Abstract
Procedure-related cardiac electronic implantable device (CIED) infections have high morbidity and mortality, highlighting the urgent need for infection prevention efforts to include electrophysiology procedures. We developed and validated a semi-automated algorithm based on structured electronic health records data to reliably identify CIED infections. A sample of CIED procedures entered into the Veterans’ Health Administration Clinical Assessment Reporting and Tracking program from FY 2008–2015 was reviewed for the presence of CIED infection. This sample was then randomly divided into training (2/3) validation sets (1/3). The training set was used to develop a detection algorithm containing structured variables mapped from the clinical pathways of CIED infection. Performance of this algorithm was evaluated using the validation set. 2,107 unique CIED procedures from a cohort of 5,753 underwent manual review; 97 CIED infections (4.6%) were identified. Variables strongly associated with true infections included presence of a microbiology order, billing codes for surgical site infections and post-procedural antibiotic prescriptions. The combined algorithm to detect infection demonstrated high c-statistic (0.95; 95% confidence interval: 0.92–0.98), sensitivity (87.9%) and specificity (90.3%) in the validation data. Structured variables derived from clinical pathways can guide development of a semi-automated detection tool to surveil for CIED infection.
Subject terms: Cardiac device therapy, Epidemiology
Introduction
Cardiovascular electronic implantable devices (CIEDs), such as pacemakers and implantable automated defibrillators, are increasing as the population ages1,2. Procedure-related CIED infections are associated with high morbidity, mortality, and medical costs. Mortality rates for deep infections involving leads implanted into the heart approach 20% and infections are estimated to cost greater than $50,0003–5. CIED infections are increasingly a concern for cardiologists, infection control departments, and patient safety programs due to a trend toward an increasing rate of procedure-related infections1. The increased rate of CIED infections is compounded by the overall increasing rates of CIED placements6. Given this trend, developing and implementing effective infection prevention programs in the electrophysiology suite is crucial for minimizing morbidity and mortality among CIED implantation recipients.
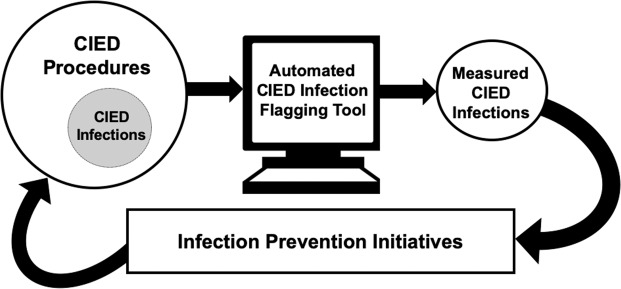
Surveillance with audit and feedback is a cornerstone of, and critical first step in, infection prevention (Fig. 1)7. Surveillance programs measure infections when they occur. This information can then be used to identify operational and patient-centered targets for prevention initiatives after infections and clusters are found. Surveillance programs can improve outcomes in several ways. They can be used to measure the effectiveness of novel infection prevention measures in real-time8 and can also provide feedback to providers to encourage uptake of evidence-based prevention practices9. Surveillance systems can also improve care through the observer, or Hawthorne, effect10. The Surgical Care Improvement Project (SCIP) infection metric bundle is an example of a surveillance and reporting system that lead to major improvements in surgical care and a reduction in post-operative infections11. However, due to the nature of the electrophysiology laboratory as a procedural area that straddles inpatient and outpatient settings, it is not encompassed by surgical site infection (SSI) quality improvement initiatives12.
Figure 1.
Automated electronic surveillance for CIED infections as a part of infection prevention and control efforts.
Given limited resources, and the increasing dissemination of electronic health records (EHRs), a promising strategy for expanding surveillance to uncovered procedural areas is the development of surveillance tools that leverage clinical data warehouses to measure and track infections. These tools could be used as stand-alone systems or could be used in a semi-automated method to augment and triage the manual review process toward cases at highest probability of having an adverse event7,13. Thus, we sought to develop and validate a surveillance tool for CIED infection surveillance using structured data elements from the Veterans Health Administration (VA)’s Corporate Data Warehouse (CDW) in order to triage cases for manual review.
Methods
Study overview
This study used structured data elements along the diagnostic and treatment pathway of CIED infection management from the VA EHR captured in the CDW to develop an algorithm to detect infections. We used a limited definition of structured data focusing on diagnostic testing orders, laboratory results, pharmacy and billing data that is structured and standardized across all EHRs in order to ensure broad potential applicability of the detection algorithm. A sample of cardiac procedures collected as part of the VA Clinical Assessment Reporting and Tracking (CART) quality program underwent manual review for the presence of infection; these cases were used in the development and validation of the semi-automated tool.
Ethical considerations
Given the retrospective nature of the study, waiver of consent was obtained. The Veterans Health Administration Boston Healthcare System Institutional Review Board and the University of Colorado Denver Anschutz Medical Campus Multiple Institution Review Board (covering the University of Colorado Denver and its affiliates: Children’s Hospital Colorado, Denver Health and Hospital Authority, University of Colorado Hospital, and the VA Eastern Colorado Health Care System) approved this study and waived the need for informed consent as part of study approval prior to data collection and analysis. The study was also approved by the VA CART-Electrophysiology (EP) program. All methods were carried out in accordance with relevant guidelines and regulations.
Cohort development
Our cohort was a subset of CIED procedures captured by the CART program. CART is a national quality initiative integrated into the VA electronic medical record, which collects data about clinical outcomes, co-morbidities, and procedural details, but does not include measurement of infections. CART reporting is mandatory for all cardiac catheterization procedures and optional for EP procedures, including device implantations and revisions. Approximately 20% of EP device procedures performed across the national VA healthcare system are captured by CART-EP. Multiple clinical and procedural variables are captured and combined with other data from the VA CDW to create a single national data repository14,15.
CIED procedures, including implantations and revisions of permanent pacemakers, implantable cardioverter defibrillators, biventricular pacemaker-implantable cardioverter-defibrillators, and biventricular pacemakers, entered into the CART-EP program during the period from 10/2007–9/2015 were considered for inclusion (N = 5753). Given the volume of CIED procedures performed and relatively low prevalence of CIED infections, an enhanced sampling approach was undertaken; details of the sampling process are described in prior published work and included in Supplementary Methods S114,15.
After development of the cohort, a sample of cases underwent manual review by a trained clinician (AA, PM, WBE) to identify CIED infections that occurred within 90 days of the index procedure, applying standard cardiac device infection definitions based on clinical symptoms, microbiology results, and/or clinician diagnosis, based on recommendations from the Centers for Disease Control National Healthcare Safety Network and multi-society guidelines16–18. Further details on cohort development and the manual review process, including how infections were measured and defined as well as the other variables collected, are available in Supplementary Methods S1 and S2 and related references14,15,17.
Diagnostic and treatment pathways and selection of potential identifying variables
Diagnostic variables
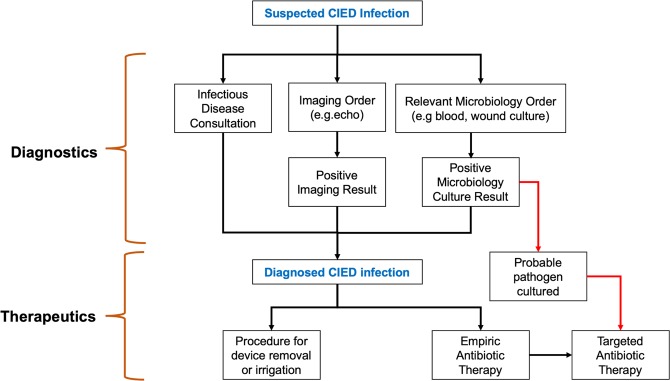
CIED infections can be diagnosed and treated in several ways (Fig. 2); these interventions generate orders and results that can potentially be leveraged for retrospective infection detection17. Many infections are first identified due to fever, which is recorded in vital signs data as a continuous numeric variable. Microbiologic testing generates an order, which is a structured data element in the EHR, and a result, which is unstructured and not standardized. Microbiologic cultures can be obtained from the site of the device (e.g., wound cultures) or can be blood cultures, used to diagnose systemic infection, such as endocarditis. Thus, there are four potential flags associated with microbiologic testing to diagnose CIED infections: 1) a blood culture order, 2) a wound culture order, 3) wound culture result and 4) a blood culture positive result. Microbiology testing is important because it is an essential part of infection diagnosis and an essential aspect of most infection surveillance definitions. If cultures are positive, they can also provide information about the organism causing the infection and guide antibiotic treatment. Diagnosis of CIED infections may also involve imaging procedures, such as echocardiography, often transesophageal echocardiography, due to how endocarditis is defined. Mechanisms of echocardiography ordering and reporting of results are highly variable and not standardized across the VA EHR.
Figure 2.
Clinical process of CIED infection cases.
Treatment variables
Treatment of CIED infections generally involves incision and drainage of fluid collections, which may generate a structured procedure code, antimicrobials, which generate an antimicrobial order, and, in many cases, removal of the CIED with later replacement, thus generating a subsequent current procedural terminology (CPT) code. Another aspect of treatment is often clinical consultation, often with either infectious diseases or cardiology. Consultation is also recorded in various places throughout the EHR (clinical notes, orders, consult section) and is thus not a standardized data element.
Development of the surveillance algorithm
Based on diagnostic and treatment pathways, we identified potentially relevant variables to develop the infection detection algorithm using structured data elements stored in CDW. Administrative billing codes, which are highly structured, were also considered for potential inclusion. Specifically, we used the Inpatient and Outpatient tables to obtain CPT and International Classification of Disease (ICD) 9 and 10 codes (Supplementary Methods S2), pharmacy tables for drug order data, Microbiology tables for laboratory orders, and vitals tables. These variables were classified as diagnostic of infection, treatment of infection, or billing for infection-related healthcare utilization. Unstructured variables, such as microbiology results, echocardiography completion, and consultation orders, where not considered for inclusion in the final identification model due to programming complexity and high facility-level variation in the text data; thus, inclusion of these types of variables was felt to limit generalizability. However, these manually extracted variables were evaluated as part of a sensitivity analysis to determine if their inclusion would improve algorithm performance if available and searchable in some EHRs. In the sensitivity analysis, microbiology results were further sub-classified into positive cultures with a likely infecting organism (probable pathogen) and positive cultures likely representing contamination.
Statistical analysis
Demographic and clinical characteristics of CIED patients by infection status were compared. Chi-squared tests were used to compare categorical variables and Mann–Whitney Wilcoxon tests were used for continuous variables.
In order to build and test the infection surveillance algorithm, we randomly split the data into a training set, including approximately two-thirds of the observations, and a validation set of the remaining one-third19. Only the electronically-available structured variables (e.g. microbiology orders, rather than results) were considered for inclusion in the final tool, to ensure scalability and portability across EHRs. Based on the univariate association with infection status and clinical reasoning, the following were evaluated for inclusion in the flagging model: presence of a microbiology order, repeat procedure status, fever, blood, wound, antibiotic post-procedure, and a limited list of antibiotics commonly used to treat CIED infections post-procedure (Supplementary Note S3) SSI code (998.x), other CPT or ICD infection code (categorized as general infection code, material infection code or procedural infection code), and elective status for the procedure, which is routinely entered into the CART database at the time of the procedure.
Binary logistic regression was used to model the probability of an infection in the training data and chose the best combination of identifiers using the R function bestglm20, based on the Akaike information criterion (AIC), which is an estimate of relative quality of statistical models for a given set of data. We then used the GLM function with the caret package21 (with 3-fold cross-validation with 5 repeats, twoClassSummary and smote sampling in order to account for the rare outcome) in order to apply the model to the training data and validate on the test data. We used Youden’s method to choose the probability threshold to define positive infection status and to estimate the model performance measures22. To ensure model quality and robustness, we assessed calibration of the model using the givitiR package; a model is well calibrated if the predicted probabilities generated by the model accurately match the observed proportions of the response23. The goodness of fit of a logistic regression model can also be expressed by pseudo R-squared statistics, and we calculated Nagelkerke’s pseudo R2 of the logistic regressions of the training and test data using the selected predictors. This statistic is based on the log likelihood for the full model compared to the log likelihood for the baseline intercept-only model and rescaled to cover the full range of possible R2 values from 0 to 124,25. We also explored a second modelling approach using elastic net regression and accounting for hospital-level correlation using the glmnet method in the caret package with a grid search to evaluate the best choice of alpha and lambda. All analyses were completed using SAS software version 9.4 (SAS Institute, Cary, NC) and R v3.5.026.
Results
The VA CART-EP database captured 5,753 CIED procedures from FY 2008–2015, representing 39 VA medical centers. 2,107 procedures among 2,068 CIED patients were manually reviewed (Table 1). The majority of patients were male (N = 2024/2068, 97.9%) and Caucasian (N = 1784/2068, 86.3%). Patients included in the cohort had a high rate of medical comorbidities; among the most prevalent were tobacco use (N = 1041, 50.3%) and diabetes (N = 977, 47.2%). Among manually reviewed cases, 97 CIED infections in 95 patients were identified within the 90-day surveillance window. Full baseline demographic details are shown in Table 1 and additional information is available in Table 1 of a related reference15.
Table 1.
Patient Characteristics for Index Device Procedure by Infection Status.
| Variable | Total (N = 2068) | No CIED infection* (N = 1973) | CIED Infection* (N = 95) | P-value |
|---|---|---|---|---|
| Training Set | 1,362 (65.9%) | 1,299 (65.8%) | 63 (66.3%) | 0.92 |
| Validation Set | 706 (34.1%) | 674 (34.2%) | 32 (33.7%) | |
| Demographics | ||||
| Age (Median (IQR)) | 71.7 (64.4–81.0) | 71.9 (64.5–81.1) | 68.6 (62.2–79.3) | 0.06 |
| Male Sex | 2,024 (97.9%) | 1,931 (97.9%) | 93 (97.9%) | >0.99 |
| Race | ||||
| White | 1,784 (86.3%) | 1,698 (86.1%) | 86 (90.5%) | 0.29 |
| Black | 248 (12.0%) | 239 (12.1%) | 9 (9.5%) | |
| Other | 36 (1.7%) | 36 (1.8%) | 0 (0.0%) | |
| Hispanic | 148 (7.2%) | 140 (7.1%) | 8 (8.4%) | 0.62 |
| Comorbidities | ||||
| Diabetes | 977 (47.2%) | 931 (47.2%) | 46 (48.4%) | 0.81 |
| Tobacco Use | 1,041 (50.3%) | 989 (50.1%) | 52 (54.7%) | 0.38 |
| Chronic Obstructive Pulmonary Disease | 631 (30.5%) | 595 (30.2%) | 36 (37.9%) | 0.11 |
| Cerebrovascular Disease | 482 (23.3%) | 452 (22.9%) | 30 (31.6%) | 0.051 |
| Peripheral Arterial Disease | 473 (22.9%) | 438 (22.2%) | 35 (36.8%) | 0.0009 |
| Chronic Kidney Disease | 675 (32.6%) | 645 (32.7%) | 30 (31.6%) | 0.82 |
| Dialysis | 66 (3.2%) | 62 (3.1%) | 4 (4.2%) | 0.54 |
Abbreviations: CIED = Cardiac Implantable Electronic Device.
*Determined through manual review.
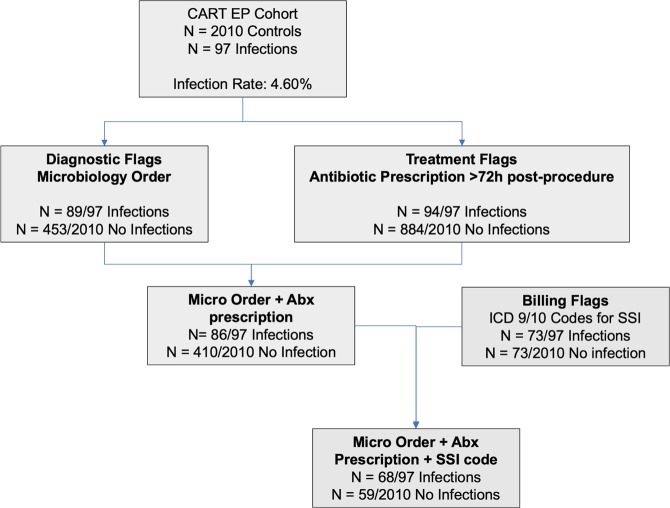
Univariate analysis identified several variables that were significantly associated with true CIED infection (Table 2). Diagnostic variables included presence of a wound or blood culture order (89/97 infections vs 453/2010 uninfected controls, p < 0.0001). Wound culture orders (55/97 vs 144/2010, p < 0.0001) and blood culture orders (78/97 vs 425/2010, p < 0.0001) were also independently associated with true infections. Therapeutic flags associated with true infection included a drug order for the limited list of antibiotics after a 72-hour window post-procedure (94/97 vs 884/2010, p < 0.0001). Billing code identifiers for SSI (73/97 vs 111/2010, p < 0.0001; list of codes in Supplementary Methods S2 as well as general infection codes (34/97 vs 109/2010, p < 0.0001) were significant identifiers; material infection and CPT infections were not. A summary of antibiotic treatment regimens against identified culprit pathogens of CIED infection cases is shown in Supplementary Table S4.
Table 2.
Procedural Characteristics for Device Procedures by Infection Status.
| Variable | Total (N = 2107) | No CIED infection* (N = 2010) | CIED Infection* (N = 97) | P-value |
|---|---|---|---|---|
| Training Set | 1,387 (65.8%) | 1,323 (65.8%) | 64 (66.0%) | 0.97 |
| Validation Set | 720 (34.2%) | 687 (34.2%) | 33 (34.0%) | |
| Type of device | ||||
| Biventricular Pacemaker | 36 (1.7%) | 34 (1.7%) | 2 (2.1%) | 0.68 |
| Biventricular Pacemaker-ICD | 282 (13.4%) | 267 (13.3%) | 15 (15.5%) | 0.54 |
| Permanent Pacemaker | 1,204 (57.1%) | 1,159 (57.7%) | 45 (46.4%) | 0.028 |
| ICD | 600 (28.5%) | 565 (28.1%) | 35 (36.1%) | 0.089 |
| Procedural variables | ||||
| Elective procedure | 1,589 (75.4%) | 1,526 (75.9%) | 63 (64.9%) | 0.014 |
| Index was revision procedure* | 787 (37.4%) | 734 (36.5%) | 53 (54.6%) | 0.0003 |
| Repeat CIED procedure | 91 (4.3%) | 85 (4.2%) | 6 (6.2%) | 0.35 |
| Diagnostic variables | ||||
| Microbiology order | 542 (25.7%) | 453 (22.5%) | 89 (91.8%) | <0.0001 |
| ID consult recorded* | 122 (5.8%) | 78 (3.9%) | 44 (45.4%) | <0.0001 |
| Fever | 23 (1.1%) | 19 (0.9%) | 4 (4.1%) | 0.019 |
| Culture positive* | 99 (4.7%) | 49 (2.4%) | 50 (51.5%) | <0.0001 |
| Wound culture order | 199 (9.4%) | 144 (7.2%) | 55 (56.7%) | <0.0001 |
| Wound culture positive* | 55 (2.6%) | 14 (0.7%) | 41 (42.3%) | <0.0001 |
| Blood culture order | 503 (23.9%) | 425 (21.1%) | 78 (80.4%) | <0.0001 |
| Blood culture positive* | 58 (2.8%) | 40 (2.0%) | 18 (18.6%) | <0.0001 |
| Probable pathogen* | 74 (3.5%) | 24 (1.2%) | 50 (51.5%) | <0.0001 |
| Type of probable pathogen* | ||||
| CONS | 14 (0.7%) | 3 (0.1%) | 11 (11.3%) | <0.0001 |
| Enterococcus | 3 (0.1%) | 2 (0.1%) | 1 (1.0%) | |
| Gram-negative Bacillus | 10 (0.5%) | 5 (0.2%) | 5 (5.2%) | |
| MRSA | 10 (0.5%) | 1 (0.0%) | 9 (9.3%) | |
| MSSA | 12 (0.6%) | 2 (0.1%) | 10 (10.3%) | |
| Pseudomonas aeruginosa | 5 (0.2%) | 1 (0.0%) | 4 (4.1%) | |
| Polymicrobial | 12 (0.6%) | 5 (0.2%) | 7 (7.2%) | |
| Streptococcal sp. | 6 (0.3%) | 4 (0.2%) | 2 (2.1%) | |
| Candida sp. | 2 (0.1%) | 1 (0.0%) | 1 (1.0%) | |
| Therapeutic Variablesa | ||||
| Any antibiotic post-procedure (>72 h)b | 978 (46.4%) | 884 (44.0%) | 94 (96.9%) | <0.0001 |
| Antibiotic from limited list post-procedure (>72 h) | 880 (41.8%) | 786 (39.1%) | 94 (96.9%) | <0.0001 |
| Type of antibiotic post-procedure (>72 h)c | ||||
| Amoxicillin | 209 (9.9%) | 187 (9.3%) | 22 (22.7%) | <0.0001 |
| Atovaquone | 1 (0.0%) | 0 (0.0%) | 1 (1.0%) | 0.046 |
| Cefazolin | 64 (3.0%) | 54 (2.7%) | 10 (10.3%) | <0.0001 |
| Cefepime | 59 (2.8%) | 50 (2.5%) | 9 (9.3%) | <0.0001 |
| Ceftriaxone | 108 (5.1%) | 93 (4.6%) | 15 (15.5%) | <0.0001 |
| Cephalexin | 184 (8.7%) | 148 (7.4%) | 36 (37.1%) | <0.0001 |
| Ciprofloxacin | 220 (10.4%) | 197 (9.8%) | 23 (23.7%) | <0.0001 |
| Clindamycin | 68 (3.2%) | 61 (3.0%) | 7 (7.2%) | 0.023 |
| Doxycycline | 123 (5.8%) | 109 (5.4%) | 14 (14.4%) | 0.0002 |
| Linezolid | 13 (0.6%) | 10 (0.5%) | 3 (3.1%) | 0.019 |
| Minocycline | 19 (0.9%) | 14 (0.7%) | 5 (5.2%) | <0.0001 |
| Nafcillin | 7 (0.3%) | 4 (0.2%) | 3 (3.1%) | 0.0029 |
| Rifampin | 8 (0.4%) | 3 (0.1%) | 5 (5.2%) | <0.0001 |
| Trimethoprim-Sulfamethoxazole | 107 (5.1%) | 85 (4.2%) | 22 (22.7%) | <0.0001 |
| Vancomycin | 274 (13.0%) | 215 (10.7%) | 59 (60.8%) | <0.0001 |
| Billing Code Variables | ||||
| Surgical Site Infection code | 184 (8.7%) | 111 (5.5%) | 73 (75.3%) | <0.0001 |
| General infection code | 143 (6.8%) | 109 (5.4%) | 34 (35.1%) | <0.0001 |
| Material infection code | 2 (0.1%) | 2 (0.1%) | 0 (0.0%) | >0.99 |
| CPT infection code | 1,726 (81.9%) | 1,648 (82.0%) | 78 (80.4%) | 0.69 |
Abbreviations: ICD = Implantable Cardioverter-Defibrillator, ID = Infectious Disease, CPT = current procedural terminology, CONS = Coagulase-negative Staphylococcus, MRSA = Methicillin-resistant Staphylococcus aureus, MSSA = Methicillin susceptible Staphylococcus aureus.
*Identified from manual review.
aAntibiotic use only after a 72-hour window period following the index CIED procedure was applied given the frequent use of prophylactic antibiotics peri-procedure.
bLimited List of antibiotics in Supplementary Note S3.
cThere was no significant difference in frequency of the following antimicrobial prescriptions between the two groups: Ampicillin, Ampicillin/sulbactam, Azithromycin, Cefpodoxime, Cefprozil, Cefuroxime, Clarithromycin, Dapsone, Demeclocycline, Dicloxacillin, Erythromycin, Gentamicin, Levofloxacin, Metronidazole, Moxifloxacin, Penicillin, Rifaximin, Tetracycline.
Sensitivity analysis of unstructured data elements
In the CART-EP manual review data, the presence of any positive microbiologic result (50/97 vs 49/2010, p < 0.0001), as well as growth from a wound culture (41/97 vs 14/2010, p < 0.0001) or growth in a blood culture (18/97 vs 40/2010), p < 0.0001) were also potential identifiers. Identification of a probable infecting organism (e.g. Staphylococcus spp) was also found to be a potential identifier (50/97 vs 24/2010, p < 0.0001). Infectious diseases consultations were also positively associated with true CIED infections (44/97 vs 78/2010, p < 0.0001). However, these unstructured and variable data elements were all excluded from consideration in the final detection algorithm due to their limited potential to be included in an operational tool.
The refined set of surveillance variables derived from the training set included in the surveillance algorithm is presented in Table 3 and Fig. 3. Among these, variables with the highest odds for identifying CIED infections included the presence of antibiotic order from the more limited set placed during the period>72 hours post-procedure (OR 17.76, 95% CI 4.11–76.79, p < 0.001), fever (OR 16.94, 95% CI 1.28–224.41, p = 0.032) billing code for SSI (OR 10.56, 95% CI 4.97–22.43, p < 0.001) and the presence of any microbiology order (OR 8.85, 95% CI 3.93–19.89, p < 0.001).
Table 3.
Regression analysis of identifiers included in detection algorithm.
| Identifiers | OR | 95% CI | P-value |
|---|---|---|---|
| CIED infection antibiotic post-procedure | 17.76 | 4.11–76.79 | <0.0001 |
| Fever | 16.94 | 1.28–224.41 | 0.032 |
| Surgical Site Infection Code | 10.56 | 4.97–22.43 | <0.0001 |
| Microbiology order | 8.85 | 3.93–19.89 | <0.0001 |
| Wound culture order | 2.95 | 1.31–6.64 | 0.0092 |
| Other ICD or CPT infection code | 2.20 | 0.59–8.26 | 0.241721 |
Figure 3.
Process of using electronic identifiers to develop a detection algorithm.
When combined in a surveillance tool, the algorithm demonstrated high sensitivity and specificity in both the training set (84.38% and 93.58%, respectively) and the validation set (87.88% and 90.25%, respectively). The c-statistic for the algorithm in the training sample was 0.95 (95% CI, 0.92–0.98, Table 4). The negative predictive value (NPV) was 99.20% and 99.36% for training and validation sets and the positive predictive value (PPV) was 38.85% (training) and 30.21% (validation, Table 4). Compared to the rates of infection in the CART-EP data, the algorithm overestimated true infections (10.0% estimated and 4.6% observed for the training set; 13.3% estimated and 4.6% observed for the validation set). The pseudo R2 values for the training and validation models were 57.9% and 58.4%, respectively (Supplementary Fig. S5). We investigated using elastic net regression, including the same set of possible identifiers and accounting for correlation of observations within a hospital, but this type of model did not significantly improve either calibration or estimation.
Table 4.
Algorithm Performance Characteristics for Training and Validation Sets.
| Characteristic | Training Set | Validation Set |
|---|---|---|
| AUC (95% CI) | 0.96 (0.94–0.98) | 0.95 (0.92–0.98) |
| Sensitivity | 84.38 | 87.88 |
| Specificity | 93.58 | 90.25 |
| Positive-predictive Value | 38.85 | 30.21 |
| Negative-predictive Value | 99.20 | 99.36 |
Discussion
Electronic, automated and semi-automated surveillance are important emerging tools in the epidemiologist’s arsenal as clinical data warehouses are increasingly available for improving bedside clinical care27. From a large sampling of VA CIED procedures, we found that a combination of clinically-oriented variables from CIED infection diagnostic and treatment pathways and administrative billing codes demonstrated clinically useful sensitivity and specificity for flagging true cases of CIED infection (Fig. 4). Algorithms based on structured data elements to retrospectively identify cases with a true infection can be used to expand infection surveillance to clinical care areas with limited infection prevention and surveillance coverage. Although the PPV was limited (~30%), the NPV was very high at 99%, demonstrating that the tool captured true infections well. This semi-automated flagging system could form the foundation of a surveillance program for measuring post-procedural CIED infections as part of a semi-automated process to expedite a manual review of cases most likely to have an infection; because only structured data elements are used, this tool has the potential to be easily implemented across many EHR systems.
Figure 4.
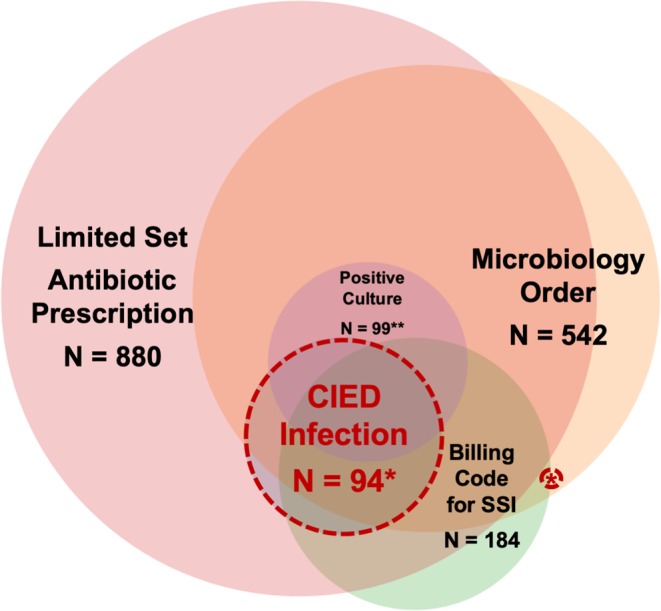
Use of electronic identifier flags in detecting CIED infection cases. Made using https://www.meta-chart.com/venn#. *3 CIED infection cases either did not have an antibiotic prescription or had antibiotics started within 72 hours of index procedure, among these three, 2 had microbiology order and positive culture and 1 had microbiology order, positive culture and billing code for SSI. **Positive growth on blood or wound culture.
Electronically augmented surveillance has great promise for expanding prevention to uncovered clinical areas, including procedural areas, such as the EP laboratory, and outpatient care. However, the best methodology to develop and implement these tools is an active area of research. Here, we used a methodology of first developing a list of potential identifiers based on clinical diagnostic and therapeutic pathways, and then mapped these potential identifiers to structured data elements in the EHR. A similar method of developing detection tools based on standard clinical processes could be used to expand detection to other areas with limited surveillance. In addition, EHR systems and clinical practice patterns may vary by institution, thus, a selection of the various elements identified could be customized to institutional-specific algorithms28. Future iterations could include advanced data extraction strategies, such as natural language processing and machine learning techniques, to further improve operating characteristics of the algorithm.
Prior work on CIED infection detection is limited. One prior single center study evaluated the utility of billing codes to measure rates of CIED infections; our data from a large, multicenter national sample suggests that this strategy may have high specificity but low sensitivity. In our study, 27.6% of true-positive cases in our analysis would not have been flagged using a system reliant on billing codes alone29. Our finding about the limited indicative utility of billing codes alone is similar to work in cardiac surgery, which similarly found that coding data had limited value for measuring post-surgical infections (PPV < 26%)30–32. The challenge of the limited sensitivity of coding data is compounded by recent data which suggests that ICD-10 codes may even lower predictive probabilities than ICD-9 codes33.
We expanded upon prior research into semi-automated electronic surveillance tools by including structured variables available in commonly used EHRs that could be leveraged for CIED identification. In combining these variables, we developed and validated an electronic surveillance tool with excellent specificity and sensitivity, high NPV, and reasonable PPV, increasing the rate of case identification from 4–5% to 30–40%. The high NPV of the tool highlights the importance of the absence of clinical identifiers in ruling out cases that do not have an infection; this NPV can be leveraged to greatly streamline a manual review process.
Prior work demonstrates that a major barrier to implementing surveillance in outpatient and procedural areas is limited time and resources34,35; a semi-automated system that is easy to program and dramatically reduces chart review has the potential to bypass some of these implementation challenges to facilitate expansion of surveillance. The substantial improvement in PPV markedly enhances the yield of a manual review process and reduces the time and resources necessary for infection detection. In addition, although the tool identified flagged some negative cases, it was calibrated to optimize case ascertainment and to triage and minimize the burden of manual review, not to replace it entirely. A combined automated and manual approach to infection detection forms a powerful tool for generating accurate data not only on infection rates but also on quality and process metrics. Identifying patient safety metrics that can be acted upon to promote improvements in infection prevention strategies is an important aspect of the response for averting additional procedure-related infections. Critical changes to patient safety and process could not be identified or implemented without the information generated during a detailed manual review and root cause analysis.
When considering implementation and adaption to other settings of care, it is useful to consider how each type of structured data element impacted the probability an adverse event occurred. For example, use of antimicrobial prescriptions post-procedure was highly sensitive but not specific. This lack of specificity is driven by the breadth of CIED infection pathogens and the diverse set of antimicrobials that can be selected to treat them15. Using a limited set of antibiotics—for example, training the algorithm to detect only antimicrobials directed toward common bacterial pathogens— might improve specificity but at the cost of potentially not detecting CIED infections caused by unusual, and potentially more severe, pathogens. Another approach to enhance the PPV of the tool might be to use combinations of identifiers such as drug-bug matches (i.e. specific antibiotic prescriptions that correspond to particular microbiology results) as a discrete variable. However, this would layer in additional complexity to programming and simultaneously reduce sensitivity. In our sample, approximately half of infections did not have positive blood or wound microbiology, thus, the process of requiring a microbiology result, let alone layering results with interaction terms, would miss more than half of the cases. In addition, our focus was on the use of structured data flags due to excellent NPV of these variables, and to ensure generalizability to a wide variety of EHRs. However, the use of typically unstructured data—such as information contained in clinical notes—may be an approach to improve PPV in future iterations.
The limitations of this study were largely due the dataset used to create and validate the algorithm and difficulty in operationalizing clinical variables into electronic identifiers. We relied on CART-EP data to develop and validate the algorithm, and the selection process for these CIED procedures may limit the generalizability of our findings to other VA CIED procedures and procedures performed outside of the closed VA healthcare system. Our reliance on data available in VA records could mean that CIED infection occurring outside the VA system may not have been captured. However, we manually reviewed all scanned in outside hospital records and these were included as cases in the study; if they did not have clinical variables input into the VA EHR, these would have been included as “false negatives” in the development and assessment of the tool and reflected in the operating characteristics of the model. Further, prior studies demonstrate that the majority of patients return to the closed VA healthcare system for subsequent procedural care.
Regarding operational challenges, we found that wound culture results had excellent promise for infection detection, however, due the nature of the variable as free-text and not standardized, it is not easily measured using an automated system across multiple sites with highly variable documentation practices.
Finally, healthcare systems and practices are not static, but rather constantly evolving. Thus, automated infection surveillance algorithms that leverage clinical pathways, including this CIED infection flagging tool, will require ongoing adaption and updates as novel therapeutics are introduced and diagnostic strategies evolve. Thus, detection algorithms must move toward a learning health system model, with constant modifications and adaptations to maintain their estimative utility36.
Conclusions
Existing surveillance structures for CIED infections are absent in many institutions, partially due to limited resources for detection and monitoring35. This study demonstrates that electronic surveillance tools based on structured data elements can be developed with high sensitivity and specificity and have the potential to expand measurement with audit and feedback to clinical areas that have not been served by traditional surveillance programs. Future improvements could include strategies to enhance detection of variables primarily captured in clinical notes.
Supplementary information
Acknowledgements
This work would not be possible without help and assistance from the VA Clinical Assessment Reporting and Tracking program. The work was supported by American Heart Association Institute for Precision Cardiovascular Medicine Award # 17IG33630052. WBE is supported by NIH NHLBI 1K12HL138049-01.
Author contributions
A.A. and W.B.E. designed the study and wrote the manuscript. M.S. and A.B. performed all statistical analyses. A.A., W.B.E. and P.M. performed manual data collection. H.M., M.S., M.H., K.G. commented on the research design, analysis and manuscript. All authors read and approved the final manuscript.
Competing interests
AA is an investigator for studies funded by Gilead Sciences and Merck since the completion of this study. WBE is a consultant for DLA Piper, LLC. All other authors declare that have no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-62083-y.
References
- 1.Greenspon AJ, et al. 16-year trends in the infection burden for pacemakers and implantable cardioverter-defibrillators in the United States 1993 to 2008. J Am Coll Cardiol. 2011;58:1001–1006. doi: 10.1016/j.jacc.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw PJ, Stobie P, Knuiman MW, Briffa TG, Hobbs MS. Trends in the incidence and prevalence of cardiac pacemaker insertions in an ageing population. Open Heart. 2014;1:e000177. doi: 10.1136/openhrt-2014-000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohail, M. R. et al. Incidence, Treatment Intensity, and Incremental Annual Expenditures for Patients Experiencing a Cardiac Implantable Electronic Device Infection: Evidence From a Large US Payer Database 1-Year Post Implantation. Circ Arrhythm Electrophysiol9, 10.1161/CIRCEP.116.003929 (2016). [DOI] [PubMed]
- 4.Sohail MR, Henrikson CA, Braid-Forbes MJ, Forbes KF, Lerner DJ. Mortality and cost associated with cardiovascular implantable electronic device infections. Arch Intern Med. 2011;171:1821–1828. doi: 10.1001/archinternmed.2011.441. [DOI] [PubMed] [Google Scholar]
- 5.Greenspon AJ, Eby EL, Petrilla AA, Sohail MR. Treatment patterns, costs, and mortality among Medicare beneficiaries with CIED infection. Pacing Clin Electrophysiol. 2018;41:495–503. doi: 10.1111/pace.13300. [DOI] [PubMed] [Google Scholar]
- 6.Greenspon AJ, et al. Trends in permanent pacemaker implantation in the United States from 1993 to 2009: increasing complexity of patients and procedures. J Am Coll Cardiol. 2012;60:1540–1545. doi: 10.1016/j.jacc.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Gastmeier P, Behnke M. Electronic surveillance and using administrative data to identify healthcare associated infections. Curr Opin Infect Dis. 2016;29:394–399. doi: 10.1097/QCO.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 8.Borer A, et al. Prevention of infections associated with permanent cardiac antiarrhythmic devices by implementation of a comprehensive infection control program. Infect Control Hosp Epidemiol. 2004;25:492–497. doi: 10.1086/502428. [DOI] [PubMed] [Google Scholar]
- 9.Powell BJ, et al. A refined compilation of implementation strategies: results from the Expert Recommendations for Implementing Change (ERIC) project. Implement Sci. 2015;10:21. doi: 10.1186/s13012-015-0209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LF, Vander Weg MW, Hofmann DA, Reisinger HS. The Hawthorne Effect in Infection Prevention and Epidemiology. Infect Control Hosp Epidemiol. 2015;36:1444–1450. doi: 10.1017/ice.2015.216. [DOI] [PubMed] [Google Scholar]
- 11.Awad SS. Adherence to surgical care improvement project measures and post-operative surgical site infections. Surg Infect (Larchmt) 2012;13:234–237. doi: 10.1089/sur.2012.131. [DOI] [PubMed] [Google Scholar]
- 12.Branch-Elliman W. A Roadmap for Reducing Cardiac Device Infections: a Review of Epidemiology, Pathogenesis, and Actionable Risk Factors to Guide the Development of an Infection Prevention Program for the Electrophysiology Laboratory. Curr Infect Dis Rep. 2017;19:34. doi: 10.1007/s11908-017-0591-8. [DOI] [PubMed] [Google Scholar]
- 13.Falen T, Noblin AM, Russell OL, Santiago N. Using the Electronic Health Record Data in Real Time and Predictive Analytics to Prevent Hospital-Acquired Postoperative/Surgical Site Infections. The health care manager. 2018;37:58–63. doi: 10.1097/hcm.0000000000000196. [DOI] [PubMed] [Google Scholar]
- 14.Asundi A, et al. Prolonged antimicrobial prophylaxis following cardiac device procedures increases preventable harm: insights from the VA CART program. Infect Control Hosp Epidemiol. 2018;39:1030–1036. doi: 10.1017/ice.2018.170. [DOI] [PubMed] [Google Scholar]
- 15.Asundi, A. et al. Real-world effectiveness of infection prevention interventions for reducing procedure-related cardiac device infections: Insights from the veterans affairs clinical assessment reporting and tracking program. Infect Control Hosp Epidemiol, 1–8, 10.1017/ice.2019.127 (2019). [DOI] [PMC free article] [PubMed]
- 16.Bongiorni MG, et al. How European centres diagnose, treat, and prevent CIED infections: results of an European Heart Rhythm Association survey. Europace. 2012;14:1666–1669. doi: 10.1093/europace/eus350. [DOI] [PubMed] [Google Scholar]
- 17.Baddour LM, et al. Update on cardiovascular implantable electronic device infections and their management: a scientific statement from the American Heart Association. Circulation. 2010;121:458–477. doi: 10.1161/CIRCULATIONAHA.109.192665. [DOI] [PubMed] [Google Scholar]
- 18.CDC. Surgical Site Infection (SSI) Event, http://www.cdc.gov/nhsn/PDFs/pscManual/9pscSSIcurrent.pdf (2015).
- 19.Dobbin KK, Simon RM. Optimally splitting cases for training and testing high dimensional classifiers. BMC Med Genomics. 2011;4:31. doi: 10.1186/1755-8794-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLeod, A. I. & Xu, C. bestglm: Best subset GLM and Regression Utilities. R package version 0.37, https://CRAN.R-project.org/package=bestglm (2018).
- 21.Kuhn, M., Contributions from Jed Wing, S. W., Andre Williams, Chris, Keefer, A. E., Tony Cooper, Zachary Mayer, Brenton Kenkel, the R & Core Team, M. B., Reynald Lescarbeau, Andrew Ziem, Luca Scrucca, Yuan Tang, Can Candan and Tyler Hunt. caret: Classification and Regression Training. R package version 6.0-81. https://CRAN.R-project.org/package=caret (2018).
- 22.Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nattino G, Finazzi S, Bertolini G, Rossi C. & Carrara, G. givitiR: The GiViTI Calibration Test and Belt. R package version. 2017;1:3. [Google Scholar]
- 24.Nagelkerke NJD. A Note on a General Definition of the Coefficient of Determination. Biometrika. 1991;78:691–692. doi: 10.2307/2337038. [DOI] [Google Scholar]
- 25.Signorell et mult. al., A. DescTools: Tools for Descriptive Statistics. R package version 0.99.32. (2020).
- 26.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/ (2018).
- 27.Branch-Elliman W, et al. Cardiac Electrophysiology Laboratories: A Potential Target for Antimicrobial Stewardship and Quality Improvement? Infect Control Hosp Epidemiol. 2016;37:1005–1011. doi: 10.1017/ice.2016.116. [DOI] [PubMed] [Google Scholar]
- 28.de Bruin JS, Seeling W, Schuh C. Data use and effectiveness in electronic surveillance of healthcare associated infections in the 21st century: a systematic review. Journal of the American Medical Informatics Association: JAMIA. 2014;21:942–951. doi: 10.1136/amiajnl-2013-002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boggan JC, et al. An Automated Surveillance Strategy to Identify Infectious Complications After Cardiac Implantable Electronic Device Procedures. Open Forum Infect Dis. 2015;2:ofv128. doi: 10.1093/ofid/ofv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jhung MA, Banerjee SN. Administrative coding data and health care-associated infections. Clin Infect Dis. 2009;49:949–955. doi: 10.1086/605086. [DOI] [PubMed] [Google Scholar]
- 31.Schweizer ML, et al. Validity of ICD-9-CM coding for identifying incident methicillin-resistant Staphylococcus aureus (MRSA) infections: is MRSA infection coded as a chronic disease? Infect Control Hosp Epidemiol. 2011;32:148–154. doi: 10.1086/657936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calderwood MS, Kleinman K, Murphy MV, Platt R, Huang SS. Improving public reporting and data validation for complex surgical site infections after coronary artery bypass graft surgery and hip arthroplasty. Open forum infectious diseases. 2014;1:ofu106. doi: 10.1093/ofid/ofu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mainor AJ, Morden NE, Smith J, Tomlin S, Skinner J. ICD-10 Coding Will Challenge Researchers: Caution and Collaboration may Reduce Measurement Error and Improve Comparability Over Time. Med Care. 2019;57:e42–e46. doi: 10.1097/MLR.0000000000001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cato KD, Liu J, Cohen B, Larson E. Electronic Surveillance of Surgical Site Infections. Surg Infect (Larchmt) 2017;18:498–502. doi: 10.1089/sur.2016.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branch-Elliman, W., Gupta, K. & Elwy, A. in IDWeek 2019.
- 36.Quality, T. A. f. H. R. a. About Learning Health Systems., https://www.ahrq.gov/learning-health-systems/about.html (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.