Abstract
Objectives
In the pathogenesis of type 2 diabetes, development of insulin resistance triggers an increase in pancreatic β-cell insulin secretion capacity and β-cell number. Failure of this compensatory mechanism is caused by a dedifferentiation of β-cells, which leads to insufficient insulin secretion and diabetic hyperglycemia. The β-cell factors that normally protect against dedifferentiation remain poorly defined. Here, through a systems biology approach, we identify the transcription factor Klf6 as a regulator of β-cell adaptation to metabolic stress.
Methods
We used a β-cell specific Klf6 knockout mouse model to investigate whether Klf6 may be a potential regulator of β-cell adaptation to a metabolic stress.
Results
We show that inactivation of Klf6 in β-cells blunts their proliferation induced by the insulin resistance of pregnancy, high-fat high-sucrose feeding, and insulin receptor antagonism. Transcriptomic analysis showed that Klf6 controls the expression of β-cell proliferation genes and, in the presence of insulin resistance, it prevents the down-expression of genes controlling mature β-cell identity and the induction of disallowed genes that impair insulin secretion. Its expression also limits the transdifferentiation of β-cells into α-cells.
Conclusion
Our study identifies a new transcription factor that protects β-cells against dedifferentiation, and which may be targeted to prevent diabetes development.
Keywords: Type 2 diabetes, Insulin resistance, β-Cell proliferation, Dedifferentiation, Transdifferentiation
Abbreviations: KLF6, Krüppel-like factor 6; GSIS, glucose-stimulated insulin secretion; HFHS, high-fat high sucrose; WGCNA, weighted gene co-expression network analysis; RT-PCR, real-time polymerase chain reaction
Highlights
-
•
Absence of Klf6 in β-cells leads to a reduction in their proliferation and mass during insulin resistance development.
-
•
Insulin receptor blockade by S961 leads to a greater dedifferentiation state in β-cell-specific Klf6 knockout mice.
-
•
Insulin receptor antagonism leads to enhanced β-cell to α-cell transdifferentiation in β-cell-specific Klf6 knockout mice.
1. Introduction
Pancreatic β-cells play a central role in glucose homeostasis by secreting insulin in response to increases in glycemia to stimulate glucose utilization by liver, fat, and muscle and to inhibit hepatic glucose production. In various physiological states, such as pregnancy, obesity, or aging, insulin target tissues become partially resistant to the action of this hormone and preserving euglycemia necessitates a compensatory increase in β-cell mass and insulin secretion capacity [[1], [2], [3]]. This functional adaptation is, however, limited and further worsening of insulin resistance may cause β-cell functional failure and reduced cellular mass, leading to development of diabetic hyperglycemia.
The mechanisms of β-cell adaptation to, or failure in insulin resistant states are not fully understood. Analysis of the pancreas of brain-dead organ donors showed that β-cell mass was increased in obesity but reduced in type 2 diabetes of long duration [4]. At the time of diabetes diagnosis, however, β-cell mass was found to be normal [5], implying that a defect in β-cell secretion capacity precedes loss of mass. The loss of β-cell function induced by insulin resistance is now recognized to be associated with, and probably caused by their dedifferentiation [6,7]. This process is characterized by the down-expression of genes controlling glucose-stimulated insulin secretion (GSIS) (Slc2a2, Gck, Kir6.2, Vddac), genes encoding mature β-cell transcription factors (Pdx1, Nkx6.1, Pax6, Foxo1), and the expression of normally disallowed genes, such as Hk1, LdhA, and Slc16a2, which modify glucose metabolism and consequently further impair GSIS [6,7]. How β-cell dedifferentiation is triggered is not fully understood. Earlier experiments have shown that transplanting control islets into diabetic mice led to a rapid loss of Glut2 expression and of GSIS, whereas the transplantation of islets from diabetic animals into control mice led to the restoration of both parameters [8,9]. Thus, disturbances in the metabolic milieu of (pre)-diabetic mice may induce β-cell dedifferentiation and failure. Hyperglycemia, elevated plasma glucocorticoids, and free fatty acids [3,6,[10], [11], [12]] may contribute to induce these deregulations.
Another aspect of the development of β-cell dysfunctions in the presence of insulin resistance is their dependence on the genetic architecture of each individual. It is indeed known that only a fraction of obese, insulin-resistant individuals progress to type 2 diabetes [13]. Similarly, mice with different genetic backgrounds display very heterogeneous deregulations of insulin secretion or insulin action when fed a high-fat, high-sucrose (HFHS) diet. To identify genes controlling β-cell functional adaptation, we recently used this mouse genetic diversity to perform a systems biology investigation of pancreatic islet adaptation to a metabolic stress. We characterized the physiological adaptation of mice from different inbred strains fed an HFHS diet for different periods of time and performed parallel islet transcriptomic analysis [14]. Weighted gene co-expression network analysis (WGCNA) [15] yielded islet gene modules that correlated with the measured phenotypes. Detailed analysis of one such module led to the identification of Elovl2, an enzyme that generates docosahexaenoic acid, an important regulator of GSIS which also confers protection against glucotoxicity-induced apoptosis [14,16].
Here, following a similar approach, we identified and characterized the role of Krüppel-like factor 6 (Klf6), a zinc finger transcription factor [17], in β-cell adaptation to insulin resistance. The study of β-cell-specific Klf6 knockout mice showed that this transcription factor is required for β-cell proliferation during pregnancy, in response to high-fat diet feeding, and following pharmacological induction of insulin resistance. Furthermore, we show that Klf6 protects β-cells against insulin resistance-induced dedifferentiation and transdifferentiation into α-cells.
2. Materials and methods
2.1. Reagents
Mouse insulin and glucagon enzyme-linked immunosorbent assays (ELISAs) were purchased from Mercodia (Uppsala, Sweden) and radioimmunoassay (RIA) kits for insulin and glucagon were from Merck Millipore (MA, USA). Exendin-4 was purchased from Bachem (Bubendorf, Switzerland), and S961 and Exendin-4 Cy3 were kind gifts from Dr. Lauge Schäffer and Dr. Jacob Hecksher-Sørensen (Novo Nordisk, Copenhagen, Denmark), respectively.
2.2. Antibodies
Rabbit anti-Ki67 was from Abcam (ab15580). Guinea pig antibodies to insulin and glucagon were purchased from Dako (AO564) and Linco (4031-01 F), respectively. Rabbit anti-Glut2 has been previously described [18]. Secondary antibodies were goat anti-rabbit immunoglobulin (IgG) antibodies coupled to Alexa-Fluor 568 (Invitrogen; A11036), Alexa-Fluor 488-labeled goat anti-guinea pig immunoglobulin antibodies (Invitrogen; A11073) and Alexa-Fluor 488-labeled anti-rabbit IgG antibodies (Invitrogen; A11008).
2.3. Mouse maintenance and diet
Male and female mice were housed in groups of 2–5 per cage on a 12-hr light/dark cycle with unlimited access to a standard rodent chow diet (Diet 3436, Provimi Kliba AG, Kaiseraugst, Switzerland) or an HFHS diet (Purified Diet 235HF, Safe Diets, Augy, France). Experiments were performed on 10- to 32-week-old mice with the respective age- and sex-matched control littermates. The mouse experiments were approved by the Veterinary Office of Canton de Vaud, Switzerland.
2.4. Generation of β-cell-specific Klf6 knockout mice
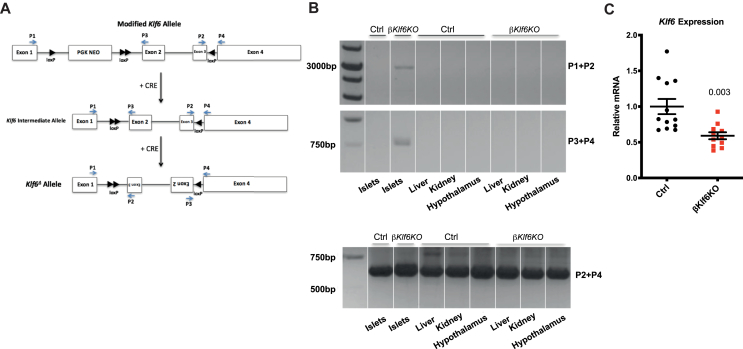
Ins1Cre/+ mice [19] were crossed with Klf6flox/flox mice [20] to generate Klf6flox/flox;Ins1Cre/+ mice (βKlf6KO) and Klf6flox/flox;Ins1+/+mice l (Ctrl). Genomic DNA (gDNA) was extracted with the Quick gDNA MiniPrep™ kit according to manufacturer's instructions (LucernaChem AG, Luzern, Switzerland), and genotyping was performed by PCR analysis using the following primers (see Figure 2) P1:5′-TGGAGGAATATTGGCAACAG-3’; P2:5′-AAGCATACCGGTGCCAAG-3’; P3:5′-GTCTTCCTGGCTGTCAAATT-3’; P4:5′-TGCCTCTTCATGTGCAGG-3’. Cell lineage tracing experiments were performed using Rosa26tdtomato mice bred with Ins1Cre/+and with Klf6flox/flox:Ins1Cre/+ to generate Klf6+/+;Rosa26tdtomato; Ins1Cre/+ and Klf6flox/flox;Rosa26tdtomato; Ins1Cre/+.
Figure 2.
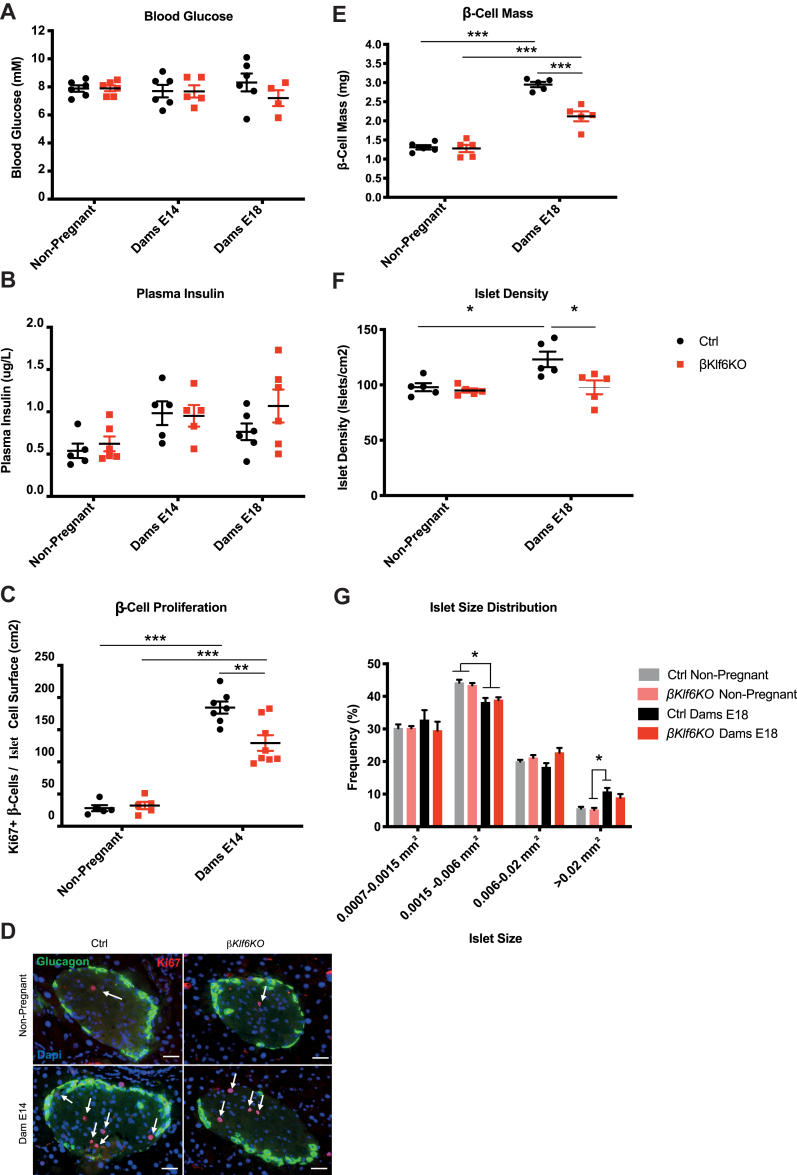
Reduced β-cell Mass Expansion During Pregnancy in βKlf6KO Mice. (a) Blood glucose and (b) plasma insulin levels of 10–12-week-old female mice taken in non-pregnant mice and mice at 14 and 18 days of pregnancy (n = 4–6). (c) Quantification of Ki67-positive β-cells over islet cell surface in non-pregnant and 14-day pregnant mice (n = 5–8). (d) Representative immunofluorescence detection of Ki67 in Ctrl and βKlf6KO mice. Scale bar: 50 μm. (e) β-cell mass in response to pregnancy (day 18). (f) Islet density and (g) size distribution following 18 days of gestation (n = 5). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
2.5. Quantitative real-time PCR and gene expression analysis
Total RNA from islets was extracted using the RNeasy Plus micro kit (Qiagen, Hombrechtikon, Switzerland). cDNA was synthesized by reversed transcription of 1 μg of total RNA using random primers (Promega) and M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. Gene expression was measured in a final volume of 10 μl by quantitative real-time PCR using 5 μl of Power SYBR Green Master Mix (Applied Biosystems, Zug, Switzerland) and 2 μl of cDNA in a 7500 Fast Light Cycler System (Applied Biosystems, Zug, Switzerland). Results were normalized to the housekeeping gene mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels. Primers were obtained from Microsynth (Balgach, Switzerland). Specific mouse primers for each sequence are listed in Supplementary Table 5.
2.6. Glucose tolerance test and in vivo insulin secretion
For glucose tolerance tests, mice were fasted overnight and placed in individual cages. Blood glucose was measured from the tail vein using a Breeze2 glucometer (Bayer, Zürich, Switzerland) before and at the indicated times following an intraperitoneal (i.p.) injection of glucose (2 g/kg). The same protocol was applied for in vivo insulin secretion, in which blood sampling from the tail vein was used for plasma insulin level measurements by ELISA.
2.7. Pancreatic insulin and glucagon content
Excised pancreata were rapidly weighed and sonicated in ethanol-acid (75% EtOH; 0.55% HCl). After overnight storage at 4 °C, samples were centrifuged at 4 °C for 5 min at 800 rpm. The supernatant was then centrifuged at 4 °C for 20 min at 4,000 rpm. The final supernatant was assessed for insulin and glucagon by radioimmunoassay (RIA). Results were normalized to initial pancreas weight.
2.8. Pancreatic islet isolation, insulin secretion, and proliferation assays
Islets from 10- to 12-week-old male mice were isolated by hand-picking following pancreata digestion by Liberase [21]. After overnight recovery in cell culture medium, insulin secretion measurements were performed on batches of 10 islets. Islets were first pre-incubated for 2 h in Krebs–Ringer Bicarbonate-HEPES buffer with bovine serum albumin (KRBH-BSA) (120 mM of NaCl, 4 mM of KH2PO4, 20 mM of HEPES, 1 mM of MgCl2, 1 mM of CaCl2, 5 mM of NaHCO3, and 0.5% BSA, pH 7.4) supplemented with 2.8 mM of glucose. Medium was then replaced with KRBH-BSA supplemented with either 2.8 mM or 16.7 mM of glucose, and incubations were continued for 1 h. Secreted insulin and islet insulin contents were measured by radioimmunoassay. For islet insulin content measurements, islets were sonicated in ethanol-acid. For proliferation measurements, 10 islets were placed on extracellular matrix (ECM)-coated cell dishes (Novamed, Jerusalem, Israel) and kept until monolayer formation (5–7 days). Proliferation assessment was performed following Exendin-4 treatment (100 nM; 72 h). ECM plates were fixed with 4% paraformaldehyde (PFA), washed with phosphate-buffered saline (PBS) (3×), and permeabilized with 0.3% Triton X-100 and 1% BSA (pH 7.4) for 30 min at room temperature. Monolayers were then incubated in the presence of rabbit anti-Ki67 antibody (diluted 1:500) for 1 h at room temperature. Following washes with PBS (3×), monolayers were incubated in the dark with a goat anti-rabbit IgG antibody coupled to Alexa-Fluor 568 (diluted 1:400) for 1 h at room temperature. Finally, after three 3 with PBS, cells were mounted on a coverslip with Vectashield-DAPI (Reactolab SA, Servion, Switzerland) and observed with a Zeiss Axiovision Fluorescence microscope. Quantification was made by counting Ki67-positive cells using the ImageJ software (http://rsbweb.nih.gov/ij/). Previous studies, using co-staining for Glut2, showed that in these culture conditions 95% of proliferating cells were beta-cells (β-cells) [22].
2.9. β-Cell purification
Pancreatic islets were incubated in RPMI 1640 supplemented with 10% fetal bovine serum (FBM), 1 mM of glutamine, and 1% pen/strep with Exendin-4-Cy3 (100 nM) for 30 min at 37 °C. Single-cell preparations were then obtained by incubating islets for 1.5 min at 37 °C, with gentle pipetting, in Ca2+ and Mg2+ free PBS containing 0.24 mM of ethylenediaminetetraacetic acid (EDTA) and 0.025% trypsin. Digestion was stopped by adding RPMI 1640 supplemented with 10% FBS, 1 mM of glutamine and 1% pen/strep. Dispersed islet cells were resuspended in 300–500 μl of KRBH-1% BSA pH 7.4 supplemented with 2.5 mM of glucose, 5 mM of EDTA and 1 μg/mL of 4′,6-diamidino-2-phenylindole (DAPI). β-cells were purified by fluorescence-activated cell sorting using a 4-laser MoFlo ASTRIOS EQ™ cell sorter (Beckman Coulter, Indianapolis, USA) using a 100-μm nozzle. After gating out the doublets and dead cells (DAPI positive), Cy3-positive cells detected using the 561 nm laser were selected.
2.10. Histomorphometric analysis and immunofluorescence detection
Histomorphometric and β-cell proliferation analysis were performed using 5-μm-thick sections from 4% paraformaldehyde (PFA)-fixed and paraffin-embedded pancreata. β-cell mass was measured by histomorphometry [21,23]. Briefly, for each condition, 6 sections per pancreas and 4–6 pancreata were analyzed, representing a total of >500 islets. β-Cell surface area was measured using the ImageJ software. β-cell mass was calculated by multiplying the β-cell area by the pancreas weight. For β-cell proliferation and α-cell mass measurements, pancreas sections were incubated overnight at 4 °C with a rabbit anti-Ki67 antibody (diluted 1:100) and guinea pig anti-glucagon antibody (diluted 1:500) and washed 3 times with PBS. The sections were then incubated for 1 h with Alexa-Fluor 568-labeled goat anti-rabbit IgG antibody and Alexa-Fluor 488-labeled goat anti-guinea pig IgG antibody, washed with PB,S and then mounted with Vectashield containing DAPI. Ki67-positive cells were counted over islet areas defined by glucagon staining in 50–100 islets. The analysis was performed using the ImageJ software. Only non-glucagon, Ki67-positive cells were counted. Because β-cells form the majority of the islets non-alpha cells, we considered the Ki67-positive cells as representing proliferating β-cells. Preceding experiments also showed that the percent of Ki67/Glut2 double-positive cells was only marginally different from the number of total Ki67-positive cells [22,24]. Glucagon staining was used for α-cell mass measurements.
For immunofluorescence detection of islet Glut2, insulin, and glucagon, mice were fixed by intracardiac perfusion with cold 4% PFA. Pancreata were then excised, incubated in % PFA for 4 h and overnight in a 30% sucrose solution before being frozen at −80 °C in optimal cutting temperature (OCT) compound. Twenty-micrometer cryosections were prepared, blocked in PBS supplemented with 3% BSA and 0.3% Triton X-10,0 and then incubated at 4 °C overnight with rabbit anti-Glut2 (diluted 1:500), guinea pig anti-insulin (diluted 1:500), and guinea pig anti-glucagon (diluted 1:500). Primary antibodies were then detected with an Alexa-Fluor 488-labeled goat anti-rabbit IgG antibody or a goat anti-guinea pig IgG antibody (diluted 1:200).
2.11. RNA sequencing analysis
Integrity of islet RNA was verified on a fragment analyzer (Agilent Technologies, Inc., Santa Clara, CA 95051, USA), and all preparations had an RNA quality number (RQN) between 8.2 and 9.3. RNA-seq libraries were prepared using 250 ng of total RNA and the Illumina TruSeq Stranded mRNA reagents (Illumina; San Diego, California, USA). Cluster generation was performed with the resulting libraries using the Illumina TruSeq SR Cluster Kit v4 reagents and sequenced on the Illumina HiSeq 2500 using TruSeq SBS Kit v4 reagents. A sequencing depth of 31–46 million reads using 125 bp single-end reads was used.
Reads were mapped with STAR-2.5.3a software [25] to the M. musculus-mm 15 reference genome. Read counts for each gene were generated with HTSeq-0.9.1 software [26] using as reference the annotation index GRCm38.83 from ENSEMBL. Counts were filtered, excluding those genes with less than one count per million in two libraries, and normalized using trimmed mean normalization method (TMM) with ‘edgeR’ [27]. Pathway enrichment analysis of the genes differentially expressed between Ctrl and βKlf6KO mice was performed using the Kyoto Encyclopedia of Genes and Genomes KEGG and gene ontology (GO) databases and the R library “clusterProfiler”. P-values were adjusted for multiple comparisons using the Benjamini Hochberg procedure (Benjamini and Hochberg 1995). Terms with an adjusted p-value ≤0.05 were considered as overrepresented.
2.12. Data representation and statistics
Results are expressed as mean ± SEM with significant p-values ranging from 0.05 to 0.001 (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Statistical analyses performed on the presented data were done with Prism 6.0 using Student's t-test, and two-way analysis of variance (ANOVA) followed by Tukey's post-hoc multiple comparison test.
3. Results
3.1. Klf6 as a potential regulator of β-cell adaptation to metabolic stress
To identify novel genes potentially involved in the control of β-cell mass and function in response to metabolic stress, we previously performed a study where mice from six different strains were fed with a regular chow (RC) or a HFHS diet for 2, 10, 30, and 90 days [14]. At each time point, mice were tested for multiple metabolic phenotypes, and their islets were isolated for RNASeq analysis. WGCNA led to the identification of gene modules, i.e., groups of genes whose expression is similarly regulated across all mouse groups. These modules were then correlated with the measured phenotypes, and the results are represented as a heat map (Supplementary Fig. 1a and Ref. [14]. Here, we further investigated the Thistle 3 module, which showed strong correlation with plasma insulin levels at 0, 15, and 90 min of an oral glucose tolerance test. This module contains 114 genes, and the transcription factor Klf6 (Krüppel-like Factor 6), is among the genes that were most negatively correlated with insulinemia and strongly associated with module membership (Supplementary Fig. 1b). To have an indication of a potential association of KLF6 with diabetes in human islets, we checked its level of expression in a large transcriptomic database of islets from brain-dead, non-diabetic and type 2 diabetic donors [28]. This data showed statistically significantly higher expression of KLF6 mRNA in type 2 diabetic islets (log2fold-change = 0.27, adj. p-value 0.015) (Supplementary Fig. 1c).
3.2. Generation of β-cell-specific Klf6 conditional knockout mice
To investigate the role of Klf6 in β-cells, we generated mice with a β-cell-specific inactivation of this gene by crossing Klf6flox/flox mice [20] with Ins1Cre/+ mice [19] to generate Klf6flox/flox;Ins1Cre/+ (βKlf6KO) mice and Klf6flox/flox;Ins1+/+ (Ctrl) mice. Figure 1 shows that Cre-mediated recombination resulted in the inversion of exons 2 and 3, leading to inactivation of Klf6. Genotyping was performed with the primers indicated in Figure 1A, and recombination was observed only in islets (Figure 1B). Quantitative RT-PCR analysis showed a 40% reduction of Klf6 expression in islets isolated from βKlf6KO mice (Figure 1C). This indicated that Klf6 was also expressed in non-β-cells. Quantitative RT-PCR analysis performed with RNA extracted from cell sorter purified β- and non-β-cells shows a 3-fold higher expression of Klf6 in non-β-cells (Supplementary Fig. 2), in agreement with published data [29]. Thus, the remaining Klf6 expression in islets from βKlf6KO mice is consistent with it being expressed at a 3-fold higher level in non-β-cells, which represent 30% of islet cells. No gross morphological difference could be observed between Ctrl and βKlf6KO mice (Supplementary Fig. 3).
Figure 1.
β-Cell-specific Inactivation of Klf6. (a) Structure of the modified Klf6 allele (top) and Klf6 allele after Ins1Cre-mediated recombination (middle and bottom). (b) PCR analysis of Klf6 recombination using primers shown in a) in the indicated tissues of 12-week-old mice. PCR of genomic DNA from islets of βKlf6KO mice yields a 3,000-bp band using Primers 1 and 2, and a 750-bp band using Primers 3 and 4. Primers 2 and 4 yield a 600-bp band. (c) qRT-PCR analysis of Klf6 expression in islets (n = 11–12). Two-tailed unpaired Student's t-test. Error bars represent SEM.
3.3. Normal glucose homeostasis, β-cell mass, and insulin secretion in βKlf6KO mice
We first assessed glucose homeostasis as well as β-cell mass and function in regular chow (RC)-fed βKlf6KO mice. These mice had similar body weight, fasted, fed, and fasted/refed glycemia, as well as plasma insulin and glucagon levels as their control littermates (Supplementary Figs. 4a–d). Their total pancreas and isolated islet insulin content (Supplementary Figs. 4e and f) were similar to that of Ctrl mice. GSIS assays using isolated islets from both types of mice yielded identical results (Supplementary Figs. 4g) and i. p. glucose tolerance tests performed on 12- and 36-week-old male and female mice were also indistinguishable (Supplementary Figs. 4h–k). Pancreatic weights (not shown) and islet morphologies (Supplementary Fig. 5) were indistinguishable between genotypes.
3.4. Reduced β-cell mass expansion in βKlf6KO mice in response to pregnancy
Because inactivation of Klf6 did not impact β-cell mass and function in non-challenged conditions, we tested whether metabolic stress would reveal a role for Klf6. In a first set of experiments, we analyzed β-cell proliferation and mass during gestation, an insulin-resistant condition associated with compensatory β-cell growth [30]. Pancreatic weights (not shown) and islet morphologies (Supplementary Fig. 5) were indistinguishable between genotypes at 14 days of pregnancy. We analyzed Ki67 staining in β-cells at day 14 and β-cell mass at day 18 of pregnancy. Ctrl and βKlf6KO mice displayed similar blood glucose and plasma insulin levels throughout gestation (Figure 2A,B) but β-cell proliferation increased by 6.5-fold in Ctrl mice as compared to 4-fold in βKlf6KO (Figure 2C,D). As a result, β-cell mass increased by 2.3-fold in Ctrl mice as compared to 1.7-fold in βKlf6KO mice (Figure 2E). The difference in β-cell mass was related to a higher number of islets per pancreas in Ctrl mice rather than to a change in islet size distribution (Figure 2F,G).
3.5. Reduced β-cell mass expansion in βKlf6KO mice in response to high-fat high-sucrose diet
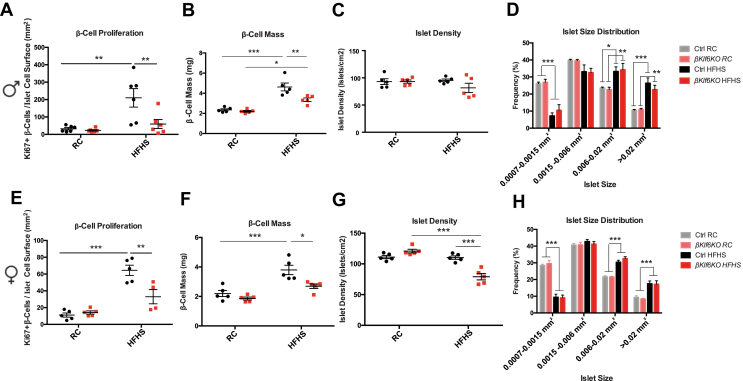
In the second approach, we fed 6-week-old Ctrl and βKlf6KO male and female mice for an additional 22 weeks with a RC or an HFHS diet. Feeding a high-energy diet induced a similar increase in body weight, glucose intolerance, and in vivo GSIS in both types of mice (Supplementary Fig. 6). Pancreatic weights (not shown) and islet morphologies (Supplementary Fig. 5) were indistinguishable between genotypes at the end of the HFHS feeding period. β-cell proliferation was, however, markedly lower in male and female βKlf6KO mice as compared to Ctrl mice (Figure 3A,E), and this was associated with reduced β-cell mass expansion (Figure 3B,F). HFHS feeding led to shift in the distribution in islet sizes to higher values (Figure 3D,H) and, in female βKlf6KO mice, a reduction in islet numbers (Figure 3G).
Figure 3.
Reduced β-cell Mass Expansion in HFHS-Fed βKlf6KO Mice. (a,e) Quantification of Ki67-positive β-cells over islet cell surface in Ctrl and βKlf6KO mice fed from 6 weeks of age for 22 weeks on regular chow diet (RC) or high-fat high-sucrose diet (HFHS) (n = 4–6). (b,f) β-cell mass in RC or HFHS fed mice. (c,g) Islet density and (d,h) size distribution following 22 weeks on respective diets (n = 5). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.6. Reduced β-cell mass expansion in βKlf6KO mice during S961 treatment
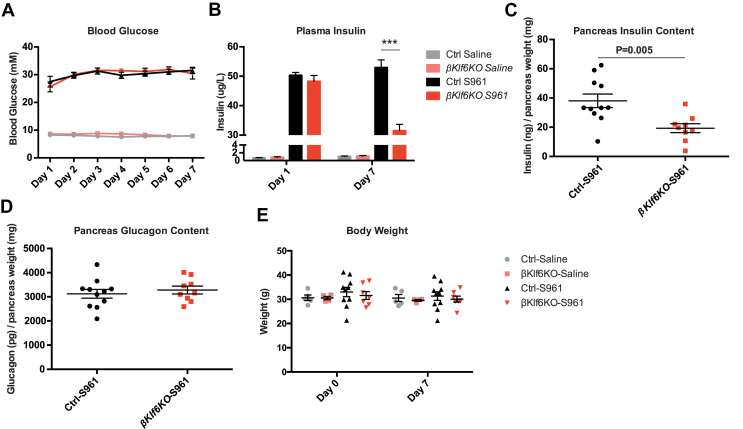
Insulin resistance induced by chronic delivery of the insulin receptor antagonist S961 [31] leads to rapid development of hyperglycemia and hyperinsulinemia. Here, we treated Ctrl and βKlf6KO male mice for 2 and 7 days with saline or S961. This induced a strong hyperglycemia from the first day of treatment until the end of the experiment in both groups of mice (Figure 4A). Insulinemia was similarly elevated in Ctrl and βKlf6KO mice at day 1 of treatment but was reduced by half in βKlf6KO mice at day 7 (Figure 4B), at which time the total pancreatic insulin content was also reduced by 50% in the pancreata of βKlf6KO mice (Figure 4C). The pancreatic glucagon contents (Figure 4D), body weights (Figure 4E), and pancreas weights (not shown) remained identical in S961-treated Ctrl and βKlf6KO mice. However, unexpectedly, the β-cell section area was reduced by 40% by S961 treatment in both groups of mice (Supplementary Fig. 7), indicating that S961 treatment also induced a reduction in β-cell volume.
Figure 4.
Impaired Insulinemic Response to Insulin Resistance in βKlf6KO Mice. Twelve-week-old male Ctrl and βKlf6KO mice were treated with saline or the insulin receptor antagonist S961 for 7 days. (a) Glycemia (n = 7–10) (b) insulinemia at days 1 (n = 9–15) and 7 (n = 9–14). (c) Pancreatic insulin and (d) glucagon contents at day 7 of treatment (n = 9–11) (e) body weight (n = 4–10). Statistical analyses have been performed by two-way ANOVA (Bonferonni's post-hoc test). ∗∗∗P < 0.001. Statistical analyses in c and d were done using a two-tailed unpaired Student's t-test. Error bars represent SEM.
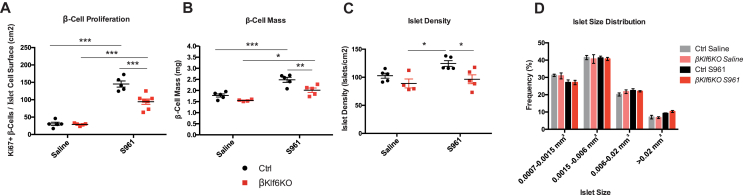
We next assessed β-cell proliferation and mass in male mice treated for 7 days with saline or S961. Figure 5A shows that the proliferation rate was increased by 5-fold in Ctrl mice and by 3-fold in βKlf6KO mice. This led to a β-cell mass increase in S961-treated mice that reached 2.5 mg/pancreas in Ctrl mice but only 2.0 mg/pancreas in βKlf6KO mice (Figure 5B). This lower β-cell mass was due to a decrease in the total number of islets, which displays the same size distribution as in S961-treated Ctrl mice (Figure 5C,D).
Figure 5.
Reduced β-cell Mass Expansion in S961-Treated βKlf6KO Mice. (a) Quantification of Ki67-positive β-cells over islet cell surface in mice treated with saline or S961 for 7 days (n = 5–7). (b) β-cell mass in response to saline and S961 treatment. (c) Islet density and (d) size distribution of islets following S961 treatment for 7 days (n = 4–5). Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. Error bars represent SEM.
Together, the above data showed impaired β-cell mass expansion in βKlf6KO mice in response to insulin resistance of pregnancy, HFHS diet feeding, or induced by pharmacological inhibition of the insulin receptor. This indicates that Klf6 is necessary for the full compensatory β-cell proliferation to insulin resistance.
3.7. Klf6 protects β-cells against dedifferentiation
To identify the changes in transcript expression resulting from Klf6 inactivation in β-cells, we performed RNASeq analysis of islets isolated from Ctrl and βKlf6KO male mice treated for 2 days with saline or S961. We selected this treatment time as we searched for early transcriptional events leading to defective β-cell adaptation and tried to minimize the changes in gene expression due to long-term metabolic alterations. In this series of treated mice, the glycemia was already markedly increased after 1 day of treatment and reached the usual maximal value of 30 mM at day 2 (Supplementary Fig. 8a). Plasma insulin levels were very high at day 1 of treatment in both Ctrl and βKlf6KO mice but, at day 2, they were already reduced by 20% in βKlf6KO mice as compared to Ctrl mice (Supplementary Fig. 8b).
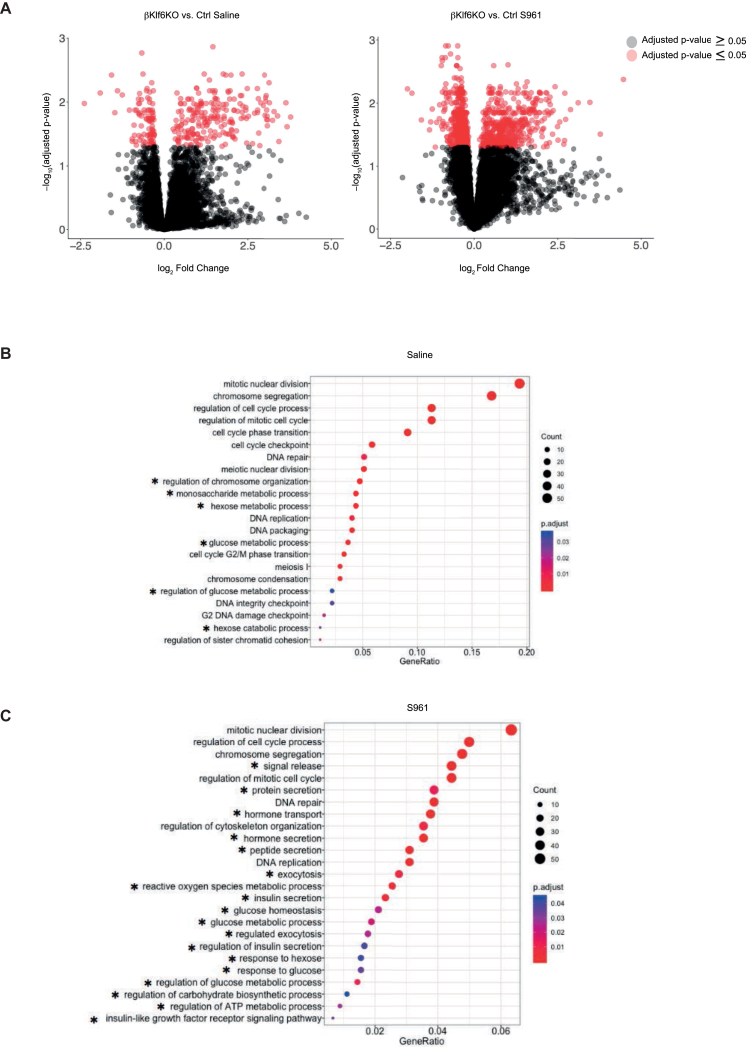
Analysis of genes differentially expressed in islets from saline or S961-treated Ctrl and βKlf6KO mice is presented as volcano plots (Figure 6A). The complete lists of differentially expressed genes are presented in Suppl. Tables 1 and 2. GO analysis of genes differentially regulated in islets from saline-treated Ctrl and βKlf6KO mice (Figure 6B, Supplementary Tables 3a and b), revealed that most pathways over-represented in knockout islets were associated with cell cycle regulation (“mitotic cell division”, “regulation of cell cycle process”, etc.) and a few with hexose metabolic processes. The absence of Klf6 did not lead to impaired proliferation under unchallenged conditions in vivo, yet islets from βKlf6KO mice had lower Ki67 staining than Ctrl islets when cultured as monolayers on an extracellular matrix in the presence or absence of Exendin-4 (Supplementary Fig. 9). In the hexose metabolic GO term, 3 glycolytic genes controlling the same step were present, Pfkfb2 (6-phosphofructo-2-kinase/Fructose-2,6-Biphosphatase 2), reduced by 34% in βKlf6KO compared to Ctrl islets, the bifunctional enzyme that produces and hydrolyzes the allosteric glycolysis activator fructose 2,6-bisphosphate (Fru-2,6-P2), Fbp2 (fructose bisphosphatase 2), increased by 2.4 fold, which catalyzes the hydrolysis of Frc-1,6-P2 into Frc-6-P, and Tigar (P53 Induced Glycolysis Regulatory Phosphatase), reduced by 30%, an enzyme that hydrolyzes both Frc-1,6-P2 and Frc-2,6-P2 into Frc-6-P thus increasing the reverse glycolytic pathway potentially leading to increased production of NADPH through the pentose phosphate shunt [[32], [33], [34]].
Figure 6.
Transcriptomic Analyses of Islets from Mice Treated with Saline and S961. Ctrl and βKlf6KO mice were treated with saline or S961 for 2 days before islet harvesting. (a) Volcano plots showing the number of islet genes differentially expressed between genotypes (Ctrl and βKlf6KO) following treatment. Selected gene ontology analysis of differentially expressed genes following (b) saline and (c) S961. ∗genes associated with metabolic processes.
GO analysis of genes differentially expressed in islets from Ctrl and βKlf6KO mice under S961 treatment (Figure 6C, Supplementary Tables 4a and b) revealed enrichment in cell division genes (“mitotic nuclear division”, “regulation of cell cycle process”) and a striking increase in terms related to glucose metabolism and insulin secretion (“hormone transport”, “hormone secretion”, “exocytosis”, “glucose metabolic process”, and others) (Figure 6C).
A more detailed analysis of the genes that were dysregulated in islets from βKlf6KO as compared to Ctrl mice following S961 treatment revealed: (Figure 7A) i) a 30% reduced expression of genes controlling different steps in GSIS, including Slc2a2, Slc30a8, Igf1r, Ffar1, Vdca1d, Atp2a2, Vamp2, and Sytl4; ii) a reduced expression of Pdx1, Nkx6.1, Foxa2, and Foxo1, essential mature β-cell transcription factors [7,[35], [36], [37]]; iii) an increased expression of several mRNAs encoding dedifferentiation markers, including Gast, Aldh1a3, Serpina7, Vim, and Slc18a2 as well as iv) expression of the “disallowed genes”, Ldha, Hk1, Slc16a2, Oat, Igfbp4, Mylk, Ly6a, Pcolce, Parp3, and Ndrg2 [1,7,[36], [37], [38], [39], [40], [41], [42], [43]].
Figure 7.
Loss of Klf6 in the β-cell Increases Dedifferentiated Phenotype Upon S961 Treatment. (a) Differential gene expression based on RNA-sequencing performed on islets (n = 8) harvested from Ctrl and βKlf6KO mice treated with S961 for 2 days. Data are represented as fold change of mRNA of specific genes. (b) RT-PCR of islets harvested from mice treated with S961 for a 7-day period (n = 5). Two-tailed unpaired Student's t-test. Error bars represent SEM. (c) Immunofluorescence of insulin, (d) Glut2 and (e) chromogranin A in Ctrl and βKlf6KO pancreata following saline or S961 treatment for 7 days (n = 4). Scale bar: 50 μm.
After 7 days of treatment with S961, the expression of Ins1, Ins2, Slc2a2, and Slc30a8, as determined by RT-QPCR analysis, was still more reduced in islets from βKlf6KO than in Ctrl mice (Figure 7B). Immunofluorescence microscopy detection of insulin and Glut2 confirmed lower expression of both proteins in islets of S961-treated βKlf6KO as compared to Ctrl mice (Figure 7C,D). The endocrine markers Foxo1 and Aldh1a3 were, however, no longer differentially expressed between Ctrl and βKlf6KO islets (Figure 7B). Immunostaining for Aldh1a3, a protein induced in a subset of dedifferentiated β-cells, was similarly increased in islets from both types of mice after S961 treatment (Supplementary Fig. 10). Finally, chromogranin A showed homogenous expression over the whole islets, although the staining intensity was lower after S961 than after saline treatment (Figure 7E). There was no difference in chromogranin A mRNA expression between islets from Ctrl and βKlf6KO mice treated with saline or S961 as indicated by the RNASeq data (not shown).
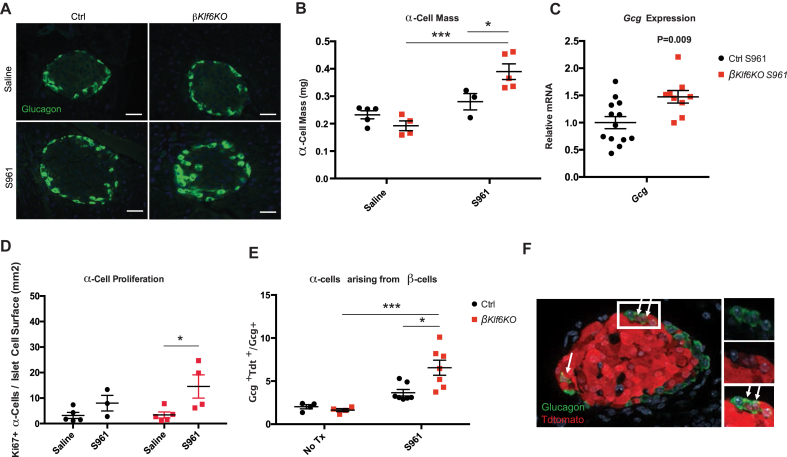
As previous reports indicated that hyperglycemia induces transdifferentiation of β-cells into α-cells [7,44], we measured α-cell mass in the pancreas of Ctrl and βKlf6KO mice after 7 days of saline or S961 treatment. Figure 8A, B shows that the α-cell mass was not increased by S961 treatment in Ctrl mice but was increased by 60% in islets from S961-treated βKlf6KO mice, and this corresponded to a 60% percent increase in islet glucagon mRNA (Figure 8C). Ki67 staining also revealed a higher proliferation rate of α-cells in S961-treated βKlf6KO mice than in Ctrl mice (Figure 8D).
Figure 8.
Increased α-cells Mass and β-cell Transdifferentiation in S961-Treated βKlf6KO Mice. Ctrl and βKlf6KO mice were administered S961 or saline for 7 days. (a) Immunofluorescence of glucagon-producing cells. Scale bar: 50 μm. (b) Measurement of α-cell mass (n = 3–5). (c) Islet gcg mRNA expression (n = 9–13). (d) Quantification of Ki67-positive α-cell over islet cell surface (n = 3–5). (e) Lineage tracing experiments were performed using Ctrl and βKlf6KO expressing tdtomato from the Rosa26 locus. Quantification of glucagon-positive cells expressing tdtomato in Ctrl and βKlf6KO following S961 treatment for 7 days with an example of immunostaining (n = 4–7). Statistical analyses were performed using two-way ANOVA (Bonferonni's post-hoc) ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. Statistical analysis in c was done using two-tailed unpaired Student's t-test. Error bars represent SEM.
To determine whether the increased number of α-cells can be accounted for by transdifferentiation of β-cells, we generated ins1Cre;Rosa26tdtomato and βKlf6KO; Rosa26tdtomato mice and treated them for 7 days with S961. We then quantified the number of glucagon positive cells also expressing tdtomato. Approximately 4% of α-cells in ins1Cre;Rosa26tdtomato mice were also positive for tdtomato and 8% in βKlf6KO; Rosa26tdtomato mice (Figure 8E). Thus, S961 treatment induced a larger number of β-cells to transdifferentiate into glucagon-producing cells in βKlf6KO mice than in Ctrl mice, suggesting that the increase in α-cell mass may result from both β-cell transdifferentiation and increased proliferation of α-cells.
4. Discussion
Here, we show that Klf6 is required for the full proliferation response of β-cells to the insulin resistance of pregnancy, HFHS diet, and induced by insulin receptor antagonism. When insulin resistance is associated with marked hyperglycemia, as in S961-treated mice, Klf6 also protects β-cells against dedifferentiation and reduces their transdifferentiation into α-cells. Klf6 is, thus, a transcription factor that is required for preserving mature β-cell functional adaptation to insulin resistance.
Klf6 is a member of the Krüppel-like family of transcription factors that contains 18 isoforms [45]. These have various functions in cell cycle progression, apoptosis, and cancer development [[46], [47], [48], [49]], and Klf6 is also associated with non-alcoholic fatty liver disease [50,51] and diabetic nephropathy [52,53]. Here, we identified Klf6 through a systems biology approach aimed at identifying novel regulators of β-cell adaptation to metabolic stress [14]. We found that Klf6 expression is not required for the normal development of adult β-cell mass and function nor for their functional stability over time when mice are fed RC. Only when an additional stress is imposed, such as during gestation when insulin resistance develops but mice remain normoglycemic, does the absence of Klf6 lead to impaired β-cell proliferation and mass expansion. In HFHS-fed mice, glucose intolerance and in vivo glucose-stimulated insulin secretion evolved similarly in βKlf6KO and Ctrl mice, but β-cell proliferation and mass were significantly less induced in βKlf6KO mouse islets. Here, we fed the mice with an HFHS diet for 22 weeks a period of time that may not be sufficient for the β-cell mass expansion defect observed to cause more severe glucose intolerance in βKlf6KO mice than in Ctrl mice. When βKlf6KO mice were treated with S961, a condition that induces rapid and massive hyperglycemia [31,54], reduced β-cell proliferation, and lower mass expansion was associated with a rapid decline in plasma insulin as compared to S961-treated Ctrl mice. Thus, combining insulin resistance with severe hyperglycemia induces stronger β-cell dysfunctions in βKlf6KO mice highlighting the important protective role of Klf6 in these conditions.
To obtain insights into the gene network controlling defective β-cell adaptation in βKlf6KO mice, we performed islet transcriptomic analysis and searched for genes differentially expressed between Ctrl and βKlf6KO mouse islets in saline or S961 treated mice. In saline treated mice, comparative transcriptomic analysis showed that the differentially expressed genes were enriched in genes controlling the cell cycle. Most of these genes were upregulated in βKlf6KO islet cells even though these cells have lower proliferation capacity than those in Ctrl islets as tested in islets cultured as monolayers. Nevertheless, Cdkn3, a cell cycle inhibitor [55], was overexpressed 6-fold in βKlf6KO islets and this may explain the reduced β-cell proliferation capacity. On the other hand, 3 glycolytic genes were also dysregulated, Pfkfb2, Fbp2, and Tigar. These genes control the production and hydrolysis of fructose-1,6-bisphosphate, a key regulatory step in glycolysis and neoglucogenesis. Activating the neoglucogenic direction of this pathway also leads to increased activity of the pentose phosphate pathway, a supplier of Nicotinamide adenine dinucleotide phosphate (NADPH) required for protection against oxidative stress and for the biosynthesis of new molecules necessary for cell proliferation [[32], [33], [34]]. Thus, in regular feeding conditions and when normoglycemia is preserved, Klf6 mainly controls the expression of proliferation genes and of a small group of glycolytic genes possibly required for supporting proliferation through redirecting glucose carbons to biosynthetic activities. In the present study, we have not analyzed whether any of these genes are direct or indirect transcriptional targets of Klf6. However, a study performed in primary mouse oligodendrocyte progenitors and immature oligodendrocytes identified genes that were direct targets of Klf6-binding [56]. Among those, the following were also found to be dysregulated in islets from βKlf6KO mice treated with S961: Ly6a, Oat, and Slc16a2 (disallowed genes), Serpina7 and Vim (dedifferentiation markers), and Slc30a8, gene linked to insulin secretion.
βKlf6KO mice and Ctrl mice under S961 treatment display severe insulin resistance and hyperglycemia. These lead to more important differences in their islets transcriptional program with several groups of genes associated with β-cell dedifferentiation in addition to the “cell cycle” and “glucose metabolism” genes as described in diabetic mice [7,57] and hyperglycemic rats [6]. These changes include down-expression of genes controlling GSIS, reduced expression of β-cell-specific transcription factors, and the overexpression of disallowed genes that negatively interfere with GSIS. These observations therefore suggest that Klf6 is a critical guardian of β-cell mass and identity when insulin resistance and hyperglycemia develop. In this context, our observation that KLF6 expression is increased in islets from type 2 diabetic organ donors suggests that, in humans, its induction aims at protecting β-cells against dedifferentiation. Another aspect of hyperglycemia-induced dedifferentiation is the capacity of β-cells to transdifferentiate into α-cells [7,44]. We show that S961 treatment induces an approximately two-fold higher rate of β-cell transdifferentiation into α-cells when Klf6 is absent.
Thus, our data demonstrates that Klf6 is required for the normal proliferative response of β-cells induced by insulin resistance to protect β-cells against dedifferentiation induced by severe insulin resistance and hyperglycemia and to limit β-cell transdifferentiation into α-cells. It is interesting to compare the role of Klf6 with that of Foxo1. In their landmark paper, Talchai et al. [7] report that β-cell specific Foxo1−/− mice display reduced β-cell mass, increased α-cell mass, impaired fasting glucose with hypoinsulinemia and hyperglucagonemia only when metabolically challenged by multiple pregnancies or aging. Inactivation of Foxo1 is associated with compensatory increased expression of Foxo3 and Foxo4 and mice with triple Foxo knockout developed glucose intolerance due to reduced insulin secretion when fed with RC [7,57]. Thus, in analogy with the Foxo transcription factor studies, inactivation of Klf6 does not induce any islet phenotype under RC, but genetic inactivation of additional Klf family members may generate a stronger diabetic phenotype. Indeed, Klf10 [58] and Klf11 [59] have been reported as potential additional regulators of β-cell function. It is also interesting to note that Klf6 expression in purified mouse β-cells has been found to decrease with age [60], potentially explaining the reduced proliferation capacity of old β-cells [61].
5. Conclusion
In conclusion, our present study identifies Klf6 as a transcription factor that controls β-cell proliferation induced by the insulin resistance of pregnancy, the consumption of high-energy-containing food, and induced by pharmacological induction of insulin resistance. In the latter condition, which is also associated with strong hyperglycemia, Klf6 also restricts β-cell dedifferentiation and transdifferentiation into glucagon-producing α-cells. Thus, Klf6 expression has a protective effect against type 2 diabetes development, and its overexpression may protect against appearance of this disease.
Author contributions
B.T. conceived the experiments and secured funding. B.T. and C.D. designed the experiments and wrote the manuscript. C.D., D.T., A.P., D.B., and X.P.B. performed the experiments. A.R.S.A and M.I. performed RNA-Seq data analysis.
Acknowledgments
This project received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreements No. 115881 (RHAPSODY) and No. 115797 (INNODIA). These Joint Undertakings receive support from the European Union's Horizon 2020 research and innovation program, European Federation of Pharmaceutical Industries Association (EFPIA), ‘Juvenile Diabetes Research Foundation (JDRF)”, and “The Leona M. and Harry B. Helmsley Charitable Trust”. This work is also supported by the Swiss State Secretariat for Education’ Research and Innovation (SERI) under contract number 16.0097 and by a grant from the Swiss National Science Foundation (310030_182496) to B.T. We also thank the Genome Technology Platform of the University of Lausanne for their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.02.001.
Contributor Information
Christopher Dumayne, Email: christopher.dumayne@unil.ch.
David Tarussio, Email: david.tarussio@unil.ch.
Ana Rodriguez Sanchez-Archidona, Email: ana.rodriguez.1@unil.ch.
Alexandre Picard, Email: alexandre.picard@unil.ch.
Davide Basco, Email: davide.basco@unil.ch.
Xavier Pascal Berney, Email: xavierpascal.berney@unil.ch.
Mark Ibberson, Email: mark.ibberson@sib.swiss.
Bernard Thorens, Email: bernard.thorens@unil.ch.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Selection of Klf6. (a) Heatmap of the correlations between metabolic traits and islet gene expressions modules. The Thistle 3 module is depicted with an arrow. (b) Correlations of Thistle 3 genes with module membership and fasted insulin levels (IS T0) and insulin levels at 15 (IS T15) and 90 min (IS T90) after an oral glucose load. (c) Boxplot showing expression of KLF6 (Affymetrix U133_Plus2 probeset 224,606_at) in human organ donor islets (N = 103). KLF6 shows increased expression in islets of individuals with type 2 diabetes (T2D, N = 19) compared to non-diabetic islets (ND, N = 84). The difference in expression between T2D and ND is statistically significant (limma moderated t-test correcting for age and gender, unadjusted p-value = 0.001, Benjamini-Hochberg adjusted p-value = 0.015, log2 fold-change = 0.27). Data were processed and analyzed as described previously [28].
Supplementary Figure 2. Klf6 Expression Distribution in Islet Cell Populations. Klf6 expression in islets, β-cells and non-β-cells of 12 weeks old male mice (one biological replicate from a pool of n = 6 C57Bl/6J mice) upon cell sorting using Exendin-4-Cy3 as a β-cell sorting marker.
Supplementary Figure 3. Ctrl and βKlf6KO mice.
Supplementary Figure 4. Characterization of Glucose Homeostasis in βKlf6KO Mice. (a) Body weight (b) Blood glucose (c) Plasma insulin, and (d) Plasma glucagon in 10–12-week-old mice (n = 12). (e) Pancreatic insulin content (n = 8). (f) Glucose-stimulated insulin secretion (GSIS) (n = 6) and (g) insulin content from isolated islets from 12-week-old male mice (n = 6–7). ∗∗∗P < 0.001. (h,j) i.p. Glucose tolerance test (GTT) in 12 (n = 6–8♂; n = 6♀) and (i,k) 36-week-old male and female mice (n = 6–9♂; n = 6♀). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗∗P < 0.01. Statistical analyses for a and e were performed using a two-tailed unpaired Student's t-test.
Supplementary Figure 5. Representative H&E stained pancreas sections from Ctrl and βKlf6KO mice. Left: pregnant and non-pregnant mice. Middle: HFHS and RC fed mice. Right: saline or S961 treated mice. Upper figures: scale bar 200 μm; bottom figures: scale bar 50 μm.
Supplementary Figure 6. Characterization of Glucose Homeostasis in HFHS-Fed βKlf6KO Mice. Cohorts of (a-c) male and (d-f) female Ctrl and βKlf6KO mice were fed a RC or a HFHS diet for 22 weeks. (a,d) Body weight (n = 9–12♂; n = 8–11♀), (b,e) i.p. glucose tolerance test (GTT) (n = 11–13♂; n = 8–11♀) and (c,f) plasma insulin levels (n = 8–10♂; n = 8–11♀) after i.p. glucose injection. Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗P < 0.05; ∗∗P < 0.01; ∗P < 0.001.
Supplemental Figure 7. β-Cell surface area in islets from 10 to 12-week-old male Ctrl and ßKlf6KO mice treated with saline or S961 for 7 days (n = 3). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗∗P < 0.01; ∗∗∗P < 0.001.
Supplementary Figure 8. Impaired Insulinemic Response to Insulin Resistance in 2-day S961-treated βKlf6KO Mice. Twelve-week-old male Ctrl and βKlf6KO mice treated with S961 for 2 days. (a) Evolution of glycemia over a 2-day period. (b) Plasma insulin levels at days 1 and 2. (n = 6–8). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗∗P < 0.01; ∗∗∗P < 0.001.
Supplementary Figure 9. Impaired Proliferation in Islet Monolayers from βKlf6KO Mice. (a) Ki67 staining measurements in isolated islets harvested from 12-week-old Ctrl and βKlf6KO mice cultured as monolayers on an extracellular matrix (ECM) plate. Islet monolayers were cultured in the presence or absence of 100 nM of Exendin-4 for 48 h (n = 5–7). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗∗P < 0.01; ∗∗∗P < 0.001. (b) Representative immunofluorescence detection. Scale bar: 50 μm.
Supplemental Figure 10. Increased expression of Aldh1a3 in islets from Ctrl and βKlf6KO mice after a 7-day S961 treatment. Scale bar: 50 μm Aldh1a3 expression is detected in islet cells only after S961 treatment.
Genes differentially expressed in islets from Ctrl and βKlf6KO mice treated with saline for 2 days.
Supplementary Table 2. Genes differentially expressed in islets from Ctrl and βKlf6KO mice treated with saline for 2 days.
Supplementary Table 3. Differential Gene Expression Analysis Under Saline Treatment. List of genes differentially in islets from Ctrl and βKlf6KO mice treated with saline for 2 days and pertaining to the gene ontology terms a) cell division and b) metabolism. Genes are identified by symbol and full name. Log2[Fold Change (βKlf6KO vs. Ctrl)] and adj.P.Value (p-value adjusted for Benjamini and Hochberg's false discovery rate).
Supplementary Table 4. Differential Gene Expression Analysis Under S961 Treatment. List of genes differentially in islets from Ctrl and βKlf6KO mice treated with S961 for 2 days and pertaining to the gene ontology terms a) cell division and b) metabolism using. Genes are identified by symbol and full name. Log2[Fold Change (βKlf6KO vs. Ctrl)], and adj.P.Value (p-value adjusted for Benjamini and Hochberg's false discovery rate).
Supplementary Table 5. List of primers used in this study.
References
- 1.Rutter G.A., Pullen T.J., Hodson D.J., Martinez-Sanchez A. Pancreatic beta-cell identity, glucose sensing and the control of insulin secretion. Biochemical Journal. 2015;466(2):203–218. doi: 10.1042/BJ20141384. [DOI] [PubMed] [Google Scholar]
- 2.Thorens B. The required beta cell research for improving treatment of type 2 diabetes. Journal of Internal Medicine. 2013;274(3):203–214. doi: 10.1111/joim.12096. [DOI] [PubMed] [Google Scholar]
- 3.Prentki M., Nolan C.J. Islet beta cell failure in type 2 diabetes. Journal of Clinical Investigation. 2006;116(7):1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 5.Rahier J., Guiot Y., Goebbels R.M., Sempoux C., Henquin J.C. Pancreatic beta-cell mass in European subjects with type 2 diabetes. Diabetes, Obesity and Metabolism. 2008;10(Suppl 4):32–42. doi: 10.1111/j.1463-1326.2008.00969.x. [DOI] [PubMed] [Google Scholar]
- 6.Jonas J.C., Sharma A., Hasenkamp W., Ilkova H., Patane G., Laybutt R. Chronic hyperglycemia triggers loss of pancreatic beta cell differentiation in an animal model of diabetes. Journal of Biological Chemistry. 1999;274(20):14112–14121. doi: 10.1074/jbc.274.20.14112. [DOI] [PubMed] [Google Scholar]
- 7.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorens B., Wu Y.J., Leahy J.L., Weir G.C. The loss of GLUT2 expression by glucose-unresponsive beta cells of db/db mice is reversible and is induced by the diabetic environment. Journal of Clinical Investigation. 1992;90(1):77–85. doi: 10.1172/JCI115858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa Y., Noma Y., Davalli A.M., Wu Y.J., Thorens B., Bonner-Weir S. Loss of glucose-induced insulin secretion and GLUT2 expression in transplanted beta-cells. Diabetes. 1995;44(1):75–79. doi: 10.2337/diab.44.1.75. [DOI] [PubMed] [Google Scholar]
- 10.Gremlich S., Bonny C., Waeber G., Thorens B. Fatty acids decrease IDX-1 expression in rat pancreatic islets and reduce GLUT2, glucokinase, insulin, and somatostatin levels. Journal of Biological Chemistry. 1997;272(48):30261–30269. doi: 10.1074/jbc.272.48.30261. [DOI] [PubMed] [Google Scholar]
- 11.Gremlich S., Roduit R., Thorens B. Dexamethasone induces posttranslational degradation of GLUT2 and inhibition of insulin secretion in isolated pancreatic beta cells. Comparison with the effects of fatty acids. Journal of Biological Chemistry. 1997;272(6):3216–3222. doi: 10.1074/jbc.272.6.3216. [DOI] [PubMed] [Google Scholar]
- 12.Poitout V., Robertson R.P. Glucolipotoxicity: fuel excess and beta-cell dysfunction. Endocrine Reviews. 2008;29(3):351–366. doi: 10.1210/er.2007-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahn S.E. Clinical review 135: the importance of beta-cell failure in the development and progression of type 2 diabetes. Journal of Clinical Endocrinology & Metabolism. 2001;86(9):4047–4058. doi: 10.1210/jcem.86.9.7713. [DOI] [PubMed] [Google Scholar]
- 14.Cruciani-Guglielmacci C., Bellini L., Denom J., Oshima M., Fernandez N., Normandie-Levi P. Molecular phenotyping of multiple mouse strains under metabolic challenge uncovers a role for Elovl2 in glucose-induced insulin secretion. Mol Metab. 2017;6(4):340–351. doi: 10.1016/j.molmet.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellini L., Campana M., Rouch C., Chacinska M., Bugliani M., Meneyrol K. Protective role of the ELOVL2/docosahexaenoic acid axis in glucolipotoxicity-induced apoptosis in rodent beta cells and human islets. Diabetologia. 2018;61(8):1780–1793. doi: 10.1007/s00125-018-4629-8. [DOI] [PubMed] [Google Scholar]
- 17.McConnell B.B., Yang V.W. Mammalian Kruppel-like factors in health and diseases. Physiological Reviews. 2010;90(4):1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorens B., Sarkar H.K., Kaback H.R., Lodish H.F. Cloning and functional expression in bacteria of a novel glucose transporter present in liver, intestine, kidney, and beta-pancreatic islet cells. Cell. 1988;55(2):281–290. doi: 10.1016/0092-8674(88)90051-7. [DOI] [PubMed] [Google Scholar]
- 19.Thorens B., Tarussio D., Maestro M.A., Rovira M., Heikkila E., Ferrer J. Ins1(Cre) knock-in mice for beta cell-specific gene recombination. Diabetologia. 2015;58(3):558–565. doi: 10.1007/s00125-014-3468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leow C.C., Wang B.E., Ross J., Chan S.M., Zha J., Carano R.A. Prostate-specific Klf6 inactivation impairs anterior prostate branching morphogenesis through increased activation of the Shh pathway. Journal of Biological Chemistry. 2009;284(31):21057–21065. doi: 10.1074/jbc.M109.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basco D., Zhang Q., Salehi A., Tarasov A., Dolci W., Herrera P. alpha-cell glucokinase suppresses glucose-regulated glucagon secretion. Nature Communications. 2018;9(1):546. doi: 10.1038/s41467-018-03034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klinger S., Poussin C., Debril M.B., Dolci W., Halban P.A., Thorens B. Increasing GLP-1-induced beta-cell proliferation by silencing the negative regulators of signaling cAMP response element modulator-alpha and DUSP14. Diabetes. 2008;57(3):584–593. doi: 10.2337/db07-1414. [DOI] [PubMed] [Google Scholar]
- 23.Tarussio D., Metref S., Seyer P., Mounien L., Vallois D., Magnan C. Nervous glucose sensing regulates postnatal beta cell proliferation and glucose homeostasis. Journal of Clinical Investigation. 2014;124(1):413–424. doi: 10.1172/JCI69154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornu M., Modi H., Kawamori D., Kulkarni R.N., Joffraud M., Thorens B. Glucagon-like peptide-1 increases beta-cell glucose competence and proliferation by translational induction of insulin-like growth factor-1 receptor expression. Journal of Biological Chemistry. 2010;285(14):10538–10545. doi: 10.1074/jbc.M109.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solimena M., Schulte A.M., Marselli L., Ehehalt F., Richter D., Kleeberg M. Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia. 2018;61(3):641–657. doi: 10.1007/s00125-017-4500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cigliola V., Ghila L., Thorel F., van Gurp L., Baronnier D., Oropeza D. Pancreatic islet-autonomous insulin and smoothened-mediated signalling modulate identity changes of glucagon(+) alpha-cells. Nature Cell Biology. 2018;20(11):1267–1277. doi: 10.1038/s41556-018-0216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rieck S., Kaestner K.H. Expansion of beta-cell mass in response to pregnancy. Trends in Endocrinology and Metabolism. 2010;21(3):151–158. doi: 10.1016/j.tem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffer L., Brand C.L., Hansen B.F., Ribel U., Shaw A.C., Slaaby R. A novel high-affinity peptide antagonist to the insulin receptor. Biochemical and Biophysical Research Communications. 2008;376(2):380–383. doi: 10.1016/j.bbrc.2008.08.151. [DOI] [PubMed] [Google Scholar]
- 32.Ros S., Schulze A. Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer & Metabolism. 2013;1(1):8. doi: 10.1186/2049-3002-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartrons R., Simon-Molas H., Rodriguez-Garcia A., Castano E., Navarro-Sabate A., Manzano A. Fructose 2,6-bisphosphate in cancer cell metabolism. Front Oncol. 2018;8:331. doi: 10.3389/fonc.2018.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 35.Swisa A., Avrahami D., Eden N., Zhang J., Feleke E., Dahan T. PAX6 maintains beta cell identity by repressing genes of alternative islet cell types. Journal of Clinical Investigation. 2017;127(1):230–243. doi: 10.1172/JCI88015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni Q., Gu Y., Xie Y., Yin Q., Zhang H., Nie A. Raptor regulates functional maturation of murine beta cells. Nature Communications. 2017;8:15755. doi: 10.1038/ncomms15755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim-Muller J.Y., Fan J., Kim Y.J., Lee S.A., Ishida E., Blaner W.S. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic beta cells in diabetic mice. Nature Communications. 2016;7:12631. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahan T., Ziv O., Horwitz E., Zemmour H., Lavi J., Swisa A. Pancreatic beta-cells express the fetal islet hormone gastrin in rodent and human diabetes. Diabetes. 2017;66(2):426–436. doi: 10.2337/db16-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cinti F., Bouchi R., Kim-Muller J.Y., Ohmura Y., Sandoval P.R., Masini M. Evidence of beta-cell dedifferentiation in human type 2 diabetes. Journal of Clinical Endocrinology & Metabolism. 2016;101(3):1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stancill J.S., Cartailler J.P., Clayton H.W., O'Connor J.T., Dickerson M.T., Dadi P.K. Chronic beta-cell depolarization impairs beta-cell identity by disrupting a network of Ca(2+)-regulated genes. Diabetes. 2017;66(8):2175–2187. doi: 10.2337/db16-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roefs M.M., Carlotti F., Jones K., Wills H., Hamilton A., Verschoor M. Increased vimentin in human alpha- and beta-cells in type 2 diabetes. Journal of Endocrinology. 2017;233(3):217–227. doi: 10.1530/JOE-16-0588. [DOI] [PubMed] [Google Scholar]
- 42.Sakano D., Shiraki N., Kikawa K., Yamazoe T., Kataoka M., Umeda K. VMAT2 identified as a regulator of late-stage beta-cell differentiation. Nature Chemical Biology. 2014;10(2):141–148. doi: 10.1038/nchembio.1410. [DOI] [PubMed] [Google Scholar]
- 43.Pullen T.J., Khan A.M., Barton G., Butcher S.A., Sun G., Rutter G.A. Identification of genes selectively disallowed in the pancreatic islet. Islets. 2010;2(2):89–95. doi: 10.4161/isl.2.2.11025. [DOI] [PubMed] [Google Scholar]
- 44.Brereton M.F., Iberl M., Shimomura K., Zhang Q., Adriaenssens A.E., Proks P. Reversible changes in pancreatic islet structure and function produced by elevated blood glucose. Nature Communications. 2014;5:4639. doi: 10.1038/ncomms5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollak N.M., Hoffman M., Goldberg I.J., Drosatos K. Kruppel-like factors: crippling and un-crippling metabolic pathways. JACC Basic Transl Sci. 2018;3(1):132–156. doi: 10.1016/j.jacbts.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andreoli V., Gehrau R.C., Bocco J.L. Biology of Kruppel-like factor 6 transcriptional regulator in cell life and death. IUBMB Life. 2010;62(12):896–905. doi: 10.1002/iub.396. [DOI] [PubMed] [Google Scholar]
- 47.DiFeo A., Martignetti J.A., Narla G. The role of KLF6 and its splice variants in cancer therapy. Drug Resistance Updates. 2009;12(1–2):1–7. doi: 10.1016/j.drup.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Sirach E., Bureau C., Peron J.M., Pradayrol L., Vinel J.P., Buscail L. KLF6 transcription factor protects hepatocellular carcinoma-derived cells from apoptosis. Cell Death & Differentiation. 2007;14(6):1202–1210. doi: 10.1038/sj.cdd.4402114. [DOI] [PubMed] [Google Scholar]
- 49.Mallipattu S.K., Horne S.J., D'Agati V., Narla G., Liu R., Frohman M.A. Kruppel-like factor 6 regulates mitochondrial function in the kidney. Journal of Clinical Investigation. 2015;125(3):1347–1361. doi: 10.1172/JCI77084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bechmann L.P., Gastaldelli A., Vetter D., Patman G.L., Pascoe L., Hannivoort R.A. Glucokinase links Kruppel-like factor 6 to the regulation of hepatic insulin sensitivity in nonalcoholic fatty liver disease. Hepatology. 2012;55(4):1083–1093. doi: 10.1002/hep.24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bechmann L.P., Vetter D., Ishida J., Hannivoort R.A., Lang U.E., Kocabayoglu P. Post-transcriptional activation of PPAR alpha by KLF6 in hepatic steatosis. Journal of Hepatology. 2013;58(5):1000–1006. doi: 10.1016/j.jhep.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qi W., Chen X., Holian J., Tan C.Y., Kelly D.J., Pollock C.A. Transcription factors Kruppel-like factor 6 and peroxisome proliferator-activated receptor-{gamma} mediate high glucose-induced thioredoxin-interacting protein. American Journal Of Pathology. 2009;175(5):1858–1867. doi: 10.2353/ajpath.2009.090263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qi W., Holian J., Tan C.Y., Kelly D.J., Chen X.M., Pollock C.A. The roles of Kruppel-like factor 6 and peroxisome proliferator-activated receptor-gamma in the regulation of macrophage inflammatory protein-3alpha at early onset of diabetes. The International Journal of Biochemistry & Cell Biology. 2011;43(3):383–392. doi: 10.1016/j.biocel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Modi H., Jacovetti C., Tarussio D., Metref S., Madsen O.D., Zhang F.P. Autocrine action of IGF2 regulates adult beta-cell mass and function. Diabetes. 2015;64(12):4148–4157. doi: 10.2337/db14-1735. [DOI] [PubMed] [Google Scholar]
- 55.Nalepa G., Barnholtz-Sloan J., Enzor R., Dey D., He Y., Gehlhausen J.R. The tumor suppressor CDKN3 controls mitosis. The Journal of Cell Biology. 2013;201(7):997–1012. doi: 10.1083/jcb.201205125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laitman B.M., Asp L., Mariani J.N., Zhang J., Liu J., Sawai S. The transcriptional activator kruppel-like factor-6 is required for CNS myelination. PLoS Biology. 2016;14(5) doi: 10.1371/journal.pbio.1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim-Muller J.Y., Zhao S., Srivastava S., Mugabo Y., Noh H.L., Kim Y.R. Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metabolism. 2014;20(4):593–602. doi: 10.1016/j.cmet.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu M.J., Wu W.C., Chang H.W., Lai Y.T., Lin C.H., Yu W.C. KLF10 affects pancreatic function via the SEI-1/p21Cip1 pathway. The International Journal of Biochemistry & Cell Biology. 2015;60:53–59. doi: 10.1016/j.biocel.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 59.Bonnefond A., Lomberk G., Buttar N., Busiah K., Vaillant E., Lobbens S. Disruption of a novel Kruppel-like transcription factor p300-regulated pathway for insulin biosynthesis revealed by studies of the c.-331 INS mutation found in neonatal diabetes mellitus. Journal of Biological Chemistry. 2011;286(32):28414–28424. doi: 10.1074/jbc.M110.215822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xin Y., Okamoto H., Kim J., Ni M., Adler C., Cavino K. Single-cell RNAseq reveals that pancreatic beta-cells from very old male mice have a young gene signature. Endocrinology. 2016;157(9):3431–3438. doi: 10.1210/en.2016-1235. [DOI] [PubMed] [Google Scholar]
- 61.Tschen S.I., Dhawan S., Gurlo T., Bhushan A. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009;58(6):1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selection of Klf6. (a) Heatmap of the correlations between metabolic traits and islet gene expressions modules. The Thistle 3 module is depicted with an arrow. (b) Correlations of Thistle 3 genes with module membership and fasted insulin levels (IS T0) and insulin levels at 15 (IS T15) and 90 min (IS T90) after an oral glucose load. (c) Boxplot showing expression of KLF6 (Affymetrix U133_Plus2 probeset 224,606_at) in human organ donor islets (N = 103). KLF6 shows increased expression in islets of individuals with type 2 diabetes (T2D, N = 19) compared to non-diabetic islets (ND, N = 84). The difference in expression between T2D and ND is statistically significant (limma moderated t-test correcting for age and gender, unadjusted p-value = 0.001, Benjamini-Hochberg adjusted p-value = 0.015, log2 fold-change = 0.27). Data were processed and analyzed as described previously [28].
Supplementary Figure 2. Klf6 Expression Distribution in Islet Cell Populations. Klf6 expression in islets, β-cells and non-β-cells of 12 weeks old male mice (one biological replicate from a pool of n = 6 C57Bl/6J mice) upon cell sorting using Exendin-4-Cy3 as a β-cell sorting marker.
Supplementary Figure 3. Ctrl and βKlf6KO mice.
Supplementary Figure 4. Characterization of Glucose Homeostasis in βKlf6KO Mice. (a) Body weight (b) Blood glucose (c) Plasma insulin, and (d) Plasma glucagon in 10–12-week-old mice (n = 12). (e) Pancreatic insulin content (n = 8). (f) Glucose-stimulated insulin secretion (GSIS) (n = 6) and (g) insulin content from isolated islets from 12-week-old male mice (n = 6–7). ∗∗∗P < 0.001. (h,j) i.p. Glucose tolerance test (GTT) in 12 (n = 6–8♂; n = 6♀) and (i,k) 36-week-old male and female mice (n = 6–9♂; n = 6♀). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗∗P < 0.01. Statistical analyses for a and e were performed using a two-tailed unpaired Student's t-test.
Supplementary Figure 5. Representative H&E stained pancreas sections from Ctrl and βKlf6KO mice. Left: pregnant and non-pregnant mice. Middle: HFHS and RC fed mice. Right: saline or S961 treated mice. Upper figures: scale bar 200 μm; bottom figures: scale bar 50 μm.
Supplementary Figure 6. Characterization of Glucose Homeostasis in HFHS-Fed βKlf6KO Mice. Cohorts of (a-c) male and (d-f) female Ctrl and βKlf6KO mice were fed a RC or a HFHS diet for 22 weeks. (a,d) Body weight (n = 9–12♂; n = 8–11♀), (b,e) i.p. glucose tolerance test (GTT) (n = 11–13♂; n = 8–11♀) and (c,f) plasma insulin levels (n = 8–10♂; n = 8–11♀) after i.p. glucose injection. Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗P < 0.05; ∗∗P < 0.01; ∗P < 0.001.
Supplemental Figure 7. β-Cell surface area in islets from 10 to 12-week-old male Ctrl and ßKlf6KO mice treated with saline or S961 for 7 days (n = 3). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗∗P < 0.01; ∗∗∗P < 0.001.
Supplementary Figure 8. Impaired Insulinemic Response to Insulin Resistance in 2-day S961-treated βKlf6KO Mice. Twelve-week-old male Ctrl and βKlf6KO mice treated with S961 for 2 days. (a) Evolution of glycemia over a 2-day period. (b) Plasma insulin levels at days 1 and 2. (n = 6–8). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗∗P < 0.01; ∗∗∗P < 0.001.
Supplementary Figure 9. Impaired Proliferation in Islet Monolayers from βKlf6KO Mice. (a) Ki67 staining measurements in isolated islets harvested from 12-week-old Ctrl and βKlf6KO mice cultured as monolayers on an extracellular matrix (ECM) plate. Islet monolayers were cultured in the presence or absence of 100 nM of Exendin-4 for 48 h (n = 5–7). Error bars represent SEM. Statistical analyses were performed using a two-way ANOVA (Bonferonni's post-hoc test) ∗∗P < 0.01; ∗∗∗P < 0.001. (b) Representative immunofluorescence detection. Scale bar: 50 μm.
Supplemental Figure 10. Increased expression of Aldh1a3 in islets from Ctrl and βKlf6KO mice after a 7-day S961 treatment. Scale bar: 50 μm Aldh1a3 expression is detected in islet cells only after S961 treatment.
Genes differentially expressed in islets from Ctrl and βKlf6KO mice treated with saline for 2 days.
Supplementary Table 2. Genes differentially expressed in islets from Ctrl and βKlf6KO mice treated with saline for 2 days.
Supplementary Table 3. Differential Gene Expression Analysis Under Saline Treatment. List of genes differentially in islets from Ctrl and βKlf6KO mice treated with saline for 2 days and pertaining to the gene ontology terms a) cell division and b) metabolism. Genes are identified by symbol and full name. Log2[Fold Change (βKlf6KO vs. Ctrl)] and adj.P.Value (p-value adjusted for Benjamini and Hochberg's false discovery rate).
Supplementary Table 4. Differential Gene Expression Analysis Under S961 Treatment. List of genes differentially in islets from Ctrl and βKlf6KO mice treated with S961 for 2 days and pertaining to the gene ontology terms a) cell division and b) metabolism using. Genes are identified by symbol and full name. Log2[Fold Change (βKlf6KO vs. Ctrl)], and adj.P.Value (p-value adjusted for Benjamini and Hochberg's false discovery rate).
Supplementary Table 5. List of primers used in this study.