Abstract
MicroRNAs (miRNAs) are small, endogenous, non-coding, single stranded RNAs which play a role in the regulation of gene expression and function. Therefore, the analysis of differentially expressed miRNAs are of great importance in disease diagnosis. This study is focussed on the differential expression of miRNAs in serum of PCOS subjects compared to control and their correlation with metabolic and endocrine parameters. Anthropometry, hormone concentrations and biochemical characteristics were measured in healthy (n = 20) and PCOS (n = 20) subjects. MiR-24, miR-29a and miR-502-3p were determined in serum by quantitative RT-PCR. The levels of miR-24 was significantly decreased in PCOS subjects (P = 0.00) compared to control. No significant difference was observed in the levels of miR-29a and miR-502-3p in PCOS and control subjects. MiR-24 showed significant inverse correlation with BMI, glucose, insulin, FIRI, HOMA, LH, testosterone, TG, and LH:FSH ratio whereas HDL levels showed significant positive association with miR-24 and miR-29a. LH showed significant negative association with miR-29a. No correlation was observed between the expression of miR-502-3p with any of the studied parameters. The receiver operating characteristic curve for miR-24 alone showed a significant discriminative capacity. The study suggests that serum miR-24 analysis in PCOS patients could be of diagnostic value that can be used as a biomarker for PCOS.
Keywords: Polycystic ovary syndrome, Body mass index, Luteinizing hormone, Follicle stimulating hormone, miRNA-24
Introduction
Polycystic ovary syndrome (PCOS) is a multifactorial, metabolic, endocrine and reproductive health disorder which affects 6–15% of global women population [1]. PCOS patients have characteristic features like acne, hirsutism, alopecia, amenorrhea, chronic oligo-anovulation, and morphologically abnormal ovaries with numerous small follicular cysts [1]. PCOS patients display hyperandrogenism, obesity, hypertension and insulin resistance.
MicroRNAs (miRNAs) are short (20–24 nucleotides long), endogenous, non-coding, single stranded RNAs that are involved in post-transcriptional regulation of gene expression. MiRNAs perform various regulatory functions and modulate biological pathways concerned with cell proliferation, differentiation, survival, apoptosis, stress response, biosynthesis and release of hormones [2]. MiRNAs are stable, with half-life of about 24 h at room temperature and can be detected easily by quantitative reverse transcriptase polymerase chain reaction [3]. Therefore, miRNA profiling serves as a potential non-invasive biomarker for disease diagnosis. Studies show that the levels of miRNAs in serum are varied in PCOS subjects compared to normal subjects [4–8]. The analysis of differentially expressed miRNAs in PCOS subjects is suggested to be useful not only as a biomarker but also in the disease prognosis and therapy [9]. Studies have found that the expression of miR-24-3p, -29a, -151-3p, and -574-3p in follicular fluid are downregulated in women with PCOS [10] while miR-502-3p is increased in type 1 diabetic patients [11], miR-502-3p targets genes which regulate lipid metabolism, adipogenesis and androgen biosynthesis [9, 12]. The present study was therefore undertaken to elucidate the serum levels of miR-24, miRNA-29a and miR-502-3p in PCOS patients and the relationship between miRNAs with hormone levels and some parameters of clinical relevance.
Materials and Methods
Subject Selection
Forty subjects (control = 20 and PCOS = 20) between 18 and 40 years visiting the Rajah Muthiah Medical College and Hospital (RMMC & H), Annamalai University, Annamalainagar, Tamil Nadu, India were recruited for this study.
Inclusion and Exclusion Criteria for Selecting Control Subjects
Twenty age-matched healthy non-PCOS women, with no medications, without hirsutism and who have predictable regular menstrual cycles were included in the study. Subjects under therapy for any other disorders were excluded from the study.
Inclusion and Exclusion Criteria for PCOS
Female subjects diagnosed with hyperandrogenism, oligomenorrhoea, amenorrhoea and ovulatory dysfunction were included in the study. Subjects who were pregnant and have used any hormonal preparations or contraceptives were excluded from the study.
The study was approved by Institutional Human Ethics Committee, Annamalai University. Written informed consents were obtained from the patients prior to the study. The subjects were diagnosed with PCOS according to the revised 2003 Rotterdam criteria, out of the listed three criteria two are required to be fulfilled. They are: (1) oligo and/or anovulation (inter-menstrual interval 35 days and or presence of eight cycles per year), (2) clinical and/or biochemical evidence of hyperandrogenism, hirsutism, presence of multiple cysts (10 small [2–8 mm in diameter each] in one or both ovaries or ovarian volume of 410 ml on USG) and (3) increased luteinizing hormone (LH)-follicle stimulating hormone (FSH) concentration ratio. Control and PCOS patients were recruited from January 25, 2016. None of the women had taken any medications, including hormonal contraceptive, vitamin supplements or any other drug therapy for the previous 6 months. None of them were alcoholics or smokers and did not suffer from any other illnesses.
Sample Collection and Preparation
Fasting blood samples were collected from healthy subjects and PCOS patients by venipuncture of antecubital area during their early follicular phase of menstrual cycle (days 3–5) undergoing normal menses. Blood samples were allowed to clot at room temperature for 1 h at 4 °C. Serum was separated by centrifugation at 3000×g for 15 min and stored at − 20 °C until further analysis.
Serum Analysis and Anthropometric Examinations
Serum FSH, LH, thyroid stimulating hormone (TSH), testosterone and prolactin were estimated by Semi Automated Chemiluminescence Plate Analyzer (Acculite CLIA kit, Lilac Medicare Pvt. Ltd., India). Packed cell volume (PCV) and haemoglobin (Hb) were analyzed using completely automated Sysmex Hematology Analyzer (Japan). Fasting blood glucose and insulin were measured using Erba chem 7 kit (Erba Mannhein, India). Primers for real time PCR were obtained from Sigma-Aldrich Pvt. Ltd. (St Louis, MO, USA).
Anthropometric parameters such as weight (kg) and height (metres) were measured to assess the body mass index (BMI). Insulin resistance was assessed by calculating the homeostasis model assessment insulin resistance (HOMA-IR) [13], quantitative insulin sensitivity check index (QUICKI) [14] and fasting insulin resistance index (FIRI) [15].
Extraction of Circulating miRNAs
Circulating miRNAs were extracted from serum using commercial miRNeasy Serum Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Briefly to 200 µL of serum sample 1 mL of QIAzol reagent was added and vortexed, followed by the addition of equal volume of chloroform. The tubes were mixed vigorously and centrifuged at 12,000×g for 15 min at 40 °C. The upper aqueous phase obtained was transferred to a new collection tube, 1.5 mL of 100% ethanol, buffer was added and centrifuged. The extracted sample was allowed to spin through column and dried. Finally, total miRNA was eluted using 14 µL of RNase-free H2O. The purity and concentration of miRNA was evaluated using nanoPhotometer P 330 (Implen, U.K, Inkarp instrument Pvt. Ltd., Hyderabad).
cDNA Synthesis and Validation of miRNA Expression Using Quantitative Real-Time PCR
One microgram of total miRNA was reverse transcribed to cDNA using the miScript II RT Kit (Qiagen, Hilden, Germany). The reaction mixtures were incubated for 60 min at 37 °C, for 5 min at 95 °C. PCR amplification of target genes was performed using real-time PCR system (Eppendorf, Hamburg, Germany) with 20 μL of total reaction mixture comprised of 2 μL reverse transcription product, 1 μL each of 50 pM forward and reverse primers and 10 μL SYBR green master mix. Nuclease free water was used for adjusting the volume. The optimum conditions for thermocycling were as follows: initial denaturation 95 °C for 3 min, annealing 60 ± 3 °C for 30 s and extension 60 °C for 30 s for 40 cycles. The miRNA primers used for real time PCR are as follows: miR-24 sense (5′ → 3′): CTCCCGTGCCTACTGAGCT, antisense (5′ → 3′): CCCTGTTCCTGCTGAACTGAG [16]. MiR-29a sense (5′ → 3′): TAGCACCATCTGAAATCGGTTA, antisense (5′ → 3′): TAACCGATTTCAGATGGTGCTA [17]. MiR-502-3p sense (5′ → 3′): CAGTGCGTGTCGTGGAGT, antisense (5′ → 3′): GGAATGCACCTGGGCA [9]. RNU6 sense (5′ → 3′): CTCGCTTCGGCAGCACA antisense (5′ → 3′): AACGCTTCACGAATTTGCGT. The relative expression of each miRNA in controls and PCOS group was calculated using ΔCT = CT miRna − CTRnu6 and ΔΔCT = ΔCT case –ΔCTcontrol. Fold change of miRNAs in PCOS with respect to control was calculated using [18].
Statistical Analysis
Statistical analyses were performed using SPSS, software version 20.0 for Windows (SPSS Inc, Chicago, IL, USA). Significant differences in means between the control and PCOS subjects for blood parameters and the levels of miRNAs were analyzed using Student’s unpaired t test. P value < 0.01 was considered to be significant. Correlation between the expression of miRNAs with metabolic parameters was determined using Pearson correlation test (P value < 0.05 was considered to be significant). Receiver operating curve (ROC) analysis was performed by plotting ‘sensitivity’ against ‘1-specificity’ for the delta threshold values (ΔCT) of control and PCOS subjects at 95% confidence interval (CI).
Results
Anthropometric Characteristics and Biochemical Parameters
The anthropometric characteristics, other parameters and biochemical findings of control and PCOS subjects are presented in Table 1. Body weight, glucose, insulin, HOMA-IR, FIRI, LH, LH:FSH ratio, testosterone and TG were found to be significantly higher whereas QUICKI and FSH values were significantly lower in PCOS subjects compared to control. No significant differences were observed in Hb, PCV, prolactin, TSH and low density lipoprotein-cholesterol (LDL-C) levels between the two groups.
Table 1.
Anthropometric characteristics, biochemical parameters and hormone levels of PCOS and control subjects
| Parameters | Control (n = 20) | PCOS (n = 20) | P value |
|---|---|---|---|
| Age (years) | 25.15 ± 4.12 | 28.35 ± 7.45 | 0.10 |
| Weight (kg) | 52.89 ± 5.76 | 74.11 ± 10.55 | 0.00 |
| Height (m) | 1.55 ± 0.06 | 1.53 ± 0.06 | 0.29 |
| BMI (kg/m2) | 22.02 ± 2.64 | 32.16 ± 4.93 | 0.00 |
| Hb (g/dL) | 15.17 ± 2.17 | 14.76 ± 1.93 | 0.53 |
| PCV | 37.46 ± 3.44 | 37.37 ± 2.81 | 0.92 |
| Glucose (mmol/L) | 4.37 ± 0.46 | 6.28 ± 1.06 | 0.00 |
| Insulin (μU/mL) | 10.00 ± 1.38 | 24.66 ± 4.07 | 0.00 |
| HOMA-IR | 1.93 ± 0.28 | 6.87 ± 1.54 | 0.00 |
| QUICKI | 0.34 ± 0.008 | 0.27 ± 0.06 | 0.00 |
| FIRI | 31.40 ± 4.57 | 110.58 ± 31.49 | 0.00 |
| LH (mIU/mL) | 5.54 ± 0.97 | 11.00 ± 2.40 | 0.00 |
| FSH (mIU/mL) | 6.24 ± 1.49 | 4.62 ± 1.99 | 0.006 |
| LH:FSH | 0.90 ± 0.14 | 2.76 ± 1.09 | 0.00 |
| Testosterone (nmol/L) | 1.61 ± 0.45 | 3.52 ± 0.90 | 0.00 |
| Prolactin (ng/mL) | 16.23 ± 2.3 | 16.69 ± 2.62 | 0.55 |
| TSH (nmol/L) | 2.40 ± 0.60 | 2.09 ± 0.99 | 0.23 |
| TG | 54.35 ± 6.95 | 303.20 ± 165.84 | 0.00 |
| LDL-C | 107.18 ± 17.92 | 119.96 ± 30.89 | 0.11 |
| HDL-C | 50.95 ± 5.75 | 33.35 ± 4.38 | 0.00 |
Statistical analysis was performed by Student’s unpaired t test. Values are expressed as mean ± SD. Parameters with significant differences in means (P < 0.01) are marked in bold
Circulating miRNAs
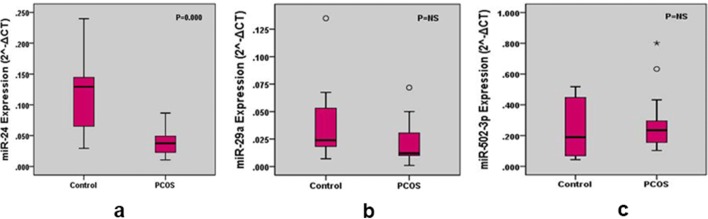
The levels of miR-24, miR-29a, miR-502-3p in PCOS and control groups are presented in Table 2. MiR-24 expression in PCOS group was significantly decreased (P =0.00) compared to the control. No significant difference was observed in the expression of miR-29a and miR-502-3p in PCOS and control subjects. The fold change of miR-24, miR-29a and 502-3p was found to be 0.35, 1.01 and 2.11 in PCOS subjects with respect to control (Table 2). Box and whisker plots represent the expression of miRNAs (Fig. 1). The upper and lower extreme line represents 5th and 95th percentiles. The horizontal line within the box represents the 50 percentile of the expression of miRNAs in control and PCOS subjects. Any data beyond these whiskers are shown as cycles and star.
Table 2.
The expressions of serum miRNAs in control and PCOS subjects and relative expression in PCOS subjects with respect to control. ∆CT and values are represented as mean ± SD
| miRNAs | Control | PCOS | Relative changes in gene expression compared to control () | P value | ||||
|---|---|---|---|---|---|---|---|---|
| ∆CT | Median | ∆CT | Median | |||||
| miR-24 | 3.29 ± 0.95 | 0.12 ± 0.06 | 1.29 × 10−1 | 4.85 ± 0.84 | 0.04 ± 0.02 | 3.74 × 10−2 | 0.53 | 0.00* |
| miR-29a | 5.16 ± 1.21 | 0.038 ± 0.03 | 2.40 × 10−2 | 6.17 ± 1.67 | 0.013 ± 0.31 | 1.19 × 10−2 | 1.01 | 0.15 (NS) |
| miR-502-3p | 2.53 ± 1.39 | 0.24 ± 0.18 | 1.89 × 10−1 | 2.05 ± 0.91 | 0.29 ± 0.21 | 2.34 × 10−1 | 2.11 | 0.56 (NS) |
miR, microRNA; ΔCT = CT miRCase − CT Rnu6; , expression of miRNAs; NS, not significant; Significant at P*< 0.01
Fig. 1.
Box and whisker plots for the expression levels of miR-24, miR-29a and miR-502-3p in control and PCOS group. The upper and lower extreme line represents 5th and 95th percentiles. The horizontal line within the box represents the 50 percentile of the expression of miRNAs in control and PCOS subjects. Any data beyond these whiskers were shown as cycles and star
Validation of miRNAs
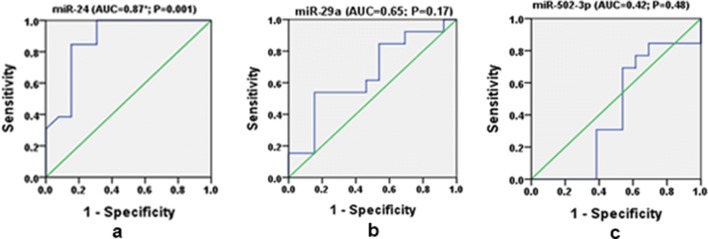
The area under the curve (AUC) was calculated using receiver operating characteristic (ROC) curve to evaluate the diagnostic value of miRNAs. Sensitivity = 1-specificity is represented by the diagonal line from bottom left to the top right. An ROC curve lying on the upper left quadrant above the diagonal line reflects the greater discriminant capacity of the diagnostic tests where as an ROC curve lying at the bottom right quadrant below the diagonal line reflects no discriminant capacity of the diagnostic tests. AUC for miR-24 was found to be 0.87 (95% CI 0.74–1.0, P = 0.001). The ROC curve lies towards the upper left quadrant and thus reflects a good diagnostic value. AUC for miR-29a was 0.65 (95% CI 0.44–0.87, P = 0.17) and the ROC curve lies almost closer to the diagonal and showed no significant difference, thus reflecting no diagnostic value and AUC for miR-502-3p observed was 0.42 (95% CI 0.18–0.65, P = 0.48), the ROC curve along the diagonal line showing no significant difference, thus reflecting no diagnostic value (Fig. 2).
Fig. 2.
Receiver operating curve (ROC) analysis by plotting ‘sensitivity’ against ‘1-specificity’ for the delta threshold values (ΔCT) of miR-24, miR-29a miR-502-3p. The Area under curve (AUC) values gives the information to discriminate PCOS from healthy control. P significant at < 0.05
Correlation Analysis Between the Expression of miRNAs and Metabolic Variables
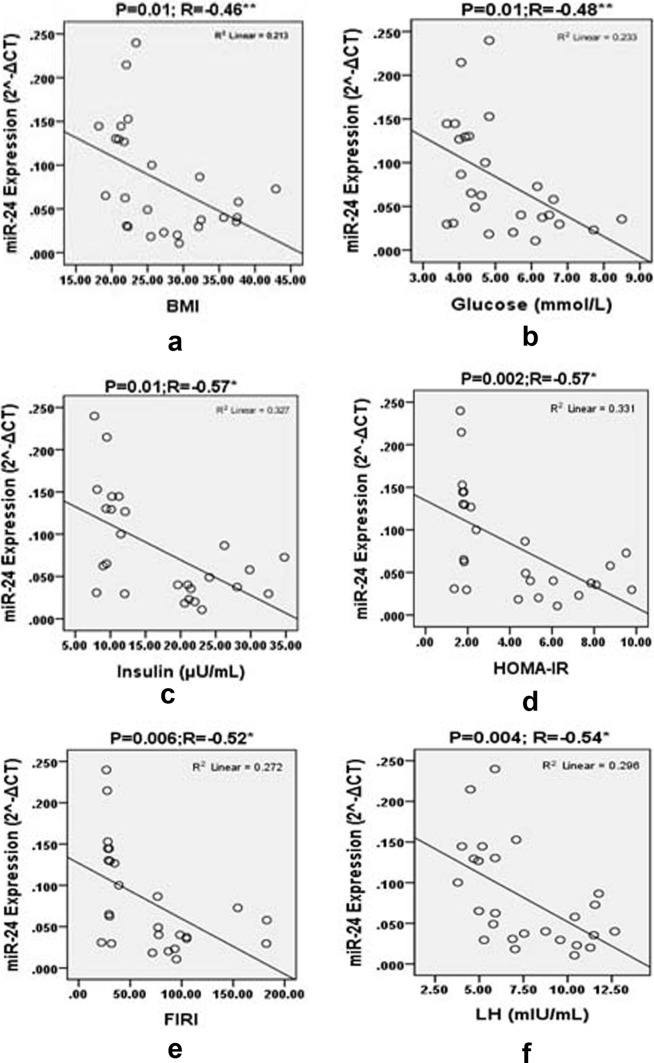
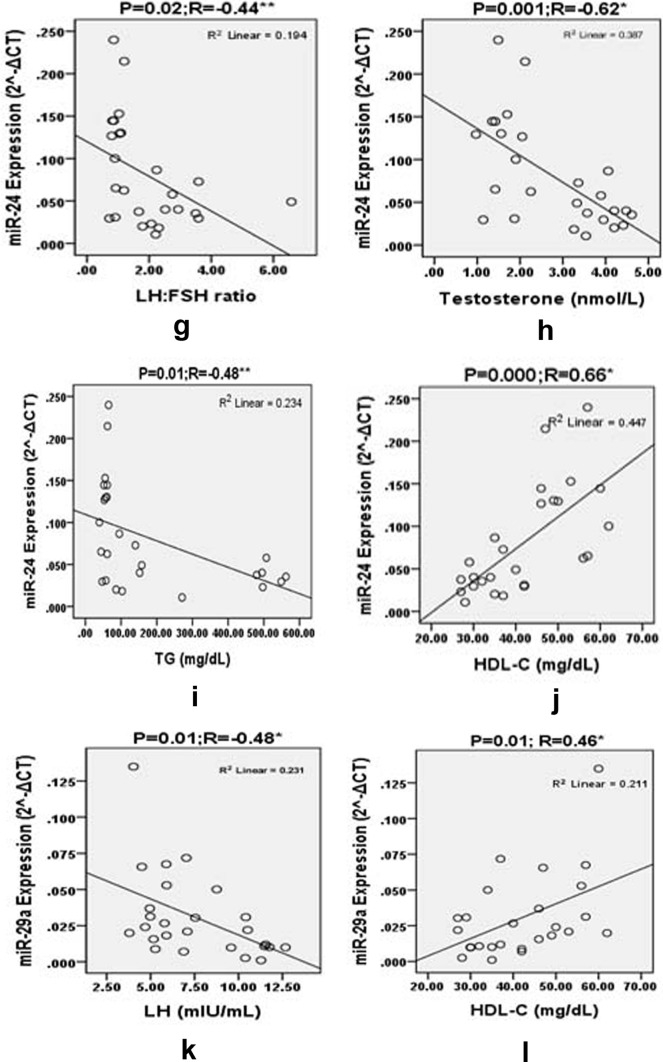
To examine the relationship between the levels of miRNAs and the clinical metabolic features of PCOS patients, correlation analysis was done. MiR-24 showed significant inverse correlation with BMI (R = − 0.46, P < 0.01), glucose (R = − 0.48, P < 0.01), insulin (R = − 0.57, P < 0.002), FIRI (R = − 0.52, P < 0.006), HOMA-IR (R = − 0.57, P < 0.002), LH (R = − 0.54, P < 0.004), testosterone (R = − 0.62, P < 0.001), TG (R = − 0.48, P < 0.012), and LH:FSH ratio (R = − 0.44, P < 0.024) (Table 3 and Fig. 3) and significant positive association with high density lipoprotein-cholesterol (HDL-C) (R = 0.66, P < 0.00). MiR-29a showed significant positive association with HDL-C (R = 0.46, P < 0.01) and significant negative association with LH (R = −0.48, P < 0.01) (Table 3 and Fig. 3). No correlation was observed between LDL-C levels and miR-29a, and miR-24. The levels of miR-502-3p did not show correlation with any of the studied biochemical parameters (Table 3).
Table 3.
Correlation analysis between the expression of miRNAs with the clinical parameters
| Parameters | miR-29a | miR-24 | miR-502-3p | |||
|---|---|---|---|---|---|---|
| r | P value | r | P value | r | P value | |
| BMI | − 0.35 | 0.07 | − 0.46 | 0.01** | 0.20 | 0.32 |
| Glucose | − 0.27 | 0.16 | − 0.48 | 0.01** | 0.17 | 0.39 |
| Insulin | − 0.29 | 0.13 | − 0.57 | 0.002* | 0.12 | 0.55 |
| HOMA-IR | − 0.31 | 0.12 | − 0.57 | 0.002* | 0.16 | 0.43 |
| QUICKI | 0.16 | 0.42 | 0.37 | 0.06 | − 0.004 | 0.98 |
| FIRI | − 0.28 | 0.16 | − 0.52 | 0.006* | 0.14 | 0.48 |
| LH | − 0.48 | 0.01* | − 0.54 | 0.004* | − 0.09 | 0.64 |
| FSH | − 0.06 | 0.76 | 0.36 | 0.07 | − 0.32 | 0.10 |
| LH:FSH ratio | − 0.20 | 0.30 | − 0.44 | 0.02** | 0.03 | 0.85 |
| Testosterone | − 0.29 | 0.14 | − 0.62 | 0.001* | 0.07 | 0.70 |
| TG | − 0.16 | 0.42 | − 0.48 | 0.01** | 0.36 | 0.06 |
| LDL-C | − 0.02 | 0.89 | − 0.002 | 0.99 | 0.24 | 0.22 |
| HDL-C | 0.46 | 0.01* | 0.66 | 0.00* | − 0.07 | 0.71 |
Pearson correlation test between the expression of selected miRNAs with metabolic parameters. P*< 0.01 and **< 0.05 were considered to be significant
Fig. 3.
Pearson correlation plots for the expression levels () of miRNA-24 with BMI, glucose, insulin, HOMA, FIRI, LH, LH:FSH ratio, testosterone, TG, HDL-C and miRNA-29a with LH and HDL-C in the groups studied. “r” values represents the correlation coefficients in the scatter plots diagram. P significant *< 0.01 and **< 0.05
Discussion
The findings of the present study reveal that the serum levels of miR-24 is lowered in PCOS subjects and could be of diagnostic value. This miRNA showed negative association with BMI, glucose, insulin, HOMA-IR, FIRI, LH, testosterone, TG and LH:FSH ratio and positive association with HDL-C levels. However, there were no significant changes in the levels of miR-29a and miR-502-3p between PCOS patients and control subjects and no significant association was found between the studied parameters with the miR-502-3p and miR-29 except for the finding that miR-29a showed negative association with LH and positive association with HDL-C.
MicroRNAs are known to regulate the target gene expression through post-transcriptional mechanisms. MiRNAs bind within the 3′-untranslated region (UTR) of the target mRNAs and either repress the translation of the target mRNA or degrade the mRNA by cleaving it. A single miRNA may have several target mRNAs and thus can regulate various cellular processes [3].
In this study, PCOS subjects had decreased levels of miR-24. MiR-24 is shown to regulate several processes like steroid hormone biosynthesis [10] and estradiol homeostasis [5]. Cytochrome P450 family 11A1 (CYP11A1) is a known target for miR-24, a key enzyme involved in biosynthesis of steroid hormones [10]. Decrease in the levels of miR-24 in PCOS patients may lead to increased expression of CYP11A1 and thereby the synthesis of androgen could be enhanced. Increased miR-24 levels has been found to be involved in regulation of estradiol homeostasis by decreasing TGF-β signaling by supressing the expression of smad proteins [19], also the ovarian folliculogenesis is regulated by TGF-β signaling [20]. The estradiol homeostasis might be dysregulated due to the decreased expression of miR-24 and thereby ovarian folliculogenesis is disrupted in PCOS.
All PCOS women in present study were found to be hyperglycemic and insulin resistant. Elevated levels of both insulin and glucose are considered a hallmark in the development of type 2 diabetes. An inverse association between the expression of miR-24 with glucose, insulin, HOMA-IR and FIRI is suggestive of the role of miRNAs in the maintenance of glucose homeostasis. The qRT-PCR validation report showed the decreased levels of miR-24 in PCOS subjects compared to control. AUC for miR-24 was found to be 0.87 (95% CI 0.74–1.0, P = 0.001) which is significant showing its potential diagnostic value. Melkman et al. [21] found that miR-24 level in plasma of type 2 diabetic patients was significantly lower than that in healthy controls and the insulin promoter activity was found to be down-regulated by miR-24 thereby resulting in the reduction in insulin mRNA levels. Taken together, it can be concluded that the reduction in miR-24 in PCOS could be responsible for the combined ovarian function and insulin resistance in them.
Members of miR-29 family are predicted to function as inhibitors of numerous mRNAs that are involved in extracellular matrix (ECM) production, fibrosis [22, 23] and cell differentiation [24]. Specifically for miR-29a, androgen receptor (AR), StAR related lipid transfer protein 3 (STARD3) and insulin receptor substrate-1 (IRS-1) are reported as targets. Among these, AR and STARD3 are involved in ovarian steroidogenesis while IRS-1 is involved in insulin signaling. In insulin resistant L6 myocytes increased miR-29a has been identified by targeting 30-UTR of IRS-1 which represses IRS-1 expression [25]. A study by Sorensen et al. [10] found that down-regulation of the miR-29a family in PCOS patients and its specific target genes are associated with folliculogenesis in PCOS patients leading to ovarian malfunction.
The levels of miR-29a in our study was found to be decreased. However significant difference between the control and PCOS group was not observed. Further no correlation was observed between the expression of miR-29a and studied glycaemic indices except that a negative association with levels of LH was found. MiR-29a may have regulatory role in the synthesis of LH which needs to be studied further.
MiR-502-3p has several target genes such as cytochrome P450 17α-hydroxylase/17, 720-lyase (CYP17) [9], cAMP response element binding protein regulated transcription coactivator 1 (CREB-CRTC1), rho/rac guanine nucleotide exchange factor (FGD1), chemokine C–C motif ligand 8 (CCL8) and StAR-related lipid transfer domain protein 8 (STARD8) which are involved in fat metabolism and/or adipogenesis [12]. Up-regulation of miR-502-3p has been reported in sera of type 1 diabetic patients [11]. In our study the levels of miR-502-3p are increased but the increase was not significant between the control and PCOS group. Also, no correlation in our study was observed between the expression of miR-502-3p with any of the studied parameters.
All the PCOS women were insulin resistant and hyperandrogenic and they were newly diagnosed (less than a year) with the disease and are in the early stage of the disease. It is possible that in the course of the progression of the disease miR-29a and miR-502-3p could be altered. Thus, the role of miR-29a and miR-502-3p in the pathophysiology and progression of the disease needs to be further investigated.
From these findings, it can be speculated that miR-24 plays a key role in the pathophysiology of PCOS and metabolic risks such as insulin resistance, type 2 diabetes and hormonal changes. The expression of genes involved in steroidogenesis and glucose homeostasis could be perturbed since the correlation analysis demonstrated the influence of LH, testosterone, LH:FSH ratio and TG on the expression of miR-24 and LH alone on miR-29a. Although hyperglycaemia is a contributor of the deranged miR-24 levels [26], miR-24 can be used as a biomarker for PCOS when correlated with other characteristic clinical parameters and phenotypic changes in PCOS women. The limitations of this study are small sample size and selection of three specific miRNAs since several miRNAs are differentially expressed in PCOS. Nevertheless, the results of this preliminary study are important which pave way for future studies with larger sample size.
Conclusion
Among the three miRNAs examined, the expression of miR-24 is decreased in PCOS subjects compared to control. The AUC for miR-24 showed a significant difference and displayed a potent diagnostic value. The association of metabolic variables and hormone levels with miR-24 suggests that this miRNA may be involved in pathophysisological changes displayed in PCOS patients.
Acknowledgements
The authors thank Dr. K. Lavanya kumari, M.B.B.S., M.D, (RMMC & H) and Dr. A. Brinda, M.D., D.G.O, Gynaecologist for their help in providing the blood samples of control and PCOS subjects and also all the participants in the study. We acknowledge DST-FIST and UGC-SAP support for the facilities developed in the Department of Biochemistry and Biotechnology, Annamalai University for executing the present study.
Funding
This study was supported by University Grants Commission (UGC) in form of Research Fellowship in Sciences for Meritorious Students (RFSMS) to Ms. Dipti Nanda.
Compliance with Ethical Standards
Conflict of interest
The authors disclose that they do not have any conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Banaszewska B, Antoni J, Pawelczyk L, Robert Z, Spaczynski Z. Lipids in polycystic ovary syndrome role of hyperinsulinemia and effects of metformin. J Obstet Gynaecol. 2006;194:1266–1272. doi: 10.1016/j.ajog.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, Du T, Huang J, Huang LL, Yang DZ. Identification of differentially expressed microRNAs in the ovary of polycystic ovary syndrome with hyperandrogenism and insulin resistance. Chin Med J. 2015;128:2. doi: 10.4103/0366-6999.149189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sang Q, Yao Z, Wang H, Feng R, Wang H, Zhao X, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab. 2013;98(7):3068–3079. doi: 10.1210/jc.2013-1715. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Fang Y, Liu Y, Yang X. MicroRNAs in ovarian function and disorders. J Ovarian Res. 2015;8:51. doi: 10.1186/s13048-015-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scalici E, Traver S, Mullet T, Molinari N, Ferrières A, Brunet C, et al. Circulating microRNAs in follicular fluid, powerful tools to explore in vitro fertilization process. Sci Rep. 2016;6:24976. doi: 10.1038/srep24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song DK, Sung YA, Lee H. The role of serum microRNA-6767-5p as a biomarker for the diagnosis of polycystic ovary syndrome. PLoS ONE. 2016;11(9):e0163756. doi: 10.1371/journal.pone.0163756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin L, Du T, Huang J, Huang LL, Yang DZ. Identification of differentially expressed microRNAs in the ovary of polycystic ovary syndrome with hyperandrogenism and insulin resistance. Chin Med J. 2015;128:169–174. doi: 10.4103/0366-6999.149189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorensen AE, Wissing ML, Englund ALM, Dalgaard LT. MicroRNA species in follicular fluid associating with polycystic ovary syndrome and related intermediary phenotypes. J Clin Endocrinol Metab. 2016;101(4):1579–1589. doi: 10.1210/jc.2015-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erener S, Marwaha A, Tan R, Panagiotopoulos C, Kieffer TJ. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight. 2017 doi: 10.1172/89656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou G, Wang X, Yuan C, Kang D, Xu X, Zhou J, et al. Integrating miRNA and mRNA expression profiling uncovers miRNAs underlying fat deposition in sheep. BioMed Res Int. 2017 doi: 10.1155/1857580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutt M, Davis CL, Spitzer SB, Llabre MM, Kumar M, Czarnecki EM, et al. Validation of the insulin sensitivity index (ISI0, 120): comparison with other measures. Diabetes Res Clin Pract. 2000;47(3):177–184. doi: 10.1016/S0168-8227(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 14.Katz A, Nambi SS, Mather K, Baron AD, Follmann DA, Sullivan G, et al. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metab. 2000;85:2402–2410. doi: 10.1210/jcem.85.7.6661. [DOI] [PubMed] [Google Scholar]
- 15.Shalam Md, Harish MS, Farhana SA. Prevention of dexamethasone- and fructose-induced insulin resistance in rats by SH-01D, a herbal preparation. Indian J Pharmacol. 2006;38:419–422. doi: 10.4103/0253-7613.28209. [DOI] [Google Scholar]
- 16.Long W, Zhao C, Ji C, Ding H, Cui Y, Guo X, et al. Characterization of serum microRNAs profile of PCOS and identifcation of novel non-invasive biomarkers. Cell Physiol Biochem. 2014;33:1304–1315. doi: 10.1159/000358698. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer SR, Grossmann KF, Cassidy PB, Yang CH, Fan M, Kopelovich L, et al. Detection of exosomal miRNAs in the plasma of melanoma patients. J Clin Med. 2015;4:2012–2027. doi: 10.3390/jcm4121957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Chan MC, Hilyard AC, Wu C, Davis BN, Hill NS, Lal A, et al. Molecular basis for antagonism between PDGF and the TGFβ family of signaling pathways by control of miR-24 expression. EMBO J. 2010;29:559–573. doi: 10.1038/emboj.2009.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trombly DJ, Woodruff TK, Mayo KE. Roles for transforming growth factor beta superfamily proteins in early folliculogenesis. Semin Reprod Med. 2009;27:14–23. doi: 10.1055/s-0028-1108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N, et al. miRNAs control insulin content in pancreatic of transcriptional repressors. EMBO J. 2011;30:835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maurer B, Stanczy J, Junge A, et al. A key regulator of collagen expression. Scler Arthritis Rheum. 2010;62(6):1733–1743. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 23.Harmanci D, Erkan EP, Kocak A, Akdogan GG. Role of the microRNA-29 family in fibrotic skin diseases. Biomed Rep. 2017;6:599–604. doi: 10.3892/br.2017.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JY, Zhang Q, Wang DD, Yan W, Sha HH, Zhao JH, et al. MiR-29a: a potential therapeutic target and promising biomarker in tumors. Biosci Rep. 2018;38:BSR20171265. doi: 10.1042/BSR20171265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol. 2007;21:2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 26.Xiang Y, et al. Hyperglycemia repression of miR-24 coordinately upregulates endothelial cell expression and secretion of von Willebrand factor. Blood. 2015;125:3377–3387. doi: 10.1182/blood-2015-01-620278. [DOI] [PMC free article] [PubMed] [Google Scholar]