCs with a high percentage (>70%) of signet-ring cells. To gain a better understanding of genomic alterations in SRCC, we performed WES on these SRCCs and paired normal tissues (Tables S2 and S3). WES data of AC and MAC from the Cancer Genome Atlas (TCGA) were analyzed for comparison (Tables S4 and S5) (TCGA, 2012).
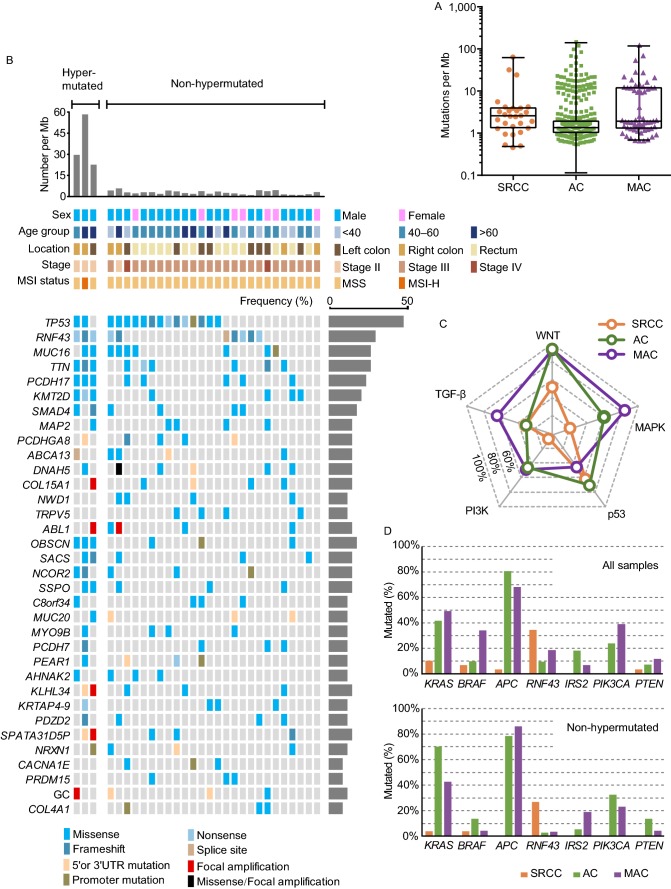
We identified 9,752 non-silent somatic mutations in SRCC samples, with dominant C>T/G>A substitutions enriched at CpG islands (Fig. S2 and Tables S6–9). The overall mutation rate of SRCC is 9.65/Mb (medium mutation rate is 3/Mb), and 3 cases (10.3%, with POLE mutation or microsatellite instability-high) were considered as hypermutated (Fig. 1A and Table S3). In total, there were 34 recurrent alterations, with TP53 (55.2%), RNF43 (34.5%), MUC16 (31.0%), TTN (31.0%), PCDH17 (27.6%), KMT2D (24.1%), and SMAD4 (20.7%) as most frequently mutated genes (Fig. 1B and Table S10). Mutations on critical cancer drivers were validated by Sanger sequencing (Fig. S3 and Table S11). By comparing mutated genes in different cancer signaling pathways, we noticed that genes in p53 (e.g., TP53) and TGF-β (e.g., SMAD4) pathways were mutated at similar frequencies cross CRC subtypes, however the mutation burden in WNT, MAPK, and PI3K pathways were dramatically lower in SRCC, suggesting subtype-specific molecular signatures (Figs. 1C and S4).
Figure 1.
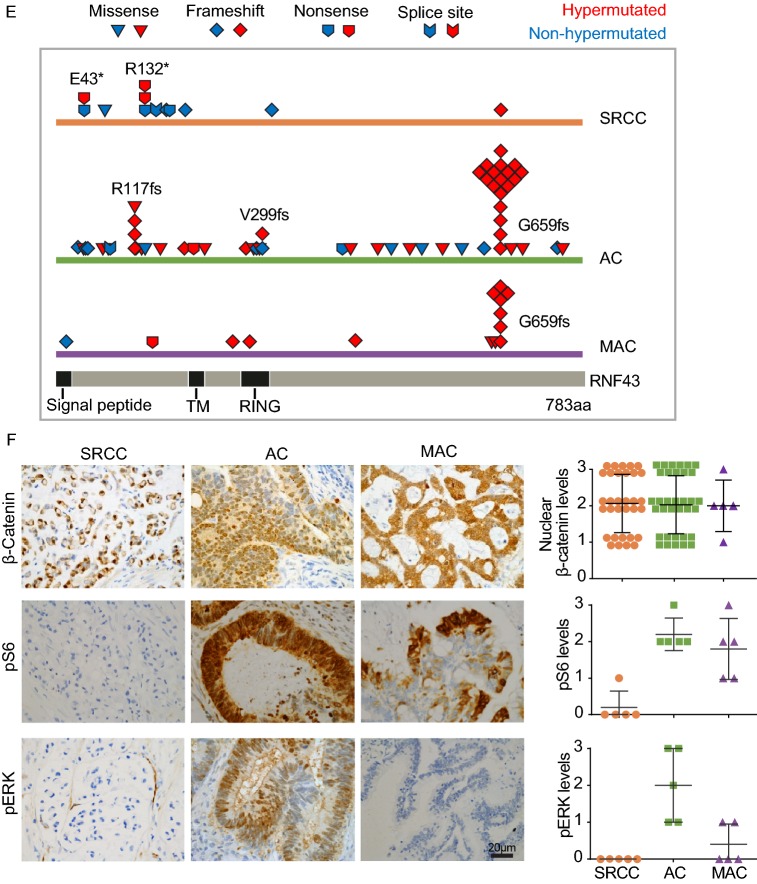
WES and pathway analysis of colorectal SRCC. (A) The tumor mutation burden (TMB) in colorectal SRCC (n = 29) from FUSCC and adenocarcinoma (AC, n = 458) and mucinous adenocarcinoma (MAC, n = 59) from the Cancer Genome Atlas (TCGA) cohort. (B) Top mutated genes and mutational landscape in 29 samples of colorectal SRCC. Different types of mutation were distinguished by colors. Frequency of each significant alteration was shown on the right of the heatmap. TMB, demographic, and clinical information for each patient were illustrated on the top of the heatmap. (C) Comparison of the alteration frequencies of top CRC-associated signaling pathways in SRCC, AC and MAC. (D) Comparison of mutation rates of cancer drivers in SRCC, AC, and MAC, all samples and non-hypermutated samples were analyzed. (E) Distribution and types of RNF43 mutations in SRCC, AC and MAC. (F) The activating status of WNT (β-catenin), MAPK (pERK Thr202/Tyr204) and PI3K (pS6 Ser240/244) pathways were assessed by immunohistochemistry (IHC, left), and quantified (right). Scale bar, 20 μm
The WNT pathway is instrumental to intestinal homeostasis and the initiation of AC (Nusse and Clevers, 2017). A striking difference between SRCC and AC/MAC was the mutated genes in the WNT pathway (Figs. 1C,1D, S4 and S5). APC mutation was most prevalent in AC (~80%) and MAC (~70%) (TCGA, 2012), whereas it only occurred in one (3.4%) hypermutated SRCC, this is consistent with recent reports (Nam et al., 2018; Korphaisarn et al., 2019). Interestingly, SRCC was associated with frequent mutations in RNF43, with nonsense mutations (p.Glu43* and p.Arg132*) enriched at the N-terminus, regardless of mutation burden (Fig. 1D and 1E). In AC/MAC, most RNF43 mutations occurred in hypermutated tumors, with a hotspot at C-terminal (p.Gly659 frameshift) (Fig. 1E and Table S12) (Giannakis et al., 2014; Yan et al., 2017). Both APC and RNF43 are key regulators of the WNT pathway, their inactivation leads to stabilization and nuclear translocation of β-catenin (Nusse and Clevers, 2017). Almost all SRCC, AC, and MAC samples showed discernable nuclear staining of β-catenin by immunohistochemistry (IHC) (Fig. 1F). It appeared that, different mechanisms are employed by CRC subtypes to activate β-catenin, in which SRCC prefers a complete inactivation of RNF43 (N-terminal nonsense mutation), whereas AC and MAC prefer APC mutation. In addition, DKK4 amplification and mutations on FZD10, AMER1, and AXIN2 were identified in SRCCs, which may also contribute to β-catenin activation in the absence of RNF43 and APC mutation (Fig. S5).
MAPK and PI3K pathways are also important in the development of AC (Fearon and Vogelstein, 1990; TCGA, 2012; Sanchez-Vega et al., 2018). SRCC presented a lower mutation load in MAPK (20.7%, 60.5%, and 84.7% in SRCC, AC, and MAC respectively) and PI3K pathways (6.9%, 46.1%, and 49.2% in SRCC, AC, and MAC respectively) (Figs. 1C, S4 and S6). The mutation rates of several cancer driver genes were dramatically different across different subtypes. For instance, mutations in PIK3CA, PTEN, IRS2 were enriched in AC and MAC, whereas almost absent in SRCC; and KRAS mutation rate in SRCC was about 4-fold lower than that in AC or MAC (Fig. 1D). Lower mutation rates of PIK3CA and KRAS in SRCC have also been reported recently (Korphaisarn et al., 2019). The phospho-ERK and phospho-S6 signals, indicators of MAPK and PI3K activities respectively, were much lower in SRCC, which was consistent with mutation data (Fig. 1F).
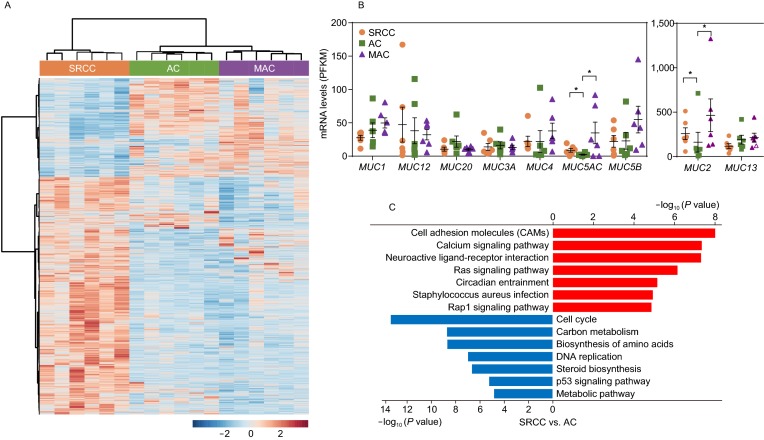
To identify subtype-specific gene expression signatures, we performed RNA-seq with SRCC, MAC and AC samples (n = 6 per subtype) (Table S13). The gene expression pattern of SRCC was drastically different from that of AC and MAC (Fig. 2A and Table S14), largely consistent with a previous study (Nam et al., 2018). Two mucin genes, MUC2 and MUC5AC, were highly expressed in SRCC and MAC, which may contribute to elevated production of mucin (Fig. 2B). Pathway enrichment analysis showed that upregulated genes in SRCC were associated with cell adhesion and calcium signaling pathways, while downregulated genes were associated with cell cycle and cellular metabolism pathways (Fig. 2C). Similar results were obtained when SRCC and MAC expression data were compared (Fig. S7).
Figure 2.
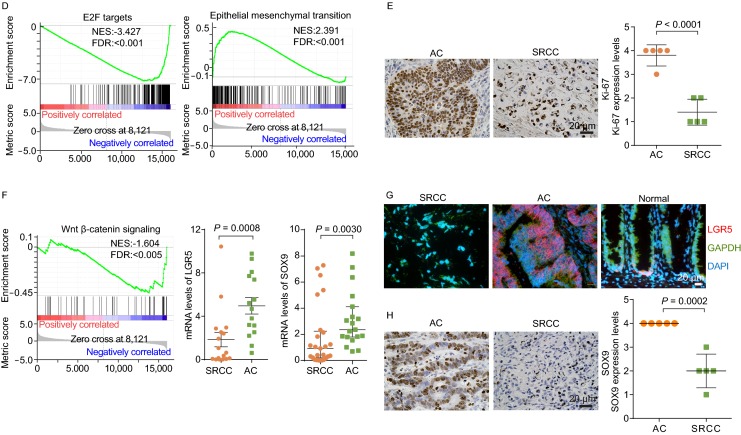
Gene expression analysis of colorectal SRCC. (A) Heatmap showing differentially expressed genes in SRCC, AC, and MAC (n = 6 per subtype, samples from FUSCC) by RNA-seq. Blue and red colors represent down- and up-regulated genes respectively. (B) Expression levels of mucin genes were compared, and MUC2 and MUC5AC, were highly expressed in SRCC and MAC. RPKM data from RNA-seq was shown, * indicate P > 0.05 (Student’s t test). For MUC2 gene, the highest data point of each group was considered as outlier and not included in analysis. (C) Kyoto encyclopedia of genes and genomes (KEGG) pathways enriched in differentially expressed genes between SRCC and AC. (D) Gene sets of E2F targets and epithelial-mesenchymal transition (EMT) were enriched in differentially expressed genes between SRCC and AC. (E) The expression (IHC, left) of proliferation marker Ki-67 in SRCC and AC, with quantification (right). (F) Targets of Wnt-β-catenin were downregulated in SRCC, and the mRNA expression of LGR5 and SOX9 was assessed in a larger cohort of SRCC and AC specimens by qPCR. (G) The mRNA expression of LGR5 in SRCC, AC, and normal colon tissue assessed by ISH. (H) The protein expression of SOX9 in SRCC and AC assessed by IHC. Scale bar, 20 μm
Using the gene set enrichment analysis (GSEA), the differentially expressed genes in SRCC (compared to AC) were negatively correlated with E2F signaling, MTOR signaling, and metabolism (Figs. 2D and S8), which may underpin the slower proliferation of SRCC, as indicated by significantly lower Ki-67 expression in SRCC (Fig. 2E). On the other hand, the expression profile of SRCC was positively associated with terms of EMT and angiogenesis, which may contribute to the aggressiveness and local metastasis of SRCC. The negative association between proliferation and metastasis appears counterintuitive, however as reported previously, slow proliferation may gain ability to spread or survive outside of original microenvironment (Anjomshoaa et al., 2009).
Another term negatively associated with genes expression in SRCC was Wnt-β-catenin signaling (Fig. 2F). Many β-catenin target genes, such as LGR5, SOX9, AXIN2, and MSI1, were expressed at lower levels in SRCC compared to those in AC (Table S14). The relatively lower expression of LGR5 and SOX9 in SRCC was further verified by quantitative PCR, in situ hybridization, or IHC (Fig. 2F–H). Comparing to APC mutations in AC or MAC, RNF43 mutations may moderately activate β-catenin, which in turn promote tumorigenesis of SRCC.
In summary, as indicated by genomic and transcriptomic profiling, SRCC represents a molecularly distinct subtype of CRC. Our findings will serve as a blueprint for dissecting pathogenic mechanisms of SRCC, and provide insights in developing molecularly-targeted therapies for SRCC patients.
FOOTNOTES
This study is supported by grants from the National Natural Science Foundation of China (81622038, 31571479, and 81772965 to F.X. Y., 31470826 and 31670858 to G. H.), the National key R&D program of China (2018YFA0800304) to F.X. Y., Science and Technology Commission of Shanghai Municipality (19JC1411100 to F.X. Y., 16411966300 to G. H., 16411966300 and 18401933402 to J. P.), Shanghai Municipal Commission of Health and Family Planning (2017BR018 to F.X. Y.) and Shanghai Sailing Program (19YF1409500 to Y. L.). We would like to thank Dr. Kang Chen for proofreading of this manuscript.
G. H., J. P., and F.X. Y. designed the experiments. Specimens and clinical data collection: Y. Li, R. W., S. C., and J. P. prepared specimens and collected clinical data. H. W., and F.X. Y. performed WES. D. Z., D. D., and Y. Liu processed WES data. S. H., J. P., and F.X. Y. performed RNAseq and analysis. Y. Li, J. L., X. W., Y. W., G. F., C. F., X. M., M. P., Q. G., X. X., X. H., Y. Z., and S. M. performed experiments. Y. Li, J. L., J. G., L. Z., Q. H., and F.X. Y. analyzed data. Y. Li, J. L., L. Z., Q. H., and F.X. Y. wrote the manuscript.
All authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Footnotes
Yaqi Li, Jian Li, and Renjie Wang have contributed equally to this work.
Contributor Information
Guoqiang Hua, Email: guoqianghua@fudan.edu.cn.
Junjie Peng, Email: pengjj67@hotmail.com.
Fa-Xing Yu, Email: fxyu@fudan.edu.cn.
References
- Anjomshoaa A, Nasri S, Humar B, McCall JL, Chatterjee A, Yoon HS, McNoe L, Black MA, Reeve AE. Slow proliferation as a biological feature of colorectal cancer metastasis. Br J Cancer. 2009;101:822–828. doi: 10.1038/sj.bjc.6605229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borger ME, Gosens MJ, Jeuken JW, van Kempen LC, van de Velde CJ, van Krieken JH, Nagtegaal ID. Signet ring cell differentiation in mucinous colorectal carcinoma. J Pathol. 2007;212:278–286. doi: 10.1002/path.2181. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- Giannakis M, Hodis E, Mu XJ, Yamauchi M, Rosenbluh J, Cibulskis K, Saksena G, Lawrence MS, Qian ZR, Nishihara R, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46:1264–1266. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugen N, van de Velde CJ, de Wilt JH, Nagtegaal ID. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. 2014;25:651–657. doi: 10.1093/annonc/mdt591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura K, Yamauchi M, Nishihara R, Kim SA, Mima K, Sukawa Y, Li T, Yasunari M, Zhang X, Wu K, et al. Prognostic significance and molecular features of signet-ring cell and mucinous components in colorectal carcinoma. Ann Surg Oncol. 2015;22:1226–1235. doi: 10.1245/s10434-014-4159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, O’Connell JB, Maggard MA, Sack J, Ko CY. A 10-year outcomes evaluation of mucinous and signet-ring cell carcinoma of the colon and rectum. Dis Colon Rectum. 2005;48:1161–1168. doi: 10.1007/s10350-004-0932-1. [DOI] [PubMed] [Google Scholar]
- Korphaisarn K, Morris V, Davis JS, Overman MJ, Fogelman DR, Kee BK, Dasari A, Raghav KPS, Shureiqi I, Trupti M, et al. Signet ring cell colorectal cancer: genomic insights into a rare subpopulation of colorectal adenocarcinoma. Br J Cancer. 2019;121:505–510. doi: 10.1038/s41416-019-0548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JY, Oh BY, Hong HK, Bae JS, Kim TW, Ha SY, Park D, Lee WY, Kim HC, Yun SH, et al. Molecular characterization of colorectal signet-ring cell carcinoma using whole-exome and RNA sequencing. Transl Oncol. 2018;11:836–844. doi: 10.1016/j.tranon.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche U, Zimmermann A, Spath C, Muller T, Maak M, Schuster T, Slotta-Huspenina J, Kaser SA, Michalski CW, Janssen KP et al. (2013) Mucinous and signet-ring cell colorectal cancers differ from classical adenocarcinomas in tumor biology and prognosis. Ann Surg 258:775–782; discussion 782-773. [DOI] [PMC free article] [PubMed]
- Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e310. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CO, Seo JW, Kim KM, Do IG, Kim SW, Park CK. Clinical significance of signet-ring cells in colorectal mucinous adenocarcinoma. Mod Pathol. 2008;21:1533–1541. doi: 10.1038/modpathol.2008.170. [DOI] [PubMed] [Google Scholar]
- TCGA Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HHN, Lai JCW, Ho SL, Leung WK, Law WL, Lee JFY, Chan AKW, Tsui WY, Chan ASY, Lee BCH, et al. RNF43 germline and somatic mutation in serrated neoplasia pathway and its association with BRAF mutation. Gut. 2017;66:1645–1656. doi: 10.1136/gutjnl-2016-311849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.