Abstract
Preanalytical errors constitute about 40–65% of laboratory errors, of which 60% are due to hemolysis. This leads to imprecise reporting and misinterpretation of the actual concentration of analytes. Hence the aim of this study was to estimate the extent of different degrees of interference by visible hemolysis. 25 hemolysed samples along with their fresh unhemolysed sample were studied. Hemolyzed serum was mixed with unhemolyzed serum in predefined serial ratios from 100%, 70%, 50%, 30% and 10% to achieve different grades of hemolysis. Each dilution was analysed for BUN, creatinine, uric acid, phosphorus, Na, K, total protein, amylase, lipase, LDH, tacrolimus and methotrexate. Percentage difference of each dilution of the hemolyzed sample as compared to the unhemolyzed sample was calculated and considered acceptable only if less than TEa. It was observed that Percentage difference of BUN, creatinine, amylase and lipase in all dilutions of hemolyzed samples were within TEa while phosphorus, Na, K, total protein and LDH were beyond the acceptance criteria. Hence It was concluded that it may be safe to analyse a hemolysed sample for BUN, creatinine, amylase, lipase, tacrolimus and methotrexate while uric acid may be estimated in a moderately hemolysed sample. Phosphorus, sodium, potassium, total protein and LDH must never be analyzed in any hemolysed sample.
Keywords: Hemolysis, Preanalytical error, Tacrolimus, Methotrexate, Total allowable error TEa
Introduction
Errors in the preanalytical phase of laboratory functioning are known to constitute about 40–65% of laboratory errors. Amongst these, about 60% are due to hemolysis of samples [1]. Hemolysis is the breakdown of red blood cells with spilling of their contents (including haemoglobin) into the plasma/serum. It is evident, therefore, that this would lead to imprecise reporting of the serum levels of various constituents whilst also increasing the haemoglobin content of the serum. Hence, it would lead to misinterpretation of the actual concentration of several constituents in circulation and also interfere with the estimation of analytes especially where the methodology involves estimation of a coloured end-product [2].
Validated reagents for chemical analysis of blood for diagnostic purposes are accompanied by a description of the performance characteristics of this reagent including the level of haemoglobin in a sample which alters the results obtained. However, when a serum sample is received for any biochemical estimation, it cannot be routinely analysed for haemoglobin content—it would require a different sample container, a separate reagent and separate process after which one would be able to ascertain whether or not it would interfere with the analysis of the required biochemical parameter. Another method of ascertaining the extent of hemolysis (or haemolytic index, as it is termed) is by visual observation of the colour and comparing it to available charts [3]. This hemolytic index is inherently subjective, and is made more so by the fact that the colour of the unhemolysed serum also varies significantly owing to protein content, intake of some drugs/antibiotics, and intake of vitamin supplements [4].
This study was designed to ascertain how much different degrees of visible hemolysis interfere with certain biochemical analytes that require urgent reporting.
Materials and Methods
Samples
The study has been conducted in a fully-automated biochemistry laboratory in a tertiary care hospital. 25 samples received in yellow-capped evacuated gel tubes and found hemolyzed after centrifugation were used for the study. As per the departmental quality policy, the fresh samples of the same patients were requested, received and processed. These, too, were used for the study. Apart from these 25 samples, also included were 5 samples received for the analysis of tacrolimus and 5 received for estimation of methotrexate, which were hemolysed and for which fresh samples were received within an hour of first sampling and before the patient had his next dose of medication.
Procedure and Data Analysis
Serum of the hemolysed samples were diluted with their unhemolysed serum of the same patient received subsequently. The dilutions were made in pre-defined ratios to achieve different grades of hemolysis. The hemolysed serum:nonhemolysed serum ratios defined were 100:0, 70:30, 50:50, 30:70, 10:90 and 0:100; these were labelled as A, B, C, D, E and F, respectively. Optical density for each dilution was measured at 440 nm on Shimadzu UV-1700 spectrophotometer which has a wavelength range of 190–1100 nm, a spectral bandwidth better than 1 nm, and a wavelength accuracy of ± 0.3 nm.
Each dilution was analysed for concentration of blood urea nitrogen (BUN), creatinine (Crt), uric acid (UA), phosphorus (Phos), sodium (Na), potassium (K), total protein (TP), amylase (Amy), lipase (Lip) and lactate dehydrogenase (LDH). All these analytes were measured on a fully automated random access chemistry analyser (Unicell DXC 800 from Beckman Coulter Inc., USA) by the methods as given in Table 1. These analytes were chosen on basis of their importance in management of patients, especially in emergency situations, in our hospital setting. In addition, tacrolimus (immunosuppressant) and methotrexate (antifolate) were also analysed in 5 hemolysed and unhemolysed samples of the same patient taken within an hour. For these two analytes dilutions of the hemolysed sample were not prepared and the analyte was measured in the highly hemolysed as well as the unhemolysed sample.
Table 1.
| Analyte | N | Method |
|---|---|---|
| Bun | 25 | Urease kinetic assay [5] |
| Creatinine | 25 | Jaffe’s kinetic assay [6] |
| Uric acid | 25 | Timed end-point uricase method [7] |
| Phosphorous | 25 | Phosphomolybdate timed rate method [8] |
| Sodium | 25 | Indirect ion-selective electrodes [9] |
| Potassium | 25 | Indirect ion-selective electrodes [9] |
| Total protein | 25 | Biuret timed rate method [10] |
| Amylase | 25 | Enzymatic-kinetic method [11] |
| Lipase | 25 | Enzymatic-kinetic method [11] |
| Lactate dehydrogenase | 25 | Enzymatic-kinetic method [11] |
| Tacrolimus | 5 | Chemiluminescent microparticle immunoassay [12] |
| Methotrexate | 5 | Chemiluminescent microparticle immunoassay [12] |
The estimated value of each analyte in each dilution of the hemolysed samples was compared to the same in the unhemolysed sample of the same patient, and the percentage difference calculated as below:
CH = Concentration of analyte in hemolysed sample
CNH = Concentration of analyte in nonhemolysed sample
The percentage difference achieved was matched to the total allowable error (TEa) as per the recommendations of Ricos and Carmen 2016 [13].
Statistical Analysis
Analysis of results were done by calculating percentage difference against non hemolysed samples and then mean percentage was matched against TEa. 95% confidence interval was also calculated for the optical density of the sample measured on the spectrophotometer.
Results
The percentage mean difference observed between the result of the unhemolysed sample and the various dilutions of the hemolysed sample are given in Table 2.
Table 2.
Percentage mean difference between results of the analytes in serial dilutions of the hemolysed sample as compared to the non-hemolysed sample
| Analyte | % Mean difference in serial dilutions | TEa | ||||
|---|---|---|---|---|---|---|
| 100:0 (H:NH) [n = 25] |
70:30 (H:NH) [n = 25] |
50:50 (H:NH) [n = 25] |
30:70 (H:NH) [n = 25] |
10:90 (H:NH) [n = 25] |
||
| Bun | 3.65 | 3.16 | 1.73 | 3.23 | 2.83 | 15.55 |
| Creatinine | 3.77 | 3.06 | 3.50 | 3.53 | 2.90 | 8.87 |
| Uric acid | 16.31* | 9.78 | 7.32 | 5.26 | 2.44 | 11.97 |
| Phos | 13.46* | 9.50 | 7.89 | 6.41 | 3.16 | 10.11 |
| Na | 0.62 | 0.81* | 0.57 | 0.35 | 0.20 | 0.73 |
| K | 30.48* | 18.34* | 13.92* | 11.22* | 5.47 | 5.61 |
| TP | 7.52* | 4.49* | 2.49 | 0.02 | 1.28 | 3.63 |
| Amylase | 0.21 | 0.05 | 0.37 | 0.69 | 2.32 | 14.6 |
| Lipase | 1.13 | 0.99 | 1.83 | 0.78 | 0.57 | 37.88 |
| LDH | 131.56* | 49.18* | 34.59* | 39.46* | 14.25* | 11.4 |
*Mean difference between hemolysed and non-hemolysed sample result > TEa. H, hemolysed sample; NH, non-hemolysed; TEa, total allowable error as per Ricos and Carmen 2016
The mean difference in results of BUN, creatinine, sodium, amylase and lipase in all dilutions of the hemolysed sample fall within the total allowable error. Uric acid and phosphorus results are unacceptable only in the 100% hemolysed sample, but are within the TEa in all other dilutions. Total protein yields an acceptable result in moderately hemolysed sample. On the other hand, difference in results of potassium and LDH are more than the TEa in all dilutions (Table 2).
To further qualify the mean difference charted in Tables 2 and 3 gives the data ascertaining the number of samples in each dilution of each hemolysed sample that yielded a result that was different from the result of the unhemolysed sample by more than the TEa. Table 3 shows that for phosphorus, sodium, potassium, total protein and LDH, more than 10% of the samples (in serial dilutions of the hemolysed sample) yield results beyond the TEa.
Table 3.
Number of samples in various dilutions of H:NH whose percentage difference of analyte concentration (as compared to the non-hemolysed sample) exceeded the TEa
| Analyte | No of samples’ results (of analytes) in various dilutions exceeding TEa | ||||
|---|---|---|---|---|---|
| 100:0 (H:NH) [n = 25] |
70:30 (H:NH) [n = 25] |
50:50 (H:NH) [n = 25] |
30:70 (H:NH) [n = 25] |
10:90 (H:NH) [n = 25] |
|
| Bun | 0 | 0 | 1 | 0 | 0 |
| Creatinine | 2 | 1 | 2 | 2 | 1 |
| Uric acid | 10 | 3 | 2 | 1 | 0 |
| Phos | 13 | 10 | 5 | 3 | 1 |
| Na | 12 | 11 | 9 | 7 | 12 |
| K | 20 | 20 | 20 | 20 | 6 |
| TP | 15 | 10 | 10 | 9 | 6 |
| Amylase | 1 | 2 | 0 | 1 | 0 |
| Lipase | 0 | 0 | 0 | 0 | 0 |
| LDH | 20 | 20 | 20 | 17 | 10 |
For tacrolimus and methotrexate, five hemolysed and unhemolysed samples were analysed and the results obtained are given in Table 4. The samples were chosen on basis of their tacrolimus/methotrexate concentration so as to cover a wide range of clinically relevant levels. It was revealed that there is no significant difference in the results of the highly hemolysed and the unhemolysed samples. Hence, further dilutions of the hemolysed samples were not performed. It also emphasized that for none of the samples did the difference in results between the hemolysed and non-hemolysed samples exceed the TEa.
Table 4.
Results of tacrolimus and methotrexate in hemolysed and non-hemolysed sera along with the difference and the percentage difference in the values
| Analyte | Sample number | Result in non hemolysed | Result in hemolysed | Difference between H and NH | % difference between H and NH | TEa | No. of results exceeding TEa |
|---|---|---|---|---|---|---|---|
| Tacrolimus (ng/mL) | 1 | 2.5 | 2.5 | 0 | 0 | 24.1 µg/L | 0 |
| 2 | 6.7 | 6.6 | 0.1 | 1.5 | |||
| 3 | 10.6 | 10.5 | 0.1 | 0.9 | |||
| 4 | 20.1 | 18.2 | 1.9 | 9.5 | |||
| 5 | 30 | 30 | 0 | 0 | |||
| Mean difference | 0.43 | 2.38 | |||||
| Methotrexate (µmol/L) | 1 | 0.619 | 0.617 | 0.002 | 0.3 | 10 µmol/L | 0 |
| 2 | 34 | 33.2 | 0.8 | 2.4 | |||
| 3 | 33.2 | 36.8 | 3.6 | 10.8 | |||
| 4 | 57.2 | 57.6 | 0.4 | 0.7 | |||
| 5 | 218.4 | 220.4 | 2.0 | 0.9 | |||
| Mean difference | 0.020 | 3.02 |
TEa is also given
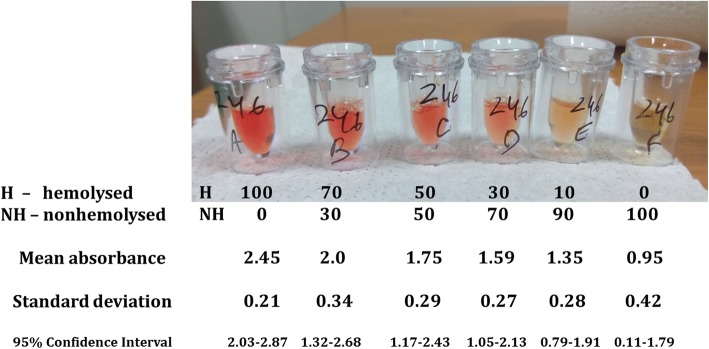
In addition, we measured the optical density of the various dilutions of all the samples at 440 nm which is used for measuring Hb. Figure 1 gives the mean ODs which we have elucidated in our laboratory against a depictive photograph of the same.
Fig. 1.
Photograph of serial dilutions of a representative hemolysed sample received in our laboratory along with their respective mean optical densities
Discussion
Clinical Situations That Pose a Dilemma and Interference Due to Hemolysis
In routine laboratory practice, we are often faced with the dilemma of whether or not to process a hemolyzed sample. Causes of hemolysis can include improper phlebotomy, improper handling of sample, faulty centrifugation, and some disease conditions (e.g. intravascular hemolysis) [14]. Theoretically, the golden rule is that no hemolyzed sample should be processed so as to prevent erroneous reporting. But, practically, there are many situations where this ‘golden rule’ is difficult to implement. For e.g.
an elderly patient with pneumonitis and electrolyte imbalance is admitted in the intensive care and three repeated attempts at phlebotomy have yielded a hemolyzed sample;
a patient with severe sepsis and shock cannot undergo repeated phlebotomy due to thin thrombosed veins;
a patient with urinary tract infection has repeatedly undergone phlebotomy for renal function tests and all the samples are hemolyzed;
a transplant recipient’s timed blood sample for estimation of immunosuppressant is hemolyzed.
A patient is on antifolates and his sample (hemolysed) has been sent for estimation of methotrexate after which he has been administered folinic acid for recovery, the next dose and time of folinic acid to be determined on basis of the methotrexate result.
In all these situations, the decision to accept and process a hemolyzed sample might be the only practical solution.
For the patient with pneumonitis and electrolyte imbalance, knowing the status of the circulating electrolytes is of paramount importance for further management.
For the patient with thin thrombosed veins, repeated sampling is not an option.
For the patient with urinary tract infection, the renal status (as assessed by renal function tests) modulates the management. For management of a patient who has undergone organ transplantation, fine-tuning of the immunosuppressant levels is the key to successful management, and as the sample has to be drawn just before the patient has his daily dose of immunosuppressant, repeat sampling is not an option (as by the time it is ascertained that the sample is hemolyzed, the patient has already had his dose of immunosuppressant).
For the patient on antifolates, the recommended regimen is to analyse the serum level of methotrexate and decide the dose of the folinic acid on basis of this result.
In all these situations, as per the manufacturer’s instructions, a hemolysed sample should not be processed. At the same time, rejecting these samples would lead to inefficient management of these patients. Hence, there is a dilemma—to receive and process the sample, or to reject it.
First let us assess why we cannot process a hemolysed sample. Theoretically, this can be answered in two steps:
Firstly, the intracellular contents of the RBCs are lost into the serum/plasma, for e.g. intracellular potassium, phosphorus and LDH (which are in much higher concentration than the extracellular contents) will spike the reported concentration of these analytes in a hemolysed sample [15].
Secondly, immunoturbidimetric or end-point colorimetric methods use the colour of the final product to calculate the result and the colour of a hemolysed sample would interfere. Where enzymatic rate reactions are used and NADP or NADPH are measured as the end-product, the colour of the hemolysed sample does not interfere with the analysis. Similarly, where immunoassays are the method of estimation, the result rarely varies between the hemolysed and unhemolysed sample [16].
Also, the manufacturer’s instructions usually mention the level of haemoglobin in the serum/plasma which would interfere with the estimation of the analyte in question. As the CBC analysers cannot analyse a serum sample for haemoglobin, it becomes difficult to assess which hemolysed sample may be analysed. There are colour charts giving an approximation of the haemoglobin in a serum sample on basis of its colour [17]. However, as mentioned earlier, this is very subjective as the colour of a sample depends on its content of not only haemoglobin but also protein, bilirubin, vitamins and some drugs. So this poses another dilemma.
Routine Analytes
We attempted to resolve these dilemmas by assessing the effect of hemolysis in different intensities on the estimation of some routine biochemical markers, viz blood urea nitrogen, creatinine, uric acid, phosphorus, sodium, potassium, total protein, amylase, lipase and lactate dehydrogenase. We also assessed the effect of hemolysis on the immunosuppressant, tacrolimus, and the antifolate, methotrexate.
As is evident in Table 2, a highly hemolysed sample may be analysed for BUN, creatinine, sodium, amylase and lipase without any clinically significant bias in the report. For estimation of uric acid and phosphorus, a highly hemolysed sample would yield a significantly altered result, though moderately and mildly hemolysed samples would yield an acceptable result as per the international guidelines of total allowable errors for these analytes. Total protein may be estimated in a moderately hemolysed sample without significant alteration of the result. However, potassium and LDH in a hemolysed sample will always yield an erroneous result.
Even though the mean difference calculated allowed for estimation of several analytes in a hemolysed samples, when individual results were assessed, the picture was not so promising. Since more than 10% of the samples yielded results beyond the TEa for phosphorus, sodium, potassium, total protein and LDH in several dilutions, it would not be advisable to measure or report these analytes in a hemolysed sample.
It could, therefore, be surmised that a hemolysed sample may be accepted under emergency situations for the estimation of BUN, creatinine, amylase and lipase. Even uric acid may be analysed in all except a highly hemolysed sample. However, where the result of the hemolysed sample is close to the decision limit, it may be advisable to test a fresh unhemolysed sample for reporting.
Specialised Analytes
In addition, though the manufacturers’ instructions state that a hemolysed sample should be avoided for estimation of tacrolimus and methotrexate, our data suggests that hemolysis does not interfere with these estimations. Hence, these precious samples, which are timed and hence not reproducible, need not be rejected.
Assessing Degree of Hemolysis
This brings us to the quantification of the degree of hemolysis within our laboratory. Though there are many charts available online to identify the degree of hemolysis, as mentioned earlier, these are very subjective as they depend on the colour of the unhemolysed serum which is dependent on several factors. To avoid the subjectiveness of these online descriptions, we measured the optical density of the various dilutions of all the samples on a spectrophotometer (Schimadzu UV-1700 Pharma Spec, Japan). Figure 1 gives the mean ODs which we have elucidated in our laboratory against a depictive photograph of the same. Our samples included serum of varying colours so as to represent a wide variety of samples. The mean ODs are well-graded as per the degree of hemolysis; however, 95% confidence intervals of the ODs are overlapping. This could be due to wide variance in the colour of the samples and their dilutions. Hence, it is suggested that, ideally, each lab should set its own ODs for comparison.
Limitations
The lack of quantitative estimation of haemoglobin as well as the inability to compare the visual estimation of hemolysis against the haemolytic index were the main limitation of the study.
Conclusions
The decision to accept/reject a hemolysed sample should be based not only on the presence of hemolysis, but also on the clinical condition of the patient. Where the sample is precious due to timing of the sample, thrombosed veins, or critical situation of the patient, this decision may also take into account the analytes to be estimated. So it may be safe to analyse a hemolysed sample for BUN, creatinine, amylase, lipase, tacrolimus and methotrexate; uric acid may be estimated in a moderately hemolysed sample. Analytes that must never be estimated in even a moderately or mildly hemolysed sample are phosphorus, sodium, potassium, total protein and LDH. In addition, each laboratory should establish their own optical density chart or colour chart for quantification of hemolysis.
Compliance with Ethical Standards
Conflict of interest
Dr.Seema Bhargava, Dr.Parul Singla, Dr.Anjali Manocha, Dr.Mamta Kankra, Dr.Anisha Sharma, Dr.Ashok Ahirwar, Ms. Rachna Ralhan, Mr. Udhavanand Thapliyal and Ms. Preet Mehra declare that they have no conflicts of interest.
Ethical Considerations
All procedures performed in this study were in accordance with the ethical standards of our institutional research committee and with the 1964 Helsinki declaration and its later amendments.
Informed Consent
Informed written consent was procured from all subjects.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Plebani M, Carraro P. Mistakes in a stat laboratory: types and frequency. Clin Chem. 1997;43:1348–1351. doi: 10.1093/clinchem/43.8.1348. [DOI] [PubMed] [Google Scholar]
- 2.Frank JJ, Bermes EW, Bickel MJ, Watkins BF. Effect of in vitro hemolysis on chemical values for serum. Clin Chem. 1978;24(11):1966–1970. doi: 10.1093/clinchem/24.11.1966. [DOI] [PubMed] [Google Scholar]
- 3.Shah JS, Soon PS, Marsh DJ. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS ONE. 2016;11(4):1–12. e0153200. 10.1371/journal.pone.0153200. [DOI] [PMC free article] [PubMed]
- 4.Dimeski G. Interference testing. Clin Biochem Rev. 2008;29(1):S43–S48. [PMC free article] [PubMed] [Google Scholar]
- 5.Talke H, Schubert GE, Wchnschr K. Enzymatic urea determination in the blood and serum in the warburg optical test. 1965;43:174. [DOI] [PubMed]
- 6.Jaffe M. Estimation of creatinine by the Jaffe Reaction. Z Physiol Chem. 1886;10:391–400. [Google Scholar]
- 7.Fossati P, Prencipe L, Berti G. Use of 3,5-dichloro-2-hydroxybenzenesulfonic acid/4-aminophenazone chromogenic system in direct enzymic assay of uric acid in serum and urine. Clin Chem. 1980;26:227–231. doi: 10.1093/clinchem/26.2.227. [DOI] [PubMed] [Google Scholar]
- 8.Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. J Biol Chem. 1925;66:375. [Google Scholar]
- 9.Scott MG, Legrys VA, Hood JL, Electrolytes and Blood Gases, Burtis CA, Ashwood ER, Bruns DE. Tietz textbook of clinical chemistry and molecular diagnostics. 5th ed. Philadelphia: Elsevier Health Sciences; 2012. p. 807–835.
- 10.Hortin GL. Aminoacids, Peptides and Proteins, Burtis CA, Ashwood ER, Bruns DE. Tietz textbook of clinical chemistry and molecular diagnostics. 5th ed. Philadelphia: Elsevier Health Sciences; 2012. p. 509–563.
- 11.Bais R, Panteghini M. Enzyme and Rate Analyses, Burtis CA, Ashwood ER, Bruns DE. Tietz textbook of clinical chemistry and molecular diagnostics. 5th ed. Philadelphia: Elsevier Health Sciences; 2012. p. 355–378.
- 12.Snozek CLH, McMillan GA, Moyer TP. Therapeutic Drugs and Their Managment, Burtis CA, Ashwood ER, Bruns DE. Tietz textbook of clinical chemistry and molecular diagnostics. 5th ed. Philadelphia: Elsevier Health Sciences; 2012. p. 1057–1108.
- 13.Lippi G, Blanckaert N, Bonini P, Green S, Kitchen S, Palicka V, et al. Haemolysis: an overview of the leading cause of unsuitable specimens in clinical laboratories. Clin Chem Lab Med. 2008;46:764–772. doi: 10.1515/CCLM.2008.170. [DOI] [PubMed] [Google Scholar]
- 14.Abd Elrouf MB, Amanullah M, Zaman GS. Interference of hemolysis in the estimation of plasma aspartate aminotransferase, potassium and phosphate. J Invest Biochem. 2014;1(1):12–15. doi: 10.5455/jib.20130611094024. [DOI] [Google Scholar]
- 15.Grafmeyer D, Bondon M, Machon M, Levillain R. The influence of bilirubin, haemolysis and turbidity on 20 analytical tests performed on automatic analyzers. Eur J Clin Chem Clin Biochem. 1995;33:31–52. doi: 10.1515/cclm.1995.33.1.31. [DOI] [PubMed] [Google Scholar]
- 16.Shah JS, Soon PS, Marsh DJ. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS ONE. 2016;11(4):1–12. doi: 10.1371/journal.pone.0153200.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippi G, Cadamuro J, Meyer AV, Simundi AM. Practical recommendations for managing hemolyzed samples in clinical chemistry testing. Clin Chem Lab Med. 2018;56(5):1–12. doi: 10.1515/cclm-2017-1104. [DOI] [PubMed] [Google Scholar]