Abstract
Many regulatory networks appear to involve partially redundant enhancers. Traditionally, such enhancers have been hypothesized to originate mainly by sequence duplication. An alternative model postulates that they arise independently, through convergent evolution. This mechanism appears to be counterintuitive to natural selection: Redundant sequences are expected to either diverge and acquire new functions or accumulate mutations and become nonfunctional. Nevertheless, we show that at least 31% of the redundant enhancer pairs in the human genome (and 17% in the mouse genome) indeed originated in this manner. Specifically, for virtually all transposon-derived redundant enhancer pairs, both enhancer partners have evolved independently, from the exaptation of two different transposons. In addition to conferring robustness to the system, redundant enhancers could provide an evolutionary advantage by fine-tuning gene expression. Consistent with this hypothesis, we observed that the target genes of redundant enhancers exhibit higher expression levels and tissue specificity as compared with other genes. Finally, we found that although enhancer redundancy appears to be an intrinsic property of certain mammalian regulatory networks, the corresponding enhancers are largely species-specific. In other words, the redundancy in these networks is most likely a result of convergent evolution.
Keywords: redundant enhancers, evolution, gene regulation, transposons
Introduction
Most phenotypic variation between individuals of the same or closely related species is assumed to result from changes in gene regulation, rather than in the genes themselves (King and Wilson 1975). In eukaryotes, gene expression is primarily regulated at the level of transcription. Transcription is initiated when the RNA polymerase II machinery recognizes and binds specific sequences in the core promoter of a gene. The resulting basal level of expression can be increased or decreased through biochemical interactions between transcription factor (TF) and cofactor proteins and cis-regulatory elements scattered throughout the genome. Cis-regulatory elements include sequences that are proximal to their target genes, such as promoters, but also distal sequences, such as enhancers. Enhancers are typically a few hundred base pairs long and harbor clusters of TF-binding sites (TFBSs). They are usually bound by tissue-specific TFs and can thereby produce highly controlled spatiotemporal gene expression patterns (Spitz and Furlong 2012).
In 2008, Mike Levine and colleagues coined the expression “shadow enhancers” to refer to enhancers with redundant regulatory activities in the fruit fly Drosophila melanogaster (Hendrix et al. 2008). Specifically, they found that the genes vnd and miR-1 are both regulated by at least a pair of enhancers each. One of the enhancers in these pairs—the “primary” enhancer—is more proximal to the transcription start site (TSS) of their target than the other—the “shadow” enhancer—, but both enhancers have similar regulatory activities and bind the same TFs. Since then, the expression “shadow enhancers” has been extended to describe two or more (possibly partially) redundant enhancers, bypassing the assignment of the labels “primary” and “shadow” (Barolo 2011; Cannavò et al. 2016). Many genes in the mammalian genome are known to be controlled by two or more redundant enhancers (Allan et al. 1995; Jeong et al. 2006; Lehoczky and Innis 2008; Bebin et al. 2010; Guerrero et al. 2010; Kunarso et al. 2010; Ghiasvand et al. 2011). Moreover, recent work has suggested that enhancer redundancy is a common feature of the mammalian genome (Osterwalder et al. 2018).
Redundant enhancers may have originated by several mechanisms, including sequence duplication and independent exaptation or co-option of transposons (Long et al. 2016). Transposons are mobile genetic sequences that behave as genomic parasites. They have been very effective in colonizing many genomes and occupy at least half of the human genome (Cannavò et al. 2016; Platt et al. 2018). Transposons harbor TFBSs and their insertion has been shown to influence the expression of nearby genes in reporter gene assays (Bejerano et al. 2006; Santangelo et al. 2007; Sasaki et al. 2008; Smith et al. 2008; Su et al. 2014; Ferreira et al. 2016; Nishihara et al. 2016) and also using the CRISPRcas9 system (Chuong et al. 2016). Consistently, many enhancers in the mammalian genome are thought to derive from transposons (Jacques et al. 2013). In particular, Franchini et al. (2011) showed that two redundant enhancers of the proopiomelanocortin gene (POMC) originated from the subsequent exaptation of two different retrotransposons: a long-terminal repeat (LTR) transposon inserted between the metatherian/eutherian split (147 Ma) and the placental mammal radiation (∼90 Ma), and a short-interspersed nuclear element (SINE) retrotransposon inserted before the origin of prototherians (166 Ma). The fact that two enhancers with redundant regulatory activities are under purifying selection could be explained by their role as regulatory buffer preventing deleterious phenotypic consequences upon the loss of one of them (Osterwalder et al. 2018). This is in agreement with the theory independently proposed by Schmalhausen and Waddington, which states that phenotypes will remain relatively invariant to genetic perturbations (Flatt 2005). Furthermore, regulatory redundancy has been suggested as a means to ensure steady gene expression (Frankel et al. 2010; Perry et al. 2010; Osterwalder et al. 2018).
Because enhancers can be located virtually anywhere relative to their target genes, their identification and characterization are challenging. Nevertheless, much progress has been made toward creating a catalog of the cis-regulatory elements in the human genome, in particular through chromatin immunoprecipitation (ChIP)-based methods. Thus, data generated by international consortia such as the Encyclopedia of DNA Elements (ENCODE; Feingold et al. 2004) have made evident the pervasiveness of multiple enhancers with similar regulatory activities near the same gene (Cannavò et al. 2016) and are starting to reveal their adaptive value (Osterwalder et al. 2018). In spite of that, it remains unknown how redundant enhancers originate. To directly assess this, we used cap analysis of gene expression (CAGE) data from the FANTOM (“functional annotation of the mammalian genome”) project (Andersson et al. 2014; Forrest et al. 2014) and a stringent approach to identify 2,117 pairs of enhancers with (possibly partially) redundant regulatory activities in the human genome. By combining their transposon annotation with phylogenetic information, we inferred that 716 of these redundant enhancer pairs were likely to derive from transposons and in 92% of such cases, the enhancer partners of the pairs had been acquired independently, from the exaptation of two different transposons. We also made similar observations for mouse, concluding that at least 31% of all redundant enhancer pairs in human and 17% of those in mouse have evolved by independent transposon exaptation. Equally important, we found that redundant enhancers are highly lineage-specific, in the sense they are not evolutionary conserved. Hence, the similar levels of redundancy observed between orthologous mammalian regulatory networks appear to be examples of convergent evolution.
Materials and Methods
Gene Annotation
Gene annotation was based on GENCODE (human: v19, mouse: vM1; Harrow et al. 2006).
Facet-Specific Quantification of Enhancer and Promoter Activity
In order to quantify the activity of a genome-wide data set of enhancers and promoters across a large number of tissue groups, we utilized data from the FANTOM5 project and an approach similar to the one presented by Andersson et al. (2014). More precisely, we first downloaded the coordinates of the enhancers and promoters (http://fantom.gsc.riken.jp/5/datafiles/phase2.5/extra/Enhancers/ and http://fantom.gsc.riken.jp/5/datafiles/phase2.5/extra/CAGE_peaks/, last accessed November 22, 2019) determined by FANTOM5, phase 2.5, in the human (hg19) and the mouse (mm9) genomes (Andersson et al. 2014; Forrest et al. 2014) together with the mapped hCAGE (cap analysis of gene expression sequencing on Heliscope single molecule sequencer) reads for the “organ-derived” samples (http://fantom.gsc.riken.jp/5/datafiles/phase2.5/basic/human.tissue.hCAGE/ and http://fantom.gsc.riken.jp/5/datafiles/phase2.5/basic/mouse.tissue.hCAGE/, last accessed November 22, 2019). We further refer to these enhancers and promoters as “FANTOM enhancers” and “FANTOM promoters,” respectively. Next, we grouped the samples from similar tissues into 44 human and 36 mouse “facets,” similarly as Andersson et al. (2014) did, and merged the corresponding hCAGE reads. The curated grouping of samples into facets (see supplementary tables S1 and S2, Supplementary Material online) was kindly provided by the FANTOM5 consortium. Enhancer activity was computed according to Andersson et al. (2014): Specifically, we took a 400-bp window around the center point of the FANTOM enhancer coordinates and removed all enhancers that overlapped with any promoter or with Ensembl gene exons. Then, for each enhancer, we counted all hCAGE reads with a 3′-end overlapping the enhancer, an edit distance ≤6 (NM flag), and a MAPQ ≥20. Finally, we computed the number of hCAGE reads (tags) per million mapped reads (TPM) associated with each enhancer. Promoter activity was computed according to Forrest et al. (2014), basically, in the same manner as described for the enhancers, but without using a window or removing any promoters. Lastly, the TPM values of the enhancers and promoters were normalized across facets. To this end, the raw library size of a facet was defined as the total number of mapped reads with a MAPQ ≥20, excluding the reads on chromosome M, and library size normalization factors were computed using the edgeR package (Robinson et al. 2009) function calcNormFactors() with parameter “method=RLE.”
Facet-Specific Binary Enhancer and Promoter Activity (“Enhancer/Promoter Usage Matrix”)
To compute binary enhancer activity (active/inactive), we followed the rationale and approach of Andersson et al. (2014). We sampled 100,000 401-bp-long regions from the human (or mouse) genome. The regions were ensured to not overlap with Ensembl exons, FANTOM promoters, or FANTOM enhancers. We counted all hCAGE reads in these random regions in the same manner as we did for the FANTOM enhancers/promoters (see Facet-Specific Quantification of Enhancer and Promoter Activity), computed an empirical P value as the fraction of random regions with equal or greater hCAGE read counts in comparison to the enhancers, and corrected the P values for multiple testing using the false discovery rate (FDR; Benjamini and Hochberg 1995). We considered an enhancer active in a specific facet if it had at least two reads, a P value ≤0.0025 and an FDR-corrected P value ≤0.1.
For the promoters, we computed the facet-specific binary activity (“promoter usage matrix”) according to Forrest et al. (2014). Specifically, we computed the percentile for the cutoff they used on the normalized activity values (0.2), and applied the corresponding value as cutoff for our data (human: 0.297, mouse: 0.319).
Assignment of Enhancer Target Genes
To assign enhancers to target genes, we used an approach relying on the correlation between enhancer and promoter activities across the facets. For the analysis, we only considered those enhancers and promoters active in at least one of the facets used in this study (see Facet-Specific Binary Enhancer and Promoter Activity (“Enhancer/Promoter Usage Matrix”)). First, we assigned all Ensembl TSS of protein-coding genes with a distance ≤500 bp to the corresponding promoters and discarded promoters without a TSS assignment. Then, we mapped the enhancer and promoter TPM values to a simplified activity scale: Although inactive enhancers (promoters) were given a value of zero, the rest was split into tertiles according to their activity values across all facets and given a 1 (weak), 2 (moderate), or 3 (strong). Finally, we computed the Pearson correlation coefficient for each pair of enhancers and promoters in the same topologically associating domain (TAD) based on their simplified activities in all facets. An enhancer (promoter) was considered to be located in a TAD if at least 50% of its sequence overlapped with it. Enhancers and promoters not located in any TAD were discarded from the analysis. The TAD coordinates were taken from Dixon et al. (2012), supplementary table S3, Supplementary Material online, for the category “hESC Combined.” The association between the enhancer and promoter activities was tested using the Pearson product-moment correlation test. P values were FDR-corrected across all tested associations. We considered all enhancer–promoter associations with r ≥ 0.5, a P value ≤0.0035, and an FDR-corrected P value of ≤0.05 as significant.
TFs Binding-Redundant Enhancer Pairs
In order to test whether the partners of our redundant enhancer pairs are both bound by the same TF, we tested them for overlap with TF ChIP-seq peaks from the ENCODE project. We used the Bioconductor package “ENCODExplorer” version 2.4.0 (Beauparlant et al. 2015) to retrieve TF ChIP-seq assay data. In particular, we downloaded the BED files for the assays that fulfilled the following requirements: “assembly=hg19,” “investigated_as” contains the string “transcription factor” (TF assay), “biosample_type=tissue” (only tissue samples), and output_type=“peaks” (standard peak calling thresholds). We further excluded CTCF assays because CTCF commonly marks boundary elements in the genome and discarded “pooled” samples (i.e., assays for which “technical replicates” contains multiple entries). In addition, we required the tissues to be mappable to our facets (see supplementary table S3, Supplementary Material online). We randomly sampled one assay per tissue and TF. If the coordinates of an enhancer overlapped with those of a peak in an assay, the enhancer was considered to be bound by the associated TF. We selected all redundant enhancer pairs with common activity in a facet and computed the number of enhancer pairs that were bound by the same TF in a tissue mapped to that facet. Specifically, for the liver facet, we retained only liver samples (biosample_name=“liver”). The corresponding assays involved five TFs: ATF3, HNF4G, YY1, NR2F2, and ZBTB33.
Genomic Properties of Redundant and Nonredundant Enhancers
Gene Ontology Enrichment Analysis
We tested the redundant enhancer target genes for enrichment in terms of the “GOTERM_MF_FAT” and “GOTERM_BP_FAT” ontologies with the tool DAVID (Huang et al. 2009). First, we analyzed each facet separately: We considered only those target genes for which both enhancer partners of a redundant enhancer pair were active in the facet. As background set, we used the target genes of all enhancers with activity in the respective facet. In addition, we conducted an analysis across all facets: All redundant enhancer target genes were compared with the target genes of all correlated enhancers.
Tissue Specificity of Enhancer Target Gene Expression
We computed the Shannon entropy (Schug et al. 2005) for each enhancer target gene. The computation was based on the TPM values across all facets of all the promoters associated with the target gene. If a target gene was associated with multiple promoters, we separately calculated the entropy for each promoter and then averaged across all promoters. We compared the entropy of nonredundant enhancer target genes with redundant enhancer target genes using a two-sided Wilcoxon rank-sum test. Each target gene was considered only once and those in both groups were excluded.
Expression Strength of Enhancer Target Genes
For each facet, we measured the expression strength of an enhancer target gene as the average of the TPM values of all its promoters. Genes with multiple promoters active in that facet such that at least one of the promoters was associated with a redundant enhancer and at least one of the promoters was associated with a nonredundant enhancer were excluded from the analysis. We compared the expression strength between redundant enhancer target genes (i.e., genes with two or more active enhancers in that facet) and nonredundant enhancer target genes (i.e., genes with only one active enhancer in that facet) with a one-sided Wilcoxon rank-sum test. P values were corrected for multiple testing using the FDR. Only facets with more than ten redundant and nonredundant enhancer target genes were considered for the analysis.
Sequence Conservation of Enhancers
We computed the sequence conservation score of each enhancer as the average of its base-wise PhastCons scores (Siepel 2005). For human the computation was based on the hg19 100way phastCons scores (http://hgdownload.cse.ucsc.edu/goldenpath/hg19/phastCons100way/hg19.100way.phastCons.bw, last accessed November 22, 2019) and for mouse on the mm9 30way placental phastCons scores (http://hgdownload.cse.ucsc.edu/goldenpath/mm9/phastCons30way/placental/, last accessed November 22, 2019).
SNP Density of Enhancers
We computed the SNP density of each enhancer as the number of SNPs per 1,000 bp of sequence. SNP data were obtained from Ensembl variation (Chen et al. 2010) (ftp://ftp.ensembl.org/pub/release-92/variation/vcf/homo_sapiens/homo_sapiens.vcf.gz, ftp://ftp.ensembl.org/pub/release-92/variation/vcf/mus_musculus/mus_musculus.vcf.gz, last accessed November 22, 2019). The enhancer coordinates were lifted over to hg38 and mm10. Enhancers that could not be lifted over were discarded from this analysis.
Facet Enhancer Enrichment
For each facet, we compared the number of active and inactive enhancers between redundant and nonredundant enhancers. We computed a P value with a two-sided Fisher’s exact test and corrected for multiple testing using the FDR.
Transposon Annotation of Enhancers
Enhancer sequences overlapping by at least 50 bp with one or more transposons of the same species (possibly interrupted by an arbitrary sequence) in the RepeatMasker database (http://www.repeatmasker.org, version 4.0.5, last accessed November 22, 2019) were annotated as transposons. For enhancers shorter than 500 bp, we used a 500-bp sequence around the center point of the original coordinates. The RepeatMasker taxonomy classifies repeats (“species”) into “families” which are, in turn, classified into “classes” (“LINE” [long-interspersed nuclear element], “SINE,” “LTR,” and “DNA”). Only the repeat classes “LINE,” “SINE,” “LTR,” and “DNA” were used in the analysis. Enhancers satisfying the overlap requirement for multiple transposon species were annotated with all of them.
We modeled the prevalence of the transposons in the human and the mouse genomes by randomly repositioning our 3,523 correlated enhancers (4,074 in mouse) in the genome and computing the transposon overlap in the same way as for the enhancers. This was repeated 1,000 times and the derived background distributions were used to test every transposon family for significant enrichment among our redundant enhancers and to compute an empirical P value. The P values were then FDR-corrected.
Enrichment of Transposon Families among Redundant and Nonredundant Enhancers
For every transposon family, we counted the number of redundant and nonredundant enhancers annotated as transposons of that family. Then, we tested the null hypothesis of equal proportions (two-sided test) among redundant and nonredundant enhancers in R with the prop.test() function.
Phylogenetic Dating of Transposon Insertions
To identify putative orthologous sequences of the human enhancers, we BLASTed their sequences against the genomes of 14 other mammalian and 4 nonmammalian vertebrate species. The assemblies we used were: panTro4, gorGor3, ponAbe2, rheMac8, calJac3, mm9, oryCun2, bosTau8, canFam3, myoLuc2, loxAfr3, dasNov3, monDom5, ornAna2, galGal4, xenTro7, fr3, and danRer10, as provided by the UCSC genome browser website. The species and assembly versions were selected based on the quality of the assemblies, the availability of Ensembl Compara data for the assemblies and in such a way as to sample all big branches of the vertebrate phylogenetic tree with focus on mammals and primates (Vilella et al. 2008). We employed BlastN from the NCBI BLAST+ suite (version 2.2.31; Camacho et al. 2009), selecting scoring parameters that promote the score of sequences with moderate similarity (target sequence similarity ∼70%; Pearson 2013): word size 7, reward 5, penalty −4, gap opening cost 8, gap extension cost 6, and an E-value cutoff of 1×10−3. For enhancers shorter than 500 bp, we used a 500-bp sequence around the center point of the original coordinates.
A BLAST hit was labeled to be the ortholog of a human enhancer if it fulfilled these three criteria: 1) It was one of the ten highest scoring BLAST hits. 2) It was located within a 2-Mb window around any target gene ortholog. For every enhancer, we identified the orthologs of all its target genes via Ensembl Compara (release 87, hg38); all gene orthology relationships (one2one, one2many, many2many) were included; Ensembl hg38 gene IDs were mapped to hg19 gene IDs (as some gene IDs are not stable across the assemblies) and the gene ortholog coordinates of some species had to be lifted over to the used assembly (bosTau6 to bosTau8, ornAna1 to ornAna2, xenTro3 to xenTro7); enhancers without any target gene orthologs were excluded from the analysis. 3) It overlapped with the same RepeatMasker transposon types and number of transposons as the human enhancer (RepeatMasker version 4.0.5; mm9: lift over from mm10, bosTau8: 4.0.5, rheMac8: 4.0.5, ornAna2: 4.0.5). In order to reduce the effect of slight annotation differences across the vertebrate genomes, we considered the transposon types identical if they belonged to the same transposon family; for enhancers overlapping with multiple transposons, all transposon families had to be there in order for the ortholog to be called in the assembly. If multiple BLAST hits fulfilled these criteria, we declared the hit with the highest E-value to be the enhancer ortholog.
We then used the identified enhancer orthologs to date the transposon insertion by reconstructing the ancestral state in a phylogenetic tree. The tree topology for the selected vertebrate species (see Identification of Transposon-Derived Enhancer Orthologs) was generated with phyloT (http://phylot.biobyte.de/, last accessed November 22, 2019), with a random breakdown of polytomies. Using binary states that represented the presence or absence of the enhancer ortholog, we reconstructed the ancestral states in the tree for every enhancer with the phangorn R package (Schliep 2011). After importing the tree topology with the read.newick() function (phytools package; Revell 2012), deleting single nodes and setting all branch lengths to one with the collapse.singles() and compute.brlen() functions (ape package; Paradis et al. 2004), respectively, we used the ancestral.pars() function (phangorn package) with type=MPR for a maximum parsimony reconstruction (Fitch–Hartigan algorithm). We defined the insertion node as the oldest node in the human (or mouse) lineage with an uninterrupted sequence of nodes (starting at the human/mouse node), where the ortholog was inferred to be present. For a few enhancers without a target gene ortholog in any of the used vertebrate species, the dating was not possible. For the enhancers overlapping with multiple transposons, the age of the youngest transposon was used as the overall transposon age (considering it as the point in time where all transposon components were present).
We dated random genomic transposons (considered were only LINE, SINE, LTR, and DNA classes) in an analogous manner. Specifically, we merged overlapping transposons from the same species, picked a random sample of 10,000 transposons, and dated them following the same steps described earlier.
For the mouse, we followed the same strategy as for human. The mm9 assembly was replaced with hg19 to search for orthologs. Ensembl Compara (release 87) was based on mm10, so the mm10 ENEMBL gene IDs were mapped to mm9 gene IDs (as some IDs are not stable).
Ages of Enhancer Target Genes
The target gene age estimates were extracted from the Ensembl Compara gene phylogenetic trees (Ensembl version 94) via the Ensembl REST API (Vilella et al. 2008).
Orthology of Target Genes and Enhancers in Human and Mouse
We used Ensembl Compara to determine the orthologs of human genes in mouse (and vice versa). The pairs of human and mouse genes with an orthology relationship targeted by at least one human and one mouse enhancer constitute the set of common enhancer target genes between the human- and the mouse-correlated enhancer data sets. Enhancer orthologs were determined as liftOver (Hinrichs 2006) hits.
Results
Redundant Enhancers Are a Common Feature of Human Regulatory Networks and Have Distinct Features
Although examples of enhancers with (possibly partially) redundant regulatory activities have been known for many years (Barolo 2011), the extent to which such regulatory redundancy contributes to the robustness of mammalian regulatory networks is only now beginning to be appreciated (Osterwalder et al. 2018). To quantify partial and absolute regulatory redundancy in the human genome, we analyzed 201,799 promoters and 54,284 enhancers identified by the FANTOM5 Consortium with the CAGE technique (Andersson et al. 2014; Forrest et al. 2014; see Materials and Methods). Active enhancers and promoters are often transcribed (Papantonis and Cook 2013), and the levels and directionality of their transcription have been shown to reflect their regulatory activities (Andersson et al. 2014). Relying on the “guilt-by-association” paradigm, cotranscription of enhancers and promoters has been proposed and successfully applied to associate enhancers with their target genes (Shen et al. 2012; Thurman et al. 2012; Sheffield et al. 2013; Andersson et al. 2014; Yao et al. 2015; Fishilevich et al. 2017). In addition, enhancers and their target genes are expected to be located within the same TAD (Dixon et al. 2012). Thus, to uncover the enhancer target genes, we computed the Pearson correlation coefficient () between the transcriptional activity profiles of pairs of enhancers and promoters in the same TADs across 44 groups of samples from related tissues. We further refer to such groups of samples as “facets” (see Materials and Methods; Andersson et al. 2014). Of the enhancers predicted by the FANTOM5 consortium, 11,582 were active in at least one facets; in turn, 10,445 of these enhancers were located within a TAD that also comprised one or more active promoters. Of the promoters, 72,272 were active in at least one facet and within 500 bp of an Ensembl protein-coding gene TSS; 55,612 were located within TADs with at least one active enhancer. This yielded a total of 314,746 possible associations between pairs of enhancers and promoters in 2,449 TADs. Approximately 3.5% of those pairs (10,952) were (positively) correlated (> 0.5, P value ≤0.0025, FDR-corrected P value < 0.05; Benjamini and Hochberg 1995), involving 3,523 enhancers and 6,474 promoters in 1,398 TADs. We regarded the transcripts (genes) corresponding to those promoters (see Materials and Methods and fig. 1A) as the putative target transcripts (genes) of the enhancers. Based on this, we identified 2,117 pairs of enhancers with the exact same target transcripts and common activity in at least one facet. For simplicity, we further refer to these putative pairs of enhancers with (possibly partially) redundant regulatory activities as “redundant enhancer pairs” and to the enhancer partners of the pairs as “redundant enhancers.” In total, 1,280 (36%) of the enhancers that were correlated with one or more promoters were redundant enhancers. This result points toward enhancer redundancy being a widespread feature of regulatory networks and is in accordance with previous studies in the Drosophila genome (Cannavò et al. 2016).
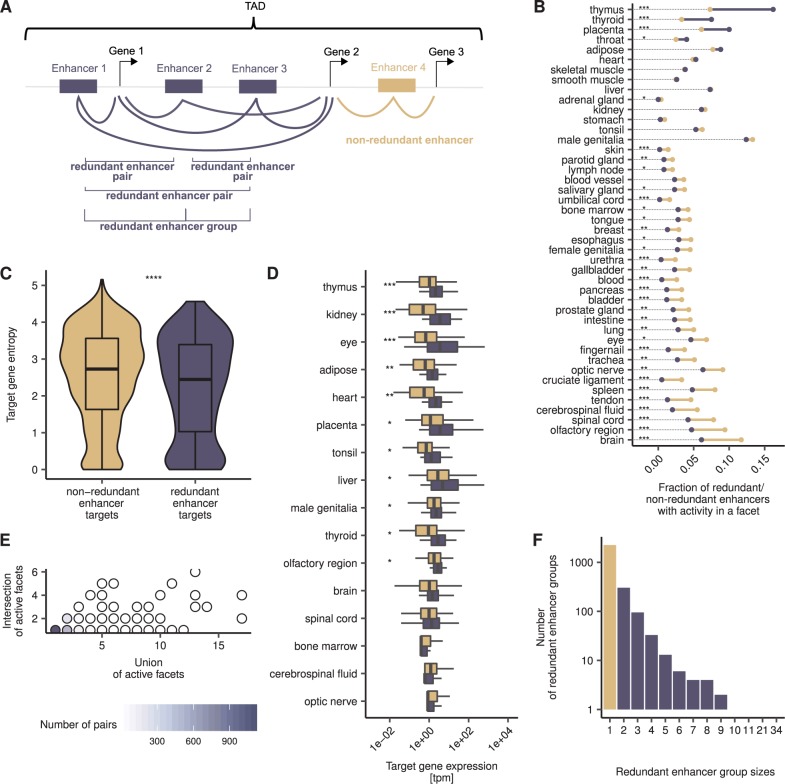
Fig. 1.—
Redundant enhancers show distinctive features. (A) Enhancers are assigned to target genes, further categorized into redundant and nonredundant enhancers, arranged into redundant enhancer pairs and redundant enhancer groups. If the activity profiles of two enhancers within a TAD are moderately significantly correlated with the activity profiles of the same set of promoters, then the two enhancers are considered a “redundant enhancer pair” and the genes associated with the promoters, their target genes. Significant correlations are shown as lines between enhancers and genes. All redundant enhancer pairs associated with the same target genes form a redundant enhancer group. (B) Some facets show strong enrichments for active redundant enhancers (e.g., thymus) and some show strong depletions (e.g., brain). The dumbbell plot shows the fractions of active nonredundant (beige) and redundant (purple) enhancers per facet. The facets are sorted according to the difference in the fractions of redundant and nonredundant enhancers; if the fraction of active nonredundant enhancers is larger than that of redundant enhancers, the line between the dots is depicted in beige; otherwise, in purple. The dotted lines serve as visual aids. Asterisks indicate FDR-corrected P values of Fisher’s exact tests: *P < 0.05, **P < 0.01, ***P < 0.001. (C) Redundant enhancer target genes are more tissue-specific than nonredundant enhancer target genes. Depicted are the entropies of nonredundant (beige) and redundant (purple) enhancer target genes. Asterisks indicate P value of Wilcoxon rank-sum test. ****P < 0.0001. (D) In most facets, redundant enhancer target genes show a stronger expression than nonredundant enhancer target genes. Facet-specific expression of nonredundant (beige) and redundant (purple) enhancer target genes with FDR-corrected P values of Wilcoxon rank-sum tests. The facets are sorted according to FDR-corrected P values. See (B) for significance code. (E) The majority of redundant enhancer pairs have a 100% overlapping activity pattern. Regulatory redundancy of redundant enhancer pairs is measured as the ratio of the number of facets with shared activity to the number of facets in which any of the partners of the pair is active. (F) Redundant enhancers form groups of different sizes. Depicted are the number of nonredundant enhancers (beige) and redundant (purple) enhancer in a group.
Both redundant and nonredundant enhancers were relatively weakly conserved (medians of average base-wise PhyloP scores of 0.04, see Materials and Methods). Nonetheless, redundant enhancers had a higher SNP density than nonredundant enhancers (medians 110 SNPs/kb and 108 SNPs/kb, respectively, P value = 0.01, two-sided Wilcoxon rank-sum test), suggesting differences in selective pressure within the human population. In addition, redundant enhancers and nonredundant enhancers had a similar number of target genes (medians = 1) and similar GC content (medians = 0.48). Furthermore, redundant enhancers were closer to the nearest TSS of their target genes than nonredundant enhancers (median distances 73 and 90 kb, respectively, P value = 0.02, two-sided Wilcoxon rank-sum test, see supplementary fig. S1, Supplementary Material online), and their target locus size (median value of all target loci of an enhancer) was larger (136 and 195 kb, P value = 1.3×10−12 two-sided Wilcoxon rank-sum test). These results hint at redundant enhancers being preferentially located in less gene-dense regions. Redundant and nonredundant enhancers also exhibited differences in their activities (see fig. 1B). Thus, compared with nonredundant enhancers, redundant enhancers were overrepresented in facets such as thymus (FDR-corrected P value = 2.4×10−14, see Materials and Methods) and underrepresented in brain (FDR-corrected P value = 4.3×10−7), cruciate ligament (FDR-corrected P value = 2.9×10−7), cerebrospinal fluid (FDR-corrected P value = 8.7×10−7), and olfactory region (FDR-corrected P value = 1.5×10−6), among other facets, indicating that redundant enhancers are particularly important for certain tissues. In addition, the target genes of redundant enhancers were enriched in transcription-related functions and processes (see supplementary figs. S2 and S3, Supplementary Material online and Materials and Methods) and more tissue-specific compared with those of nonredundant enhancers (median entropies 2.4 and 2.7, respectively; P value = 4.2×10−5, two-sided Wilcoxon rank-sum test, see fig.1C and Materials and Methods). Indeed, although redundant enhancers have been hypothesized to be associated with tissue-specific expression patterns, nonredundant enhancers have been linked to house-keeping genes (Osterwalder et al. 2018). Equally remarkably and in agreement with findings in other tissues (Denisenko et al. 2019), we observed that the target genes of redundant enhancers were generally more strongly expressed than those of nonredundant enhancers (see fig. 1D). Specifically, this was the case for 69% facets (11 out of the 16 facets with more than ten redundant and nonredundant enhancer target genes; FDR-corrected P values ≤0.05; see Materials and Methods); in the remaining facets, there was no significant difference. The increase in expression specificity and strength—also known as superfunctionalization (Majunder and Biswas 2006)—confers robustness to the regulatory network (Frankel et al. 2010) and could explain enhancer redundancy.
Based on ChIP-seq data, 67% of all redundant enhancer pairs were bound by the same TF (see Materials and Methods), providing evidence for both enhancer partners participating in the same regulatory networks. On an average, the partners of a redundant enhancer pair had common activity in 1.2 facets and 1,328 pairs (63%) had 100% identical activity profiles (see fig. 1E). Consistent with previous studies (Barolo 2011; Cannavò et al. 2016) and supporting the idea that regulatory redundancy is scattered across more than two enhancers, we found that 672 redundant enhancers (52%) were partners of multiple redundant enhancer pairs. To gain insight into such complex relationships, we arranged redundant enhancers into groups, such that all enhancers in a group have the same target genes and common activity with at least another enhancer in the group. On an average, each group comprised 2.7 enhancers (see fig. 1F). Two redundant enhancer groups consisting of 34 and 21 enhancers were exceptionally large. An explanation for such a high number of redundant enhancers could be a very complex spatiotemporal expression profile of the targets in these groups. The group of 35 enhancers was associated with eight transcripts of the thyroglobulin (ENSG00000042832) gene, which according to the Ensembl expression atlas is mainly expressed in the thyroid gland (Petryszak et al. 2016). Interestingly, all enhancers in this group have highly redundant regulatory activities, with all of them being active in the “thyroid” facet. This suggests that thyroglobulin is transcribed in a complex condition-specific manner, rather than having a complex spatial expression pattern. Similarly, the group of 21 enhancers apparently regulates the expression of three transcripts of the ADAM metallopeptidase domain 12 (ENSG00000148848) gene in the placenta. In addition to demonstrating differences in sizes, redundant enhancer groups differed in the number of associated target transcripts: Although 47% of the groups had only one target transcript, 53% had multiple ones; the average number of associated target transcripts over all groups was 2.3. This result is supported by other genetic and genome-wide studies (van Arensbergen et al. 2014; Quintero-Cadena and Sternberg 2016) and indicates a relatively high level of regulatory complexity, with multiple enhancers being associated with multiple promoters. Finally, although not every pair of enhancers in a group was required to have common activity, almost all of them did (2,117 out of a total of 2,134 pairs). In summary, most redundant enhancer pairs in our data set are active in a small number of facets and appear to be perfectly redundant.
A Large Fraction of Human-Redundant Enhancer Pairs Has Independent Origins
Redundant enhancers have been proposed to arise by duplication (Hendrix et al. 2008). Nevertheless, this hypothesis has not been systematically tested and remains speculative. Indeed, it has been clearly established that a large number of cis-regulatory elements are derived from transposons (Feschotte 2008), and examples of independent exaptation of transposons into redundant enhancers exist (Franchini et al. 2011). In order to quantify the contribution of transposons to the genesis and evolution of redundant enhancers, we annotated each enhancer based on its overlap with RepeatMasker elements (see Materials and Methods). Forty-eight percent (1,686) of the 3,523 enhancers that were correlated with at least one promoter were annotated as transposons; hence, they may derive from transposons. This fraction is lower than expected from the prevalence of transposons in the genome (log2-fold difference in observed transposon overlap compared with the random expectation = −0.5, see supplementary fig. S4, Supplementary Material online), and depends on the facet, ranging from 24% (adipose) to 66% (fingernail, see supplementary fig. S5, Supplementary Material online). These findings are in agreement with previous studies (Chuong et al. 2017; Simonti et al. 2017; Trizzino et al. 2018). We further refer to the enhancers annotated as transposons as “transposon enhancers.”
Most of the transposon enhancers (67%, 1,132 out of 1,686) were annotated with only one transposon species (see Materials and Methods), showing a clear origin. In contrast, 33% (554) was annotated with multiple (two to four) transposon species and may comprise cases of coordinated co-option of transposons (Nishihara et al. 2016). The five most represented transposon families among the enhancers were L2s (21%), Alus (20%), MIRs (18%), L1s (13%), and ERVL-MaLRs (5%). Although these are also the most prominent transposon families in the human genome, their frequencies among the enhancers did not directly reflect their prevalence in the genome. Thus, whereas L2s and MIRs were overrepresented among the enhancers, L1s, Alus, and ERVL-MALRs were depleted (empirical P values <0.05, fig. 2A, see Materials and Methods). Therefore, certain transposon families appear to show a higher propensity toward exaptation into transposons.
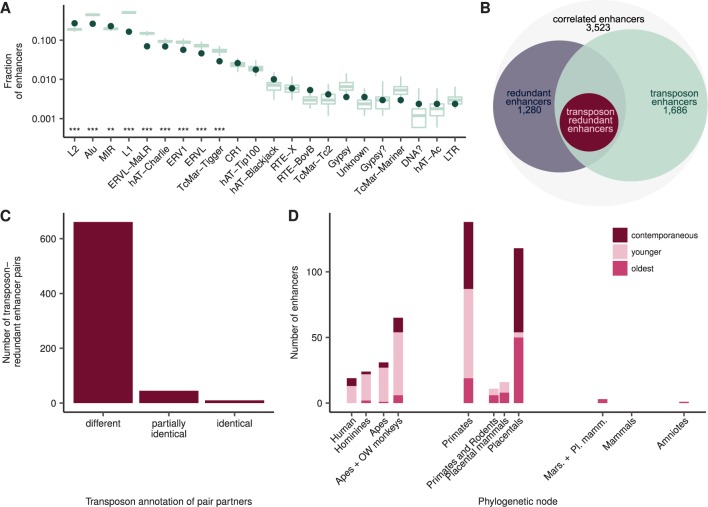
Fig. 2.
—Most partners of transposon-redundant enhancer pairs originated independently. (A) Transposon families contribute differently to the evolution of redundant enhancers. The green dots indicate the fraction of transposon enhancers annotated with each transposon family. The green boxplots depict the distribution of the fraction of random genomic sequences annotated with the same transposon family, estimated based on 1,000 random sets. Asterisks indicate FDR-corrected empirical P values. *P < 0.05, **P < 0.01, ***P < 0.001. (B) The scheme depicts the connection of the redundant enhancer, transposon enhancer, and transposon-redundant enhancer sets. The 3,523 enhancers that are significantly correlated to a promoter are referred to as “correlated enhancers.” About 1,686 correlated enhancers annotated as transposons and are referred to as “transposon enhancers.” About 1,280 correlated enhancers form one or more redundant enhancer pairs and are referred to as “redundant enhancers.” Finally, if both partners of a redundant enhancer pair are annotated as transposons, they are referred to as “transposon-redundant enhancers”; these pairs comprise a total of 432 enhancers. (C) In the majority of transposon-redundant enhancer pairs, the partners have different transposon annotation. Depicted are the number of pairs where partners have different, partially identical (i.e., some transposons are identical), or identical transposon species annotation. (D) Redundancy in human transposon-redundant enhancers seems to have originated throughout the evolution of placentals. Shown is the age of transposon-redundant enhancers, estimated based on the insertion times of the transposons with which they are annotated. Redundancy can arise by 1) the evolution of a new (“younger”) enhancer with regulatory activities that are (possibly partially) redundant to those of an already existing (“oldest”) enhancer or 2) by the simultaneous (“contemporaneous”) evolution of two or more enhancers with (possibly partially) regulatory activities.
The fraction of transposon enhancers did not differ significantly between redundant and nonredundant enhancers. Moreover, we observed no differences between redundant and nonredundant enhancers when comparing the fraction of enhancers annotated with a particular transposon family. Out of the 2,117 redundant enhancer pairs, 34% (716) were transposon-redundant enhancer pairs, in the sense both enhancer partners in these pairs were annotated as transposons. These transposon-redundant enhancer pairs involved 432 enhancers, to which we further refer as “transposon-redundant enhancers” (see table 1 and fig. 2B). The number of transposon-redundant enhancer pairs is consistent with the expectation based on the tissue-specific activities of our redundant enhancers and the rates of transposon exaptations into enhancers that have been reported in the past (Simonti et al. 2017). Remarkably, the partners of the vast majority (92%, 661) of transposon-redundant enhancer pairs had different transposon species annotation (see fig. 2C), indicating that they mostly derived from independent transposon insertions. In order to account for inaccuracies in the transposon annotation, we also compared the annotation of the enhancers on the family level of the transposon taxonomy (see Materials and Methods). Although the number of transposon-redundant enhancer pairs with different transposon annotation decreased (466, 65%), it was still the majority. Furthermore, increasing the stringency to annotate enhancers as transposon enhancers (see Materials and Methods) resulted in at most a moderate decrease in the fraction of transposon-redundant enhancer pairs with different transposon annotation, both on the species and family levels of the transposon annotation (see supplementary fig. S6, Supplementary Material online). These results suggest that in the majority of transposon-redundant enhancer pairs the two enhancers have independent origins.
Table 1.
Number of Enhancers, Enhancer Pairs, and Enhancer Groups in the Human Genome, for Multiple Sets
| Set | Number of Enhancers | Number of Pairs | Number of Groups | Coloring in Main Figures |
|---|---|---|---|---|
| Correlated enhancers | 3,523 | — | — | Gray |
| Redundant enhancers | 1,280 | 2,117 | 465 | Purple |
| Transposon enhancers | 1,686 | — | — | Green |
| Transposon-redundant enhancers | 432 | 716 | 157 | Red/raspberry |
note.—The 3,523 enhancers that are significantly correlated to a promoter are referred to as “correlated enhancers.” About 1,686 correlated enhancers overlap with transposons and are referred to as “transposon enhancers.” About 1,280 correlated enhancers form one or more redundant enhancer pairs and are referred to as “redundant enhancers.” Finally, if both partners of a redundant enhancer pair are annotated as transposons, they are referred to as “transposon-redundant enhancers”; these pairs comprise a total of 432 enhancers. Analogously to redundant enhancer groups, transposon-redundant enhancer pairs can also be arranged into transposon-redundant enhancer groups.
To obtain a more comprehensive picture of the origin of our transposon-redundant enhancers, we inferred the evolutionary age of the corresponding transposons using a maximum parsimony approach (see Materials and Methods). Specifically, we first searched for candidate orthologous sequences for the transposon-redundant enhancers in the genomes of 18 mammalian and vertebrate species using BLAST (see supplementary fig. S7, Supplementary Material online). Next, we required the candidate orthologs to be in the neighborhood of a target gene ortholog and to overlap with the same transposon types as the human enhancer. Thus, we found that 96% of our transposon-redundant enhancers had an ortholog among other primate species, 22% in rodents, 47% in the Laurasiatheria, and 1% in the opossum branch. In general, we observed the expected trend: The more distantly related a clade was to human, the fewer enhancers had an ortholog in that clade. The only exception was the rodent clade, which had relatively few orthologs despite its close relationship to human. This can be explained by the fact that rodents have a faster molecular clock than primates (Drost and Lee 1995; Bromham 2011), which leads to a faster loss of the transposon signatures (Glusman et al. 2001). Subsequent phylogenetic reconstruction (see Materials and Methods) showed that most of the transposon insertions in the transposon-redundant enhancers date back to the common ancestor of placentals (25%, 105.5 Ma) or primates (32%, 73.8 Ma). This finding is supported by the estimated activity periods of the corresponding transposons. Consistent with previous studies (Simonti et al. 2017), we inferred that the transposons in the transposon-redundant enhancers were older than the average transposon in the human genome (mean ages: 69.16 vs. 76.0 Ma; P value = 6.26×10−4, two-sided Wilcoxon rank-sum test, see supplementary fig. S8, Supplementary Material online). Indeed, it has been reported that this holds true even when correcting for the transposon type and hypothesized that the likelihood of a transposon being co-opted into an enhancer increases with its age in the genome (Simonti et al. 2017). Overall, the median age difference between the transposons of the partners of the transposon-redundant enhancer pairs was 31.7 Myr. Assuming that this is a reasonable estimate for the time elapsed between their co-option into enhancers, the introduction of redundancy in mammalian regulatory networks appears to have taken place over long periods. Interestingly, we found that among transposon-redundant enhancer pairs in which one or both of the partners derived from a transposon insertion in the common primate ancestor or its successors—but only among them—, the younger partners had a significantly higher SNP density than their older counterparts (medians 116 vs. 106 SNPs/kb, FDR-corrected P value = 3.6×10−5). That is, the younger partners of those pairs are systematically less constrained to evolve than the older partners. In addition, the regulatory networks of both the placental and the primate common ancestors seem to have been already highly redundant (see fig. 2D). In particular, we observed that 3% (4) of the enhancers derived from a transposon insertion in the common ancestor of placentals were redundant to others originating earlier, whereas 42% (50) represents cases in which redundancy possibly evolved in a contemporaneous manner. Hence, the transposon-derived regulatory network in the common ancestor of placentals apparently had a redundancy level of at least 45%. This is even more prominent in the primate ancestor: Although 49% (68) of the enhancers derived from a transposon insertion in the common primate ancestor were redundant to others originating earlier, 37% (51 enhancers) evolved contemporaneously. Together, these findings point to the introduction of redundancy through independent transposon insertions as a common event in mammalian evolution.
To investigate to which degree the introduction of regulatory redundancy in mammalian regulatory networks is driven by the acquisition of new genes, we estimated the age of the transposon-redundant enhancer target genes and compared it with that of the corresponding transposons (see Materials and Methods). The majority of the genes (2,473 out of 2,976 target genes, see supplementary fig. S9, Supplementary Material online) date back to the “bony vertebrates” or older clades and, thus, to a large extent, can be assumed to be older than their enhancers. This indicates that newly originating genes are not the main driver for the introduction of regulatory redundancy in our data. Only seven transposon-redundant enhancer pairs had gene targets that were younger than one of the partners in the pair, and among those, there were five—all belonging to the same redundant enhancer group—for which one of the target genes originated after the older enhancer, but before the younger one: DRICH1, which is broadly expressed but highly expressed in testis (Petryszak et al. 2016). Hence, with a few exceptions, the emergence and fixation of regulatory redundancy are not associated with the acquisition of new genes.
Regulatory Redundancy Is Mostly Lineage-Specific
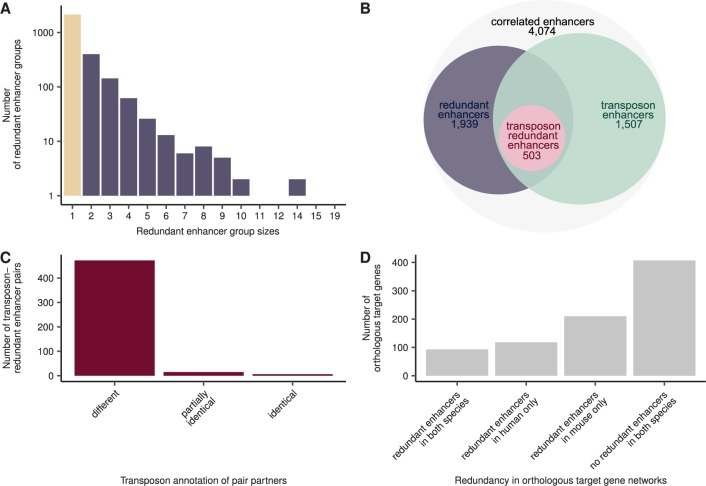
In order to establish to what extent our findings hold true for the genomes of other mammalian species, we conducted the same analyses in the mouse genome (see Materials and Methods). From originally 44,459 mouse FANTOM enhancers and 158,965 promoters, we identified 4,074 enhancers with a significant correlation coefficient to 8,082 promoters (see supplementary tables S4 and S5, Supplementary Material online). Among those, we distinguished 1,939 redundant enhancers, forming 2,787 redundant enhancer pairs (see fig. 3A and B; supplementary table S6, Supplementary Material online). In contrast to their human counterparts, mouse-redundant enhancers did not differ in their SNP density from nonredundant enhancers, but they had a slightly lower GC content (with medians of 0.50 and 0.49, respectively, P value = 3.4×10−6, two-sided Wilcoxon rank-sum test). Analogously to what we observed in the human genome, the target genes of redundant enhancers in the mouse genome were more tissue-specific (median Shannon entropy of 2.3 for redundant enhancer target genes and 2.5 for nonredundant enhancer target genes, P value = 1.1×10−3, two-sided Wilcoxon rank-sum test, see Materials and Methods and supplementary fig. S10, Supplementary Material online) and tended to be more strongly expressed (in 53% of the evaluated facets, with no difference in the remaining 47%, FDR-corrected P values ≤0.05, see Materials and Methods) than those of nonredundant enhancers. We further observed that mouse-redundant enhancers were, like human-redundant enhancers, overrepresented in thymus (FDR-corrected P value = 1.2×10−29, see Materials and Methods) and depleted in brain-related facets such as spinal cord (P value = 1.1×10−21), brain (P value = 7.8×10−15) and pituitary gland (P value = 2.8×10−10). Mouse-redundant enhancer pairs also showed a very high level of redundancy, sharing common activity in an average of 1.1 facets. Moreover, 1,961 pairs (70%) had 100% identical activity profiles. Taken together, these results indicate that mouse- and human-redundant enhancers fulfill similar functions, despite differences in their evolutionary divergence and genomic location.
Fig. 3.
—Redundant enhancers in the mouse genome show similar properties to those in the human genome, but redundancy in orthologous regulatory networks is mainly lineage-specific. (A) Redundant enhancers in the mouse genome form groups of different sizes, similar as in human. Depicted are the number of nonredundant enhancers (beige) and redundant-enhancer groups (purple). (B) The scheme depicts the connection of the redundant enhancer, transposon enhancer, and transposon-redundant enhancer sets. See figure 2B for details. (C) The enhancers of redundant enhancer pairs show the same transposon annotation pattern as in human. Depicted are the number of transposon-redundant enhancer pairs where partners have different, partially identical (some transposons are identical), or identical transposon species annotation. (D) A large fraction of target genes that are orthologous between human and mouse are regulated by redundant enhancers in just one of the two species. Shown are the common orthologous enhancer target genes between the human- and mouse-correlated enhancer data sets and their regulation by redundant enhancers in human and mouse.
Compared with human, a substantially smaller fraction of mouse enhancers were transposon enhancers (37%, log2-fold difference in observed transposon overlap compared with the random expectation = −0.77, see supplementary fig. S11, Supplementary Material online). This may be partially explained by the faster molecular clock of mouse (Bromham 2011) and the ensuing difficulty to recognize transposon-derived sequences as such (see supplementary fig. S12, Supplementary Material online). The five most prominent transposon families among mouse enhancers were the Alu (17% of transposons), B4 (16%), ERVL-MaLR (12%), L1 (11%), and ERVK (8%) families (see supplementary fig. S13, Supplementary Material online). Thus, although Alus were the most prominent family in both human and mouse, the ERVK family was much more frequent in mouse and MIRs and L2s were less frequent (7% and 4%, ranking in the seventh and eighth positions, respectively, after B2 with 7%) in mouse than they were in human. As observed for human, L2s and MIRs were overrepresented among the enhancers as compared with their frequencies in the genome, whereas Alus, L1s, ERVKs, and ERVL-MaLRs were depleted (empirical P values ≤0.05, see Materials and Methods). Finally, although the rodent-specific family B4 made up a large fraction of the mouse enhancers, its frequency matched the genomic expectation. Hence, many transposon families display the same trend toward exaptation into enhancers in both human and mouse. Further, we did not find any difference between the transposon families represented among redundant and nonredundant enhancers. Reflecting the lower number of transposon-derived enhancers in mouse, only 18% (493) of redundant enhancer pairs were transposon-redundant enhancer pairs, involving 503 transposon-redundant enhancers. Nevertheless, similarly to what we observed in the human genome, the majority of them had different transposon species (96%, see fig. 3C) and family (76%) annotations, providing further support for the hypothesis that transposon-redundant enhancers in mammalian genomes mainly originate by independent exaptation events. Interestingly, only few (17%) transposon-redundant enhancers in mouse had an ortholog in other mammalian species, mainly among primates and Laurasiatheria (15% and 13%, respectively); only 6% had an ortholog in rabbit, the only other member of the glires considered in the analysis. Consequently, 87% (438) of transposon-redundant enhancers—and, in general, a vast fraction of enhancers—appear to be mouse-specific. This can only be partly justified by the fact that most of the species included in the phylogenetic analysis were only distantly related to mouse (see Materials and Methods, supplementary fig. S7, Supplementary Material online) and is in line with rapid evolution of enhancers in the mammalian genome (Villar et al. 2015). In summary, the transposon-redundant enhancers in mouse are composed of other transposon families than in human, but still the vast majority (96%) of transposon-redundant enhancer pairs consists of partners that are apparently derived from different transposon species (76% on transposon family level)—and thus 17% (13% on transposon family level) of all redundant enhancer pairs in the mouse genome—appear to have evolved independently and in parallel.
Out of 870 enhancer target genes that were common in human and mouse (see Materials and Methods), only 96 (11%) were regulated by redundant enhancers in both species, whereas 354 (40%) were regulated by redundant enhancers in one species, but not in the other (see fig. 3D). The number of genes regulated by redundant enhancers in both species is small, but greater than expected by chance (odds-ratio = 1.4, P value = 0.02, one-sided Fisher’s exact test), which hints at a requirement of redundancy in the regulatory networks of both species. However, redundant enhancers appear to be mainly species-specific. Indeed, only 36 (1%) of human enhancers had an ortholog in the mouse genome (see Materials and Methods), and only 9 (2%) out the 556 human-redundant enhancers with orthologous target genes in the mouse genome were orthologous to the corresponding mouse-redundant enhancers. Conversely, only 11 (0.3%) mouse enhancers had an ortholog in the human genome, and only 4 (0.5%) out of the 880 mouse-redundant enhancers with orthologous target genes in the mouse genome had orthologs in the human genome. This indicates that the enhancers driving the expression of the genes that are regulated in a redundant manner in both human and mouse have evolved independently. Together, our results suggest that, with some exceptions, evolution of regulatory redundancy is a largely species-specific process among mammals, both concerning the target genes and the enhancers, and that orthologous regulatory networks with similar levels of redundancy are likely to result from convergent evolutionary processes.
Discussion
It has long been hypothesized that redundant enhancers originate by duplication (Hendrix et al. 2008). However, many enhancers in the mammalian genome are thought to have evolved by exaptation or co-option of transposons (Bejerano et al. 2006; Rebollo et al. 2012; Chuong et al. 2016). Emerging evidence suggests that this might also be the case for redundant enhancers (Franchini et al. 2011). Our study shows that a large number of redundant enhancers appear to have arisen from independent transposon exaptations.
Our analysis relies on an enhancer data set generated by the FANTOM5 consortium using CAGE. Thus, our results are conceivably limited to enhancers that produce bidirectional eRNA. It is still a matter of debate whether this is a property that every active enhancer shows. Certainly, it is likely that not all active enhancers have this property (Rahman et al. 2017). In any case, the features distinguishing these two alleged classes of enhancers remain unclear, and therefore, it is difficult to assess to what degree our findings can be extended to all the enhancers in the genome. The FANTOM5 consortium quantified enhancer activity at single base-pair resolution on a remarkably large number of tissue samples. The latter was important for our study, since we used a correlation-based strategy similar to the one employed by Andersson et al. (2014) to assign the enhancers to the promoters of their target genes.
Correlation-based approaches are often used to identify enhancer–promoter associations (Shen et al. 2012; Thurman et al. 2012; Sheffield et al. 2013; Andersson et al. 2014; Yao et al. 2015). They work on the premise that functionally linked enhancers and promoters tend to show common activity patterns. Despite being widely used, correlation-based approaches are affected by confounders that can result in false positives. For example, two enhancers may target two different genes but still exhibit exactly the same activity patterns; in that case, our correlation-based approach may incorrectly infer that the enhancers constitute a redundant enhancer pair. Yet, most (73%, 940) of our redundant enhancers were associated with a single target gene, suggesting that the cutoff set for was high enough to minimize such cases. Naturally, the number of (positively) correlated enhancers and promoters decreases with the increase of the cutoff value for , but even at , 18% (1,942) of the enhancer–promoter associations remain. To further assess the significance of our correlations, we compared the value of obtained for each enhancer–promoter pair within a TAD to those computed between the same enhancer and all promoters in 20 neighboring TADs (see supplementary materials and methods, Supplementary Material online). As enhancers and promoters in different TADs are unlikely to interact (Ron et al. 2017), the corresponding correlations constitute an appropriate null model to generate an empirical P value. In this manner, we found 4,895 pairs of (positively) correlated enhancers and promoters (> 0.5, empirical P value ≤0.0025, FDR-corrected P value < 0.05). Of these pairs, 4,887 were among our original pairs of correlated enhancers and promoters. Hence, a substantial fraction of our enhancer–promoter associations fulfill more stringent requirements. Although false negatives are generally of less concern than false positives, our correlation-based approach also led to false-negative associations: Only 3,523 (out of 10,445 tested) enhancers were correlated with one or more promoters in the human genome, effectively limiting the analysis to the regulatory networks of 2,976 genes. In any event, a correlation merely indicates an association and not a functional relationship. Time and resource expensive functional assays such as reporter gene assays are required to validate or reject the associations (Shlyueva et al. 2014). There is currently no large database of experimentally validated enhancer–promoter interactions. For example, GeneHancer (Fishilevich et al. 2017) contains 63 experimentally validated enhancer–promoter interactions, involving 63 enhancers and 59 genes. Of those enhancers, 16 are represented in the FANTOM data set, making it virtually impossible to draw any meaningful conclusion. An alternative to functional assays could be chromatin conformation capture assays, such as Hi-C. Hi-C is a proximity ligation method to identify genome-wide chromatin–chromatin interactions. Spatial proximity between distal genomic sequences does not imply functional interactions between them, and the technology still suffers from relatively low resolution and high false-positive rates (Yardımcı et al. 2019). Moreover, there are simply no available Hi-C data sets for many of the tissues in our facets. Nevertheless, because we acknowledge their value, our approach does take Hi-C data into account: As interactions between promoters and enhancers do not normally cross TAD boundaries (Ron et al. 2017), we only tested enhancer–promoter pairs where both partners were located within the same TAD for correlation, as opposed to testing within windows of a fixed size. TAD organization is known to be largely conserved across tissues and even mammalian species (Dixon et al. 2012; Nora et al. 2012; Rao et al. 2014; Battulin et al. 2015). Indeed, the TADs maps computed for different tissues are highly similar (Li et al. 2019). Although a more stringent strategy would require the use of facet-specific TAD maps, such data are not available; furthermore, given the overall stability of the TADs, it is not expected to be a major error source.
In the human genome, 2,117 pairs of enhancers were associated with the promoters of the exact same target transcripts and exhibited common activity in at least one facet. We required redundant enhancers to be associated with the exact same set of target transcripts as opposed to only at least one of them. This led to a slightly smaller number of redundant enhancer pairs than when pairing all enhancers with at least one common target transcript but makes the pairing more conservative. Although our redundant enhancer pairs are of a putative nature, they rely on data that have been extensively validated over the past 5 years. In particular, the enhancer predictions in the FANTOM5 data set have been: 1) shown to overlap by at least 71% with predictions based on DNase-I-Hypersensitivity, H3K27ac and H3K4me1 histone modifications (Andersson et al. 2014); 2) successfully tested in zebrafish reporter gene assays (Andersson et al. 2014); and 3) scrutinized to demonstrate other enhancer attributes (Denisenko et al. 2017, 2019). Consistently, based on ChIP-seq data from the ENCODE project, the enhancer partners of most (67%) redundant enhancer pairs were bound by the same TF (see Materials and Methods). As only nine facets could be mapped to tissues assayed by ENCODE, this analysis involved only 202 redundant enhancer pairs and should be interpreted cautiously. Nevertheless, even for individual facets such as liver, for which a relatively large number of TFs had been assayed, the enhancer partners of 80% (47 out of 59) of the redundant enhancer pairs were bound by at least one common TF; 76% (45) of those were bound by HNF4G and/or NR2F2, which are nonubiquitously expressed TFs and important for liver expression (Fagerberg et al. 2014). ChIP-seq cannot demonstrate by itself the functional significance of the TFs found to be bound to the enhancers. Still, the results 1) provide additional support for both enhancer partners being truly redundant in the considered facets and 2) point toward enhancer partners sharing a common regulatory logic, similar to the shadow enhancers described in Drosophila (Hendrix et al. 2008).
Our results indicate that redundant enhancers are common features of regulatory networks not only in Drosophila (Cannavò et al. 2016) but also in mammals. In contrast to some of the previous studies (Franchini et al. 2011; Cannavò et al. 2016; Osterwalder et al. 2018), we found that the majority of our redundant enhancers were completely redundant in terms of their activities. However, this result depends on the number of tissue samples available. Although the number of tissues that was involved in the current analysis is relatively large, our redundant enhancers may still diverge in their activity in other tissues or under different conditions. In addition, some of our facets comprise several distinct cell types, which could lead to an overestimation of the number of (completely) redundant enhancers. Further, it has been hypothesized before that the partners of highly redundant enhancer pairs might be subjected to different selective pressures, with one enhancer being less conserved than the other and free to evolve new—ultimately, nonredundant—functions. Although this is supported by the slightly higher SNP densities that we noted for relatively young enhancer partners, it is not consistent with the fact that redundant enhancers were indistinguishable from nonredundant enhancers in terms of sequence conservation. This has also been observed in Drosophila (Cannavò et al. 2016). On the contrary, our data suggest that regulatory redundancy provides a selective advantage that would contribute to the fixation of redundant enhancers. Indeed, in agreement with previous findings (Perry et al. 2010; Alberga et al. 2011; Lam et al. 2015), we show that redundant enhancers are associated with more precise (i.e., lower Shannon entropy) and stronger target gene expression.
Approximately 50% of the human (and 40% of the mouse) redundant enhancers were annotated as transposons and, as such, represent possible cases of transposon exaptation. Although we only required 50 bp of the enhancer sequence (10% of the sequence for most enhancers) to overlap with a transposon to consider it transposon-derived, the vast majority of the enhancers displayed a substantially larger overlap (see supplementary fig. S14, Supplementary Material online). This indicates that the threshold is high enough to identify a permissive set of transposon-derived enhancers. In agreement with observations for nonredundant enhancers (Chuong et al. 2017; Simonti et al. 2017), our redundant enhancers were generally depleted of transposons compared with the entire genome. This does not imply that the number of enhancers derived from transposons is small; it is just smaller than expected from the fraction of the total genome that is derived from transposons. Several factors could account for this. Specifically, given the low-sequence conservation of many enhancers, a fair number of enhancers might be derived from ancient transposons for which the transposon signature is not visible anymore. Furthermore, it appears plausible that only some transposon families are well suited to be co-opted into enhancers. Certainly, our enhancers were only enriched among particular transposon families. In addition, transposons carry TFBS that lead to activity in certain tissues and, therefore, the different enrichment levels could be partially explained by the tissue specificities of the enhancers represented in the data set. The number of redundant enhancer pairs active in a given tissue varied greatly, as did the total number of enhancers active in that tissue. Accordingly, some of our findings may reflect the tissue-specific activities of the redundant enhancers in our data set. Interestingly, the vast majority of our transposon-redundant enhancer pairs in both human and mouse comprised enhancers that were most presumably derived from different transposon species, and are, thus, likely to have evolved from independent transposon exaptation events. Increasing the stringency in the identification of enhancers annotated as transposons led to only a mild decrease in the number of transposon-redundant enhancer pairs involving different transposon species, if any. Moreover, we observed hardly any changes when restricting the analysis to redundant enhancer pairs for which the enhancer–promoter associations had been computed using a cutoff for of 1 (see supplementary fig. S15, Supplementary Material online) or based on the empirical P value described earlier (see supplementary fig. S16, Supplementary Material online).
Our phylogenetic analysis suggests that most transposon insertions associated with the origination of human-redundant enhancers took place in the primate or placental ancestors, and that in at least 77% of transposon-redundant enhancer pairs, the partners originated several tens of millions of years after one another. In the remaining 23% of the cases, the partners of the pairs might have evolved in a contemporaneous manner. However, it must be noted that the primate and placental common ancestor nodes in our phylogenetic tree cover relatively large periods. Consequently, what we consider contemporaneous, could actually be several tens of million years apart. This cannot be completely circumvented because there are no recent species that would allow to estimate the ancestral state for certain periods. Furthermore, this analysis disregards redundancy introduced from nontransposon-redundant enhancers. As some nontransposon enhancers likely represent ancient transposon-derived enhancers, redundancy is likely to have been first introduced earlier than the estimated date. The mouse transposon-redundant enhancers were mostly mouse-specific and, thus, substantially younger than those in the human genome. This is expected due to the faster molecular clock of mouse (Drost and Lee 1995; Glusman et al. 2001; Bromham 2011), but is also impacted by the degree of relatedness of the species included in our phylogenetic analysis. On top of that, the quality of the genome assembly of the most closely related species (rabbit) is not as high as that for the assemblies of the primate species, and we may have failed to detect some orthologs in rabbit due to gaps in its assembly. In any case, even if the transposon insertions had been contemporaneous, there could be millions of years between the transposon insertions and their exaptation into enhancers. Finally, many of the redundant enhancers that were not annotated as transposons and, hence, disregarded in these calculations, could actually be ancient transposon-derived sequences for which the transposon signature is not traceable anymore. Our results therefore presumably underestimate the extent to which independent transposon exaptation has contributed to redundant enhancer origination.
We found hardly any orthologs among human- and mouse-redundant enhancers, which is consistent with rapid enhancer turnover (Kunarso et al. 2010; Vierstra et al. 2014; Lynch et al. 2015; Villar et al. 2015; Trizzino et al. 2018). Still, we observed that among orthologous target genes, the number of genes with redundant enhancers in both species was larger than expected by chance. As the corresponding redundant enhancer pairs were not orthologous, the redundancy must have developed independently. Hence, certain regulatory networks apparently have a higher propensity—or at least a higher tolerance—toward regulatory redundancy. Nevertheless, most orthologous regulatory networks showed differential redundancy, in the sense not all the genes that were redundantly regulated in one species were redundantly regulated in the other. Moreover, in the cases in which they were, the regulation was actualized by nonorthologous cis-regulatory elements. In the light of the enhancer turnover model and the low-sequence conservation of many known enhancers (Villar et al. 2015), regulatory redundancy in some of these networks may be merely temporal and not exerting an essential function. Yet, the clear association of redundant enhancers with gene expression specificity and strength suggests that regulatory redundancy per se is an important property of regulatory networks.
In summary, the redundant partners in 92% of all transposon-redundant enhancer pairs—31% of all redundant enhancer pairs—in the human genome have been acquired independently, in successive waves of transposon expansions. The mouse genome displays comparable numbers (95% of mouse transposon-redundant enhancer pairs—17% of all redundant enhancer pairs—have independent origins). Considering that many enhancers without a transposon signature are likely to derive from ancient transposons, transposon exaptation may even be the predominant mechanism of redundant enhancer origination. In addition, despite redundant enhancers being a common feature of many mammalian regulatory networks, redundant enhancers are poorly conserved and mostly lineage-specific, with hardly any orthologous redundant enhancers between different lineages. Hence, most regulatory redundancy in mammalian networks appears to have developed independently by convergent evolution. In any event, given its widespread distribution, understanding the evolutionary processes by which regulatory redundancy arises is key to understanding how networks respond to perturbations, in particular, those associated with disease.
Supplementary Material
Literature Cited
- Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M.. 2011. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development 111:983–992. [DOI] [PubMed] [Google Scholar]
- Allan CM, Walker D, Taylor JM.. 1995. Evolutionary duplication of a hepatic control region in the human apolipoprotein E gene locus. Identification of a second region that confers high level and liver-specific expression of the human apolipoprotein E gene in transgenic mice. J Biol Chem. 270(44):26278–26281. [DOI] [PubMed] [Google Scholar]
- Andersson R, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507(7493):455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S. 2011. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 72:181–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battulin N, et al. 2015. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 16(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauparlant CJ, Lemaçon A, Deschenes L-L, Droit A.. 2015. ENCODExplorer: a compilation of ENCODE metadata. R package version 2.4.0.
- Bebin A-G, et al. 2010. In vivo redundant function of the 3’ IgH regulatory element HS3b in the mouse. J Immunol. 184:3710–3717. [DOI] [PubMed] [Google Scholar]
- Bejerano G, et al. 2006. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441(7089):87–90. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 57:289–300. [Google Scholar]
- Bromham L. 2011. The genome as a life-history character: why rate of molecular evolution varies between mammal species. Philos Trans R Soc B. 366(1577):2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavò E, et al. 2016. Shadow enhancers are pervasive features of developmental regulatory networks. Curr Biol. 26(1):38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. 2010. Ensembl variation resources. BMC Genomics 11(1):293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C.. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351(6277):1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C.. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 18(2):71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko E, et al. 2017. Genome-wide profiling of transcribed enhancers during macrophage activation. Epigenet Chromatin. 10(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko E, et al. 2019. Transcriptionally induced enhancers in the macrophage immune response to Mycobacterium tuberculosis infection. BMC Genomics 20(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, et al. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485(7398):376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost JB, Lee WR.. 1995. Biological basis of germline mutation: comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ Mol Mutagen. 25(S2):48–64. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 13(2):397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold E, et al. 2004. The ENCODE (ENCyclopedia Of DNA Elements) project. Science 306:636–640. [DOI] [PubMed] [Google Scholar]
- Ferreira LMR, et al. 2016. A distant trophoblast-specific enhancer controls HLA-G expression at the maternal–fetal interface. Proc Natl Acad Sci U S A. 113(19):5364–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 9(5):397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich S, et al. 2017. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford):bax028. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T. 2005. The evolutionary genetics of canalization. Q Rev Biol. 80(3):287–316. [DOI] [PubMed] [Google Scholar]
- Forrest ARR, et al. 2014. A promoter-level mammalian expression atlas. Nature 507(7493):462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini LF, et al. 2011. Convergent evolution of two mammalian neuronal enhancers by sequential exaptation of unrelated retroposons. Proc Natl Acad Sci U S A. 108(37):15270–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, et al. 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466(7305):490–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiasvand NM, et al. 2011. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat Neurosci. 14(5):578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, et al. 2001. Comparative genomics of the human and mouse T cell receptor loci. Immunity 15(3):337–349. [DOI] [PubMed] [Google Scholar]
- Guerrero L, Marco-Ferreres R, Serrano AL, Arredondo JJ, Cervera M.. 2010. Secondary enhancers synergise with primary enhancers to guarantee fine-tuned muscle gene expression. Dev Biol. 337(1):16–28. [DOI] [PubMed] [Google Scholar]
- Harrow J, et al. 2006. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 7(Suppl 1):S4.1–S4.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Levine MS, Hong JW.. 2008. Shadow enhancers as a source of evolutionary novelty. Science 321:1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34(90001):D590–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- Jacques P-É, Jeyakani J, Bourque G.. 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 9(5):e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ.. 2006. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133(4):761–772. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC.. 1975. Evolution at two levels in humans and chimpanzees. Science 188(4184):107–116. [DOI] [PubMed] [Google Scholar]
- Kunarso G, et al. 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 42(7):631–634. [DOI] [PubMed] [Google Scholar]
- Lam DD, et al. 2015. Partially redundant enhancers cooperatively maintain mammalian Pomc expression above a critical functional threshold. PLoS Genet. 11(2):e1004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky JA, Innis JW.. 2008. BAC transgenic analysis reveals enhancers sufficient for Hoxa13 and neighborhood gene expression in mouse embryonic distal limbs and genital bud. Evol Dev. 10(4):421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Barth NKH, Pilarsky C, Taher L.. 2019. Cancer is associated with alterations in the three-dimensional organization of the genome. Cancers (Basel) 11(12):1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J.. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167(5):1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, et al. 2015. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep. 10(4):551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majunder A, Biswas B.. 2006. Biology of inositols and phosphoinositides: subcellular biochemistry. Boston (MA): Springer. [Google Scholar]
- Nishihara H, et al. 2016. Coordinately co-opted multiple transposable elements constitute an enhancer for wnt5a expression in the mammalian secondary palate. PLoS Genet. 12(10):e1006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485(7398):381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, et al. 2018. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554(7691):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Cook PR.. 2013. Transcription factories: genome organization and gene regulation. Chem Rev. 113(11):8683–8705. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290. [DOI] [PubMed] [Google Scholar]
- Pearson WR. 2013. Selecting the right similarity-scoring matrix. Curr Protoc Bioinformatics. 43:3.5.1–3.5.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M.. 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 20(17):1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, et al. 2016. Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 44(D1):D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN, Vandewege MW, Ray DA.. 2018. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 26(1–2):25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Cadena P, Sternberg PW.. 2016. Enhancer sharing promotes neighborhoods of transcriptional regulation across eukaryotes. G3 (Bethesda) 6:4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, et al. 2017. Single-cell profiling reveals that eRNA accumulation at enhancer–promoter loops is not required to sustain transcription. Nucleic Acids Res. 45(6):3017–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Farivar S, Mager DL.. 2012. C-GATE – catalogue of genes affected by transposable elements. Mob DNA. 3(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3:217–223. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2009. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron G, Globerson Y, Moran D, Kaplan T.. 2017. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat Commun. 8(1):2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo AM, et al. 2007. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 3(10):e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, et al. 2008. Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci U S A. 105(11):4220–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27(4):592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, et al. 2005. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6(4):R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, et al. 2013. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 23(5):777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488(7409):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A.. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 15(4):272–286. [DOI] [PubMed] [Google Scholar]
- Siepel A. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonti CN, Pavličev M, Capra JA.. 2017. Transposable element exaptation into regulatory regions is rare, influenced by evolutionary age, and subject to pleiotropic constraints. Mol Biol Evol. 34(11):2856–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, et al. 2008. A novel mode of enhancer evolution: the Tal1 stem cell enhancer recruited a MIR element to specifically boost its activity. Genome Res. 18(9):1422–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong E.. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 13(9):613–626. [DOI] [PubMed] [Google Scholar]
- Su M, Han D, Boyd-Kirkup J, Yu X, Han J.. 2014. Evolution of Alu elements toward enhancers. Cell Rep. 7(2):376–385. [DOI] [PubMed] [Google Scholar]
- Thurman RE, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489(7414):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Kapusta A, Brown CD.. 2018. Transposable elements generate regulatory novelty in a tissue-specific fashion. BMC Genomics 19(1):468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arensbergen J, van Steensel B, Bussemaker HJ.. 2014. In search of the determinants of enhancer–promoter interaction specificity. Trends Cell Biol. 24(11):695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346(6212):1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, et al. 2008. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19(2):327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160(3):554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Berman BP, Farnham PJ.. 2015. Demystifying the secret mission of enhancers: linking distal regulatory elements to target genes. Cox Crit Rev Biochem Mol Biol. 50:1549–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardımcı GG, et al. 2019. Measuring the reproducibility and quality of Hi-C data. Genome Biol. 20(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.