Abstract
In the related research article, entitled “Identification of novel triazole-based nicotinamide phosphoribosyltransferase (NAMPT) inhibitors endowed with antiproliferative and antiinflammatory activity” [1], we reported the in vitro hepatic metabolism data for compounds 30c, 48b, and 31b (here named as E5, A6, and T1), in comparison with the reference compounds GPP78 and FK866 [1–3]. In this article, we retrieved the available data about the hepatic microsomal stability and metabolites structural characterization of the entire library of triazole-based NAMPT inhibitors, also implementing the given information with data regarding aqueous solubility and CYP inhibition. Compounds are divided in subclasses based on the hydrolytic resistant groups replacing the amide function of GPP78 [1, 2].
Keywords: Triazole-based NAMPT inhibitors, In vitro metabolism, Aqueous solubility, LC-MS
Specifications Table
| Subject area | Drug Discovery |
| More specific subject area | ADME investigation in Drug Discovery |
| Type of data | Table, text file, product ion MS spectra |
| How data was acquired | Liquid chromatography coupled to UV–Vis and mass spectrometry, UV–Vis spectrophotometry |
| Data format | Raw and analyzed |
| Experimental factors | In vitro microsomal incubations followed by samples protein precipitation before analysis. |
| Experimental features | Metabolic stability and metabolite profiling data have been determined by incubations in rat and human liver microsomes supplied with NADPH-regenerating system. Samples analysis have been performed by LC-UV and LC-ESI-MS equipment. Compounds have been ranked in terms of kinetic aqueous solubility by using the MultiScreen® Solubility protocol provide by Merck. For three selected compounds, identified in the related research article as potent inhibitor of NAMPT, CYP inhibitory activity has been determined by means of the aminopyrine N-demethylase assay. |
| Data source location | Department of Pharmaceutical Sciences, University of Piemonte Orientale, Largo Donegani 2, 28100 Novara, Italy |
| Data accessibility | Data are presented in this article |
| Related research article |
Travelli C. et al.; Identification of Novel Triazole-Based Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors Endowed with Antiproliferative and Antiinflammatory Activity. J. Med. Chem. 2017, 60, 1768–1792https://doi.org/10.1021/acs.jmedchem.6b01392 |
Value of the Data
|
1. Data
For our novel class of triazole-based nicotinamide phosphoribosyltransferase (NAMPT) inhibitors, data on metabolic stability and metabolite profiling in rat and human liver microsomes are presented. Besides, compounds are ranked in terms of kinetic aqueous solubility. For three selected compounds (A6, E5, T1), identified in the related research article as potent inhibitors of NAMPT, CYP inhibitory activity has been determined.
1.1. In vitro metabolic stability and metabolite structural characterization
Table 1 lists the chemical structures and the in vitro hepatic metabolism data collected for a panel of triazole-based NAMPT inhibitors previously tested in terms of cytotoxicity (SH-SY5Y cell line) and inhibitory activity on NAMPT [[1], [2], [3]]. Compounds are presented in classes based on the modification of the tail group and classified as follows: A – inverse amides (A1-A6), E – ethers (E1-E7), C – carbamates (C1–C3), U – ureas (U1–U3), S – sulfonamides (S1–S3), others (the amine N1 and the bis-triazole T1). Examples of the LC-ESI-MSn data acquired alongside with the chemical structures of the revealed metabolites are reported in the supplementary material S1, S2.
-
•
A - Inverse amides
Table 1.
In vitro hepatic metabolism data for a panel of triazole-based NAMPT inhibitors.
| Compound | Structure | [M+H]+ | Metabolic stabilitya | Revealed metabolic pathwaysb | |||||
|---|---|---|---|---|---|---|---|---|---|
| RLM | HLM | Pyridine N-oxidation |
Aromatic oxidation |
Aliphatic oxidation | Hydrolysis | Other | |||
| A1 |  |
441 | 51 | 72 | ● | ◌ | ● | ◌ | |
| A2 | 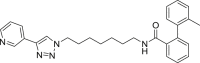 |
454 | 30 | 84 | ● | ◌ | ● | ◌ | |
| A3 | 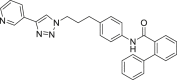 |
460 | 21 | 78 | ● | ● | ● | ◌ | |
| A4 | 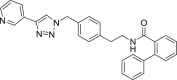 |
460 | 46 | 91 | ● | ● | ● | ◌ | |
| A5 |  |
426 | 62 | 92 | ● | ● | ● | ◌ | |
| A6 |  |
440 | 45 | 88 | ● | ● | ● | ◌ | Oxidative N-dealkylation |
| E1 | 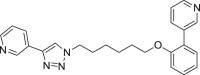 |
400 | 23 | 60 | ● | ● | ◌ | ◌ | |
| E2 |  |
413 | 8 | 32 | ◌ | ● | ● | ◌ | |
| E3 |  |
433 | 70 | 94 | ● | ● | ● | ◌ | |
| E4 | 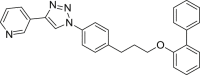 |
433 | 77 | >99 | ◌ | ● | ● | ◌ | |
| E5 | 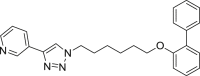 |
399 | 31 | 49 | ● | ● | ● | ◌ | |
| E6 | 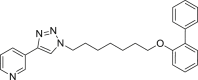 |
413 | 14 | 61 | ● | ● | ● | ◌ | |
| E7 |  |
427 | 16 | 84 | ● | ● | ● | ◌ | |
| C1 |  |
442 | 66 (61)c | 67 (87)c | ● | ● | ● | ● | |
| C2 |  |
476 | 69 (92)c | 90 | ● | ● | ◌ | ● | |
| C3 |  |
456 | 60 (89)c | 86 (92)c | ● | ● | ● | ● | |
| U1 |  |
441 | 79 | 95 | ● | ● | ● | ◌ | |
| U2 |  |
455 | 60 | 85 | ● | ● | ● | ◌ | |
| U3 | 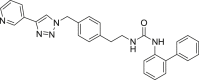 |
475 | 95 | 96 | ● | ● | ● | ◌ | |
| S1 | 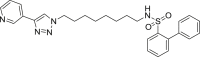 |
490 | 62 | 78 | ● | ● | ● | ◌ | Oxidative N-dealkylation |
| S2 | 499 | 45 | 80 | ● | ◌ | ● | ◌ | Oxidative N-dealkylation |
|
| S3 | 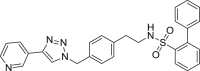 |
496 | 76 | 84 | ● | ◌ | ● | ◌ | Oxidative N-dealkylation |
| N1 |  |
446 | 38 | 74 | ● | ◌ | ● | ◌ | Oxidative N-dealkylation |
| T1 |  |
464 | 47 | 87 | ● | ● | ● | ◌ | |
The metabolic stability was assessed by incubating each compound at 50 μM concentration with rat (RLM) or human (HLM) liver microsomes in the presence or absence of NADPH-regenerating system [4]. Residual substrate percentage was evaluated by LC-UV analysis measuring the remaining substrate after incubation.
For structural characterization of the metabolites, all samples were analyzed by LC-ESI-MSn.
Residual substrate in incubations without the presence of NADPH-regenerating system.
Compounds A1-A6 showed a variable substrate depletion, ranging the residual substrate from 21% (A3) to 62% (A5) in rat liver microsomes (RLM) incubations, and from 72% (A1) to 92% (A5) in human liver microsomes (HLM). A5 resulted slightly more stable in both species respect to A6 suggesting that the C6 aliphatic linker might enhance the microsomal stability. Compounds A3 and A4 bear a benzene ring embedded in the aliphatic linker. While A4 underwent a similar depletion compared to A6, A3 resulted less stable toward microsomal oxidation (only being the residual substrate 21% in RLM and 78% in HLM) thus suggesting that the insertion of a benzene ring adjacent to the amide function might increase the susceptibility to metabolism. Both the presence of a biphenyl pyridine (A1) or a methyl substituent on the biphenyl moiety (A2) could represent a metabolic soft spot. Although the presence of an additional site, theoretically prone to undergo microsomal N-oxidation, the metabolic stability of A1 did not substantially change in comparison to A6. Conversely, for A2, beside the abundant pyridine N-oxide (A2-m3) and the aliphatic hydroxylated (A2-m1) metabolites, LC-ESI-MSn analysis revealed the presence of an additional intense ion (m/z 470) related to the biphenyl hydroxymethyl metabolite A2-m2, whose fragmentation gave rise to an intense fragment ion at m/z 452 attributable to the loss of water (see supplementary material S2 for mass spectral data and metabolites structure).
In our previous investigation (where the presence of the pyridine N-oxide metabolite for compounds A6 and E5 was confirmed synthetizing the reference compounds) [1], and in this work, we noticed that for all the compounds the chromatographic peaks related to the pyridine N-oxide metabolites eluted at a retention time close to that of the parent compounds.
-
•
E - Ethers
Among the seven inhibitors of NAMPT bearing the ether function, E5 showed a quite low metabolic stability being the residual substrate 31% and 49% after RLM or HLM incubation respectively. Lengthening the aliphatic linker of one (E6) or two (E7) carbon atoms, the corresponding compounds resulted less stable in RLM (14–16% residual substrate) than in HLM (61–84%) thus suggesting the occurrence of interspecies differences. The introduction of a benzene ring in the aliphatic linker (compounds E3 and E4), increased the metabolic stability both in RLM and HLM incubations. Similarly to the amide A2, the introduction of a methyl substituent on the biphenyl moiety (E2) strongly reduced its metabolic stability, especially in RLM being 8% the residual substrate. We reported in supplementary material S2 the spectral data for compound E3.
-
•
C - Carbamates
Carbamates C1–C3 showed an overall good stability. Also, for carbamates, lengthening the hydrocarbon chain of one unit, (compound C3) did not change the metabolic stability in RLM but increased it in HLM incubation with respect to C1. LC-ESI-MSn analysis of carbamate derivatives demonstrated that the involved metabolic pathways were: pyridine N-oxidation, aromatic and aliphatic hydroxylation, and the additional hydrolysis of the carbamate function. Indeed, when C1–C3 were incubated in liver microsomes without activating the mixed-function oxidase system by NADPH, all the compounds experienced a small substrate depletion due to the formation of the hydrolytic metabolites. However, the carbamate hydrolysis only occurred in a limited extent, especially in HLM where the residual substrate after incubation in the absence of NADPH ranges from 87 to 95% (Table 1). See supplementary material S2 for C3 mass spectral data.
-
•
U - Ureas
Overall, compounds U1 and U3 resulted more stable than the corresponding carbamates toward microsomal metabolism. Once again, the presence a C7 aliphatic linker (U2) promoted the microsomal oxidation with respect to U1, whereas the metabolic stability is increased by the insertion of a benzene ring in the paraffinic linker being the compound U3 metabolized very little. Spectral data for compound U1 are reported in supplementary material S2. Ureidic compounds underwent aromatic and aliphatic hydroxylation, whereas no hydrolytic products have been detected.
-
•
S - Sulfonamides
Sulfonamides S1-3 showed similar metabolic stability ranging the residual substrate from 60% to 76% and 78%–84% respectively in RLM or HLM incubations. We reported in supplementary material S2 the mass spectral data for S3. Beside to pyridine N-oxidation and carbon hydroxylation, oxidative N-dealkylation (metabolite S3-m1) occurred as additional metabolic pathway.
-
•
Others
Amine N1 metabolism involved the following pathways: pyridine N-oxidation, hydrocarbon tail hydroxylation and, similarly to sulfonamides, oxidative N-dealkylation. LC-ESI-MSn data for N1 were reported in supplementary material S2 whereas metabolic data of T1 were already provided in the related research article [1].
1.2. Apparent kinetic aqueous solubility data
As summarized in Fig. 1, only the compounds A1 and A5 gave kinetic solubility values similar to those of both the target concentrations (200 and 500 μM), demonstrating their aqueous solubility in this range. Differently, since their minor solubility, both FK866 and the other triazole-based NAMPT inhibitors, gave solubility values in the 3.13–200 μM concentration range (see experimental section). Moreover, since the impossibility to obtain a consistent calibration curve even in the range 3.13–200 μM, for compounds E6, E7, E4 and E3 the apparent solubility was not determined indicating their very limited aqueous solubility.
Fig. 1.
Kinetic aqueous solubility data.
Inverse amides with a paraffinic linker (e.g. A5, A6) showed approximately a two/three-fold increase of aqueous solubility than compounds with a benzene ring embedded in the linker (A3, A4). A significant increase of aqueous solubility was observed both by replacing the biphenyl moiety with biphenylpyridine (A1 vs A6) or shortening to six carbon atoms the paraffinic linker (A5 vs A6). Overall, despite their lower intrinsic aqueous solubility, esters, carbamates, ureas, and sulfonamides, followed the same trend. The double triazole T1 (considered the isostere of amide A6) showed a reduced kinetic solubility (67 μM) with respect to its counterpart A6 (201 μM). This confirmed our previous findings about the negative contribution of the triazole nucleus to the aqueous solubility [4].
1.3. CYP inhibition
In Fig. 2, we reported the CYP inhibition data for our most active NAMPT inhibitors, namely: A6, E5, and T1 compared to FK866 [1] evaluated as their potential to inhibit the aminopyrine N-demethylase (AND) activity over a concentration range of 1–100 μM [5].
Fig. 2.
Inhibition of aminopyrine N-demethylase activity (nmol/min/mg ± S.E.) in rat liver microsomes.
FK866 showed a mild AND inhibitory activity only at 100 μM concentration while, the tested NAMPT inhibitors, have been demonstrated to be more potent CYP inhibitors yet at 1–10 μM concentrations. Inverse amide A6 showed the greatest AND inhibitory activity compared to ether E5 and bis-triazole T1; however, A6 approximatively exhibited double aqueous solubility compared to E5 and T1 (Fig. 2) possibly reflecting enhanced AND inhibitory effect compared to the other compounds. Moreover, with the aim of maximizing the solubility of the tested compounds in the assay media so reducing differences in activity due to their insolubility, we added to the incubation mixtures both a cosolvent (acetonitrile) and a surfactant (Tween 80) (see experimental section).
2. Experimental design, materials, and methods
2.1. Metabolic stability in rat and human liver microsomes
In vitro metabolic stability was evaluated using the same procedure and materials reported in the related research article [1].
Briefly, the standard incubation (0.5 ml final volume), was carried out in a TRIS-HCl buffer (50 mM, pH 7.4) containing 3.3 mM MgCl2, 1.3 mM β-NADPNa2, 3.3 mM glucose 6-phosphate, 0.4 Units/ml glucose 6-phosphate dehydrogenase, 5 μl of acetonitrile (1% of total volume), and the substrate (final concentration of 50 μM). After pre-equilibration of the mixture, an appropriate volume of rat (RLM) or human (HLM) microsomal suspension was added to give a final protein concentration of 1.0 mg/ml. The mixture was shaken for 60 minutes at 37 °C. Each incubation was stopped by the addition of 0.5 ml of ice-cold acetonitrile, vortexed and centrifuged at 12,500 rpm for 10 min. The supernatants were analyzed by LC-UV to evaluate the metabolic stability, which was expressed as % residual substrate (mean of two replicates) with respect to control incubation without microsomes. Substrate depletion below 5% was considered not significative. Metabolite structural characterization was performed by LC-ESI-MS analysis. The chromatographic conditions are reported in the supplementary material S1.
2.2. Kinetic aqueous solubility
The apparent aqueous solubility was determined by a filtration-based assay previously described [5] and using MultiScreen® Solubility filter plates (Merck) [6].
Calibration protocol: Five calibrators (500, 200, 50, 12.5, and 3.13 μM) for each compound were made by dilution of a 10 mM DMSO stock solution in an 80:20 PBS/acetonitrile mixture maintaining the level of DMSO at 5%. The plate was covered and then gently shaken at room temperature for 30 min, centrifuged at 2800 rpm (1600 g) for 10 min, and the absorbance of 200 μl per well aliquots were measured at 260 and 280 nm using a multiplate reader (Victor3V, PerkinElmer). For each compound, if the standard calibration curve is linear (in the range of 3.13–500 μM and at least at one of the two wavelengths) with a coefficient of determination (absorbance vs concentration) r2 greater than 0.85, aqueous solubility can be determined at the 500 and 200 μM target concentrations. Conversely, if r2 was only greater than 0.85 in the range of 3.13–200 μM, solubility can be determined at the 200 μM target concentration.
Solubility filter plate: Solutions (nominal concentrations 500 and 200 μM) in PBS (pH 7.4) were prepared in a MultiScreen® solubility 96-well filter plate by dilution of a 10 mM DMSO stock solution for each compound to be tested. Samples were covered and gently shaken at room temperature for 90 min. The plate was then filtered, and 160 μl per well of filtrate were diluted by adding 40 μl acetonitrile. The absorbance of the diluted filtrate was measured at 260 and 280 nm.
Data analysis: The filtrate concentration (μM) was determined by dividing the absorbance by the slope of the calibration curve and multiplying by a factor of 1.25 to account for dilution with acetonitrile prior to the absorbance measurement. All determinations were performed in duplicate and all calculation performed with a Microsoft Excel® electronic spreadsheet.
2.3. CYP inhibition
Aminopyrine N-demethylase activity (AND) was determined by detecting the amount of formaldehyde produced by RLM according to the procedure described in reference [7]. The incubation was carried out in Tris-HCl buffer (50 mM, pH 7.4) supplemented with 150 mM KCI and 10 mM MgCl2. The incubation contained 2 mM aminopyrine, acetonitrile (1% final volume), Tween 80 (0.8 mg/ml), and 1.0 mg/ml of RLM in a total volume of 0.5 ml. Increasing concentrations (1–100 μM) of FK866 and the tested compounds A6, E5, and T1 were added to the incubation mixture to inhibit AND activity. After 3 min pre-incubation at 37 °C, the reaction was initiated by adding the NADPH-regenerating system (1.3 mM β-NADPNa2, 3.3 mM glucose 6-phosphate, 0.4 U/ml glucose 6-phosphate dehydrogenase) and carried out at 37 °C for 15 min with moderate shaking. The reaction was then quenched by addition of 0.25 ml of 10% (w/v) cold TCA solution. After centrifugation at 12,500 rpm for 10 min, a 650 μl aliquot of the protein-free supernatant was treated with 325 μl of Nash reagent and incubated in a water bath at 37 °C for 40 min. The absorbance of the resultant solution was determined at 412 nm, subtracting the blank sample absorbance (without NADPH-regenerating system). The concentration of formaldehyde was calculated (nmol/min/mg) by comparison with a standard curve prepared from commercially available formaldehyde solution freshly standardized by iodometric titration. All incubations were performed in triplicate.
Nash reagent: 3.75 g ammonium acetate, 50 μl glacial acetic acid, and 75 μl acetylacetone were dissolved in 25 ml deionized water. The solution was freshly prepared and used for one day only.
Acknowledgments
This work was supported by the Italian Ministry of Education (Grant PRIN-2010W7YRLZ_002). Financial support from University of Piemonte Orientale (Vercelli, IT) is acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.105034.
Contributor Information
Silvio Aprile, Email: silvio.aprile@uniupo.it.
Ubaldina Galli, Email: ubaldina.galli@uniupo.it.
Gian Cesare Tron, Email: giancesare.tron@uniupo.it.
Erika Del Grosso, Email: erika.delgrosso@uniupo.it.
Cristina Travelli, Email: cristina.travelli@unipv.it.
Giorgio Grosa, Email: giorgio.grosa@uniupo.it.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Travelli C. Identification of novel triazole-based nicotinamide phosphoribosyltransferase (NAMPT) inhibitors endowed with antiproliferative and antiinflammatory activity. J. Med. Chem. 2017;60(5):1768–1792. doi: 10.1021/acs.jmedchem.6b01392. [DOI] [PubMed] [Google Scholar]
- 2.Genazzani A.A. 2014. Inhibitors of Nicotinamide Phosphoribosyltransferase, Compositions, Products and Uses Thereof. WO2014178001 (A1) [Google Scholar]
- 3.Colombano G. A novel potent nicotinamide phosphoribosyltransferase inhibitor synthesized via click chemistry. J. Med. Chem. 2010;53:616–623. doi: 10.1021/jm9010669. [DOI] [PubMed] [Google Scholar]
- 4.Aprile S. In vitro metabolism study of 2-isopropyl-9H-thioxanthen-9-one (2-ITX) in rat and human: evidence for the formation of an epoxide metabolite. Xenobiotica. 2011;41(3):212–225. doi: 10.3109/00498254.2010.532887. [DOI] [PubMed] [Google Scholar]
- 5.Massarotti A. Are 1,4-and 1,5-disubstituted 1,2,3-triazoles good pharmacophoric groups? ChemMedChem. 2014;9(11):2497–2508. doi: 10.1002/cmdc.201402233. [DOI] [PubMed] [Google Scholar]
- 6.http://www.merckmillipore.com/IT/it/product/MultiScreenHTS-PCF-Filter-Plates-for-Solubility-Assays,MM_NF-C8875#anchor_DS
- 7.Niwa T. Contribution of human hepatic cytochrome P450s and steroidogenic CYP17 to the N-demethylation of aminopyrine. Xenobiotica. 1999;29(2):187–193. doi: 10.1080/004982599238731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




