Abstract
Humans are infected with many viruses, and the immune system mostly removes viruses and the infected cells. However, certain viruses have entered the human genome. Of the human genome, ∼45% is composed of transposable elements (long interspersed nuclear elements [LINEs], short interspersed nuclear elements [SINEs] and transposons) and 5-8% is derived from viral sequences with similarity to infectious retroviruses. If integration of retrovirus occurs in a germline, the integrated viral sequences are heritable. Accumulation of viral sequences has created the current human genome. This article summarizes recent studies of retroviruses in humans and bridges clinical fields and evolutionary genetics. First, we report the repertories of human-infective retroviruses. Second, we review endogenous retroviruses in the human genome and diseases associated with endogenous retroviruses. Third, we discuss the biological functions of endogenous retroviruses and propose the concept of accelerated human evolution via viruses. Finally, we present perspectives of virology in the field of evolutionary medicine.
Key Indexing Terms: Retrovirus, Human endogenous retrovirus, Disease, Human genome, Evolutionary medicine
INTRODUCTION
Currently there are 5,630 confirmed virus species on the earth,1 and Anthony et al estimated that at least 320,000 undiscovered viruses infect mammalian hosts.2 RNA viruses are a major threat to human health, and severe acute respiratory syndrome (SARS) coronavirus, Ebola virus and Middle East respiratory syndrome (MERS) coronavirus are recently known to cause pandemics.3 Of the discovered virus species, 214 viruses are known human-infective RNA viruses and 9 viruses are retroviruses (Table 1 ).3 The human-infective retroviruses are human T-lymphotropic viruses (HTLV), human immunodeficiency viruses (HIV) and simian foamy viruses (SFV), and consist of 3 genuses (Deltaretrovirus, Lentivirus and Spumavirus, respectively; Table 1). The range of hosts of retroviruses is narrow, and HIV can be acquired only from humans although HTLV and SFV are from suspected zoonotic origin and intraspecies transmission to humans after first infection in nonhuman primates.3 , 4 Of the human genome, 5-8% are endogenous retroviruses derived from viral sequences with similarity to the infectious retroviruses. Understanding the endogenous retroviruses is clinically important because the accumulation of viral sequences has created the current human genome, and it can cause diseases associated with endogenous retroviruses.
TABLE 1.
Human-infective retroviruses (Retroviridae species).
| Species | Genus | References |
|---|---|---|
| PTLV-1 | Deltaretrovirus | 45 |
| PTLV-2 | Deltaretrovirus | 46 |
| PTLV-3 | Deltaretrovirus | 47 |
| HIV-1 | Lentivirus | 48 |
| HIV-2 | Lentivirus | 49 |
| SIV | Lentivirus | 50 |
| SFVagm | Spumavirus | 51 |
| SFVmac | Spumavirus | 52 |
| SFV | Spumavirus | 53 |
The table shows human-infective retroviruses: Primate T-lymphotropic virus (PTLV), human immunodeficiency virus (HIV), simian immunodeficiency virus (SIV), African green monkey simian foamy virus (SFVagm), macaque simian foamy virus (SFVmac) and simian foamy virus (SFV).
HUMAN-INFECTIVE RETROVIRUSES
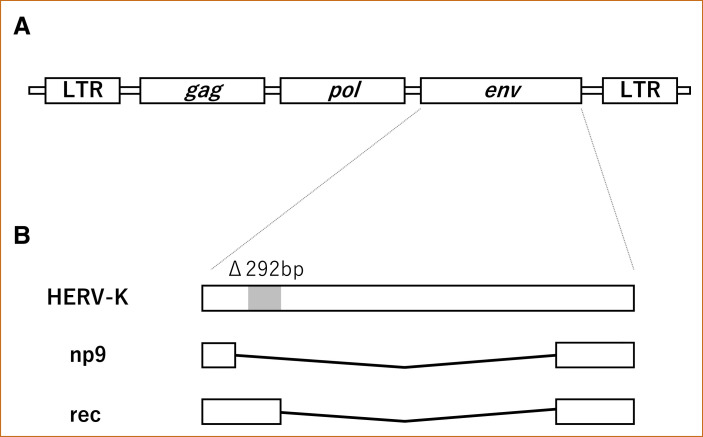
Retroviruses consist of a dimer of single-stranded positive sense RNA enclosed in a capsid of a lipid bilayer envelope and contain a reverse transcriptase (RT) enzyme.5 The retrovirus genome is composed of 3 genes (gag, pol and env) enclosed between 2 long terminal repeats: pol (polymerase) has RT and integrase function, gag (group antigens) is a polyprotein with processed matrix and core proteins, and env encodes envelope proteins (Figure 1 A).5
FIGURE 1.
Structure of a retrovirus genome and HERV-K. A, The retrovirus genome has at least 3 genes (gag, pol and env) enclosed between 2 long terminal repeats. B, HERV-K env genes originate 2 proteins (np9 and rec) with a different length, and np9 proteins present 292bp deletion.
Primate T-lymphotropic viruses (PTLVs) are composed of 3 distinct groups (PTLV-1, -2 and -3) and are also called HTLVs in the case of infections in humans. PTLVs are known to cause adult T-cell lymphoma/leukemia, and inflammation due to T lymphocytes in several tissues presents as bronchitis, uveitis and demyelinating diseases called HTLV-1 associated myelopathy/tropical spastic paraparesis. PTLV-1 is the first retrovirus discovered from T-cell lymphoma, and PTLV-2 was found originally in a patient with hairy cell leukemia.6 , 7 Lately, PTLV-3 was discovered in Central Africa,8 and Wolfe et al suggested that hunting and eating bushmeat is one of the transmission routes of PTLV beyond species. PTLVs are transmittable via sexual and maternal contact.9
HIV destroys lymphoid CD4 T cells by pyroptosis with caspase-1 activation and causes acquired immunodeficiency syndrome.10 , 11 HIV-1 and -2 are classified into 4 groups (M-P) and 8 groups (A-H), respectively, and are further finely grouped into subtypes whose configuration changes frequently.12 The origin of HIV-1 and -2 is believed to be independent transmission from different primates, since the HIV-1 and -2 sequences are similar to those of chimpanzee simian immunodeficiency virus (SIV) and Old World monkey SIV, respectively.13 HIVs are transmittable via direct contact by sexual, iatrogenic, broken skin and maternal routes, and SIV can be transmitted to humans from simian blood cells through broken skin.13, 14, 15
The association of SFV with specific diseases has not been clarified, although the prevalence of SFV in humans has been well studied.16 After infection by SFV, it has been observed that foamy degeneration and vacuolization occurs in the infected cells, with formation of numerous cytoplasmic vacuoles, and glycoproteins produced are expressed at the cell surface and result in fusion of cells.16 SFVs are known to be transmittable among simians only via direct contact with broken skin.17, 18, 19
ENDOGENOUS RETROVIRUSES IN HUMAN GENOME
The reverse transcripted retroviral genome can integrate into a host genome, and the integrated genome is heritable if the retrovirus infects a germ line of the host. The integrated retroviral sequence, which is 7-11 kb, is called a provirus.5 Of the human genome, 5-8% is believed to comprise proviruses with sequence similarity to genes and fragments of retroviruses, and the provirus sequences are also called human endogenous retroviruses (HERVs).20 Some HERVs still have open reading frames with the possibility of protein expression. There have been 3,173 HERV sequences identified from the human genome, and 39 canonical types of HERVs are categorized as classes I, II and III on the basis of sequences similar to different genera of infectious retroviruses (Gammaretrovirus/Epsilonretrovirus, Betaretrovirus and Spumaretrovirus, respectively).5 , 21, 22
It is believed that HERVs are associated with physiological functions and certain diseases based on model animal studies, but the role of HERVs is still under debate.5 Multiple sclerosis (MS) is caused by the destruction of myelin and oligodendrocytes, leading to axonal disruption in the brain and spinal cord. In MS patients, it has been reported that expression or abnormal representation of HERV-H, -K and -W and a polymorphism in HERV-Fc1 occurs.23 Secretion of env proteins coded by HERV-W in oligodendroglial precursor cells reduces oligodendroglial differentiation via activation of Toll-like receptor, and blocking their differentiation results in demyelinated and degenerating axons.24 HERV env proteins are related to complex pathological disorders and can be one of the targets for therapeutic approach.25 A monoclonal antibody against a HERV-W env protein is under clinical trial as a therapeutic approach for MS.26 , 27
Upregulation of HERV-W was also reported in blood cells and peripheral nerve lesions of chronic inflammatory demyelinating polyradiculoneuropathy patients, and chronic inflammatory demyelinating polyradiculoneuropathy might be caused by Toll-like receptor-mediated effects of env proteins on primary Schwann cells.28 In sporadic amyotrophic lateral sclerosis patients, env proteins of HERV-K are specifically expressed in cortical and spinal neurons and can cause cellular degeneration.29 RT proteins encoded by the pol gene of HERV-K have been detected in brain and blood tissues, but transmission of amyotrophic lateral sclerosis has not yet been demonstrated.23 , 30
HERVs are believed to be pathogenic in autoimmune rheumatic diseases and cancers, with evidence of increased expression of HERVs at the protein and RNA levels. Increased expression of HERVs was reported in patients with rheumatoid arthritis, systemic lupus erythematosus, juvenile idiopathic arthritis and Sjogren's syndrome.31 Nakkuntod et al reported hypomethylation of HERV-E and -K in systemic lupus erythematosus patients, and a lower methylation level can allow higher expression of HERV genes.32 HERV activation has also been reported in breast cancer, lymphoma, melanoma, ovarian cancer and prostate cancer, and the expression of env proteins can be involved in tumorigenesis via inducing cell-cell fusion.33 In addition, it is known that sequences derived from HERVs can be a trigger of cancers if partial sequences of HERVs or HERV itself translocates to regulatory regions of oncogenes.33 Tomlins et al reported that a translocation of HERV-K upstream of ETS translocation variant 1 caused cancerogenesis via enhanced expression of ETS translocation variant 1.34 HERV-K env genes originate 2 proteins (np9 and rec) with a different length by alternative splicing, and np9 proteins present 292bp deletion but rec is not (Figure 1B).25 It has been reported that rec and np9 contribute to tumorigenesis.25 Viral infections induced HERVs transactivation, and the HERVs transactivation causes enhancement of several signal transductions and transcription factors.35 HERV-K transactivated by Kaposi's sarcoma-associated herpes virus is suggested the involvement of tumorigenesis in Kaposi's sarcoma.36
EVOLUTIONARY MEDICINE OF ENDOGENOUS RETROVIRUSES
The endogenous retroviruses remaining in the genome should be neutral or advantageous. If all endogenous retroviruses were harmful, all of the retroviruses would have been excluded from the genome during evolution leading to humans. HERV-K is a case of beneficial endogenous retroviruses and has been integrated stepwise during primate evolution.37 It is known that HIV-1 infection stimulates a T cell response to HERV-K antigens because of protein similarity.38 Monde et al showed that gag proteins encoded by HERV-K changed the size and morphology of HIV particles, and these changes caused significant diminishing of the release efficiency and infectivity of HIV viruses.39
Vargiu et al estimated that HERVs diverged in the host genome from 6 to 100 million years ago, and this means the integration of HERVs occurred from after the divergence of Eutheria to the divergence between chimpanzees and humans.22 Syncytin-1 and -2 belong to the HERV-W and -FRD families, respectively, and both of them express env proteins with fusogenic activity and are involved in fusion of trophoblast cells.40 The syncytin-1 and -2 sequences are believed to have integrated 12-80 million years ago, and at the same time these sequences obtained mammalian-specific placental function.41 As in this case, HERVs acquired essential functions that can be evolutionarily preserved.
Retroviral sequences can accelerate the evolution of host genomes. In the host genome, the integrated retroviral elements can be a promotor or enhancer and provide alternative and aberrant sites for splicing of transcripts.5 Endogenous retroviruses can enhance recombination and rearrangement of the host genome via long terminal repeats, while crossover between HERV-I loci on the Y chromosome is a cause of male infertility due to deletion of an azoospermia factor-a region.42
CONCLUSIONS
In this review, we focused on known human-infective retroviruses and endogenous retroviruses in the human genome. It was previously believed that only retroviruses can integrate into the host genome, but Horie et al showed that sequences derived from nonretroviral RNA exist in the human genome. Bornaviruses, negative sense single stranded RNA viruses, encode 6 genes (M, X, P, N, G and L), and 2 proteins with high similarity to N genes, called endogenous borna-like N -1 and -2, have been found.43 It is believed that N genes of bornaviruses integrated by RT activation of long interspersed nuclear element, and this mechanism can enhance the integration of any viruses without dependence on retroviruses.43 The mechanism is the same as a system of making a processed gene, which was the evolutionary force to increase a member of a gene family in the human evolution.44 If the activation of long interspersed nuclear element enhances the integration of any viruses, there should be many more known and unknown viruses in the human genome. Whole genome sequencing of viruses and infected hosts will help to discover new viral sequences in the human genome, and the number of viral sequences in the genome could be higher than the current estimation based on the sequence similarity of only retroviruses (5-8%). Most viruses can infect specific tissues or cells in the human body, and it is not clear how a virus can recognize specific cells and invade cells. If there is affinity between the virus and cells, the virus might tend to get into the genome of specific cells with high affinity. If there is a niche in the localization of the virus, the niche might allow an entry route of the virus and thereby determine integration.
AUTHOR CONTRIBUTIONS
Y.K. and S.A. conceived and designed the analysis, and Y.K. wrote the paper.
Footnotes
This work was supported by JSPS KAKENHI 18K14766.
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.International Committee on Taxonomy of Viruses. https://talk.ictvonline.org/taxonomy/ [Accessed December 20, 2018].
- 2.Anthony SJ, Epstein JH, Murray KA, et al. A strategy to estimate unknown viral diversity in mammals. mBio. 2013;4 doi: 10.1128/mBio.00598-13. e00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woolhouse MEJ, Brierley L. Epidemiological characteristics of human-infective RNA viruses. Sci Data. 2018;5 doi: 10.1038/sdata.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 6.Poiesz BJ, Ruscetti FW, Gazdar AF, et al. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalyanaraman VS, Sarngadharan MG, Robert-Guroff M, et al. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science. 1982;218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 8.Calattini S, Chevalier SA, Duprez R, et al. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in Central Africa. Retrovirology. 2005;2:30. doi: 10.1186/1742-4690-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfe ND, Daszak P, Kilpatrick AM, et al. Bushmeat hunting, deforestation, and prediction of zoonotic disease. Emerg Infect Dis. 2005;11:1822–1827. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doitsh G, Galloway NL, Geng X, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monroe KM, Yang Z, Johnson JR, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharp PM, Bailes E, Chaudhuri RR, et al. The origins of acquired immune deficiency syndrome viruses: where and when? Philos Trans R Soc Lond B Biol Sci. 2001;356:867–876. doi: 10.1098/rstb.2001.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van't Wout AB, Kootstra NA, Mulder-Kampinga GA, et al. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visseaux B, Damond F, Matheron S, et al. Hiv-2 molecular epidemiology. Infect Genet Evol. 2016;46:233–240. doi: 10.1016/j.meegid.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Fouchet D1, Verrier D, Ngoubangoye B, et al. Natural simian immunodeficiency virus transmission in mandrills: a family affair? Proc Biol Sci. 2012;279:3426–3435. doi: 10.1098/rspb.2012.0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meiering CD, Linial ML. Historical perspective of foamy virus epidemiology and infection. Clin Microbiol. 2001;14:165–176. doi: 10.1128/CMR.14.1.165-176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweizer M, Schleer H, Pietrek M, et al. Genetic stability of foamy viruses: long-term study in an African green monkey population. J Virol. 1999;73:9256–9265. doi: 10.1128/jvi.73.11.9256-9265.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calattini S, Wanert F, Thierry B, et al. Modes of transmission and genetic diversity of foamy viruses in a Macaca tonkeana colony. Retrovirology. 2006;3:23. doi: 10.1186/1742-4690-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soliven K, Wang X, Small CT, et al. Simian foamy virus infection of rhesus macaques in Bangladesh: relationship of latent proviruses and transcriptionally active viruses. J Virol. 2013;87:13628–13639. doi: 10.1128/JVI.01989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belshaw R, Pereira V, Katzourakis A, et al. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci USA. 2004;101:4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths DJ. Endogenous retroviruses in the human genome sequence. Genome Biol. 2001;2:1017.1–1017.5. doi: 10.1186/gb-2001-2-6-reviews1017. reviews 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vargiu L, Rodriguez-Tomé P, Sperber GO, et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Küry P, Nath A, Créange A, et al. Human endogenous retroviruses in neurological diseases. Trends Mol Med. 2018;24:379–394. doi: 10.1016/j.molmed.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kremer D, Schichel T, Förster M, et al. Human endogenous retrovirus type W envelope protein inhibits oligodendroglial precursor cell differentiation. Ann Neurol. 2013;74:721–732. doi: 10.1002/ana.23970. [DOI] [PubMed] [Google Scholar]
- 25.Grandi N, Tramontano E. HERV envelope proteins: physiological role and pathogenic potential in cancer and autoimmunity. Front Microbiol. 2018;9:462. doi: 10.3389/fmicb.2018.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diebold M, Derfuss T. The monoclonal antibody GNbAC1: targeting human endogenous retroviruses in multiple sclerosis. Ther Adv Neurol Disord. 2019;12 doi: 10.1177/1756286419833574. 1756286419833574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niegowska M, Wajda-Cuszlag M, Stepien-Ptak G, et al. Anti-WENV antibodies are correlated with seroreactivity against Mycobacterium Avium subsp. paratuberculosis in children and youths at T1D risk. Sci Rep. 2019;9:6282. doi: 10.1038/s41598-019-42788-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faucard R, Alexandra Madeira A, Gehin N, et al. Human endogenous retrovirus and neuroinflammation in chronic inflammatory demyelinating polyradiculoneuropathy. EBioMedicine. 2016;6:190–198. doi: 10.1016/j.ebiom.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Lee MH, Henderson L, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7:307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douville RN, Liu J, Rothstein J, et al. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tugnet N, Rylance P, Roden D, et al. Human endogenous retroviruses (HERVs) and autoimmune rheumatic disease: is there a link? Open Rheumatol J. 2013;7:13–21. doi: 10.2174/1874312901307010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakkuntod J, Sukkapan P, Avihingsanon Y, et al. DNA methylation of human endogenous retrovirus in systemic lupus erythematosus. J Hum Genet. 2013;58:241–249. doi: 10.1038/jhg.2013.6. [DOI] [PubMed] [Google Scholar]
- 33.Bannert N, Hofmann H, Block A, et al. HERVs New role in cancer: from accused perpetrators to cheerful protectors. Front Microbiol. 2018;9:178. doi: 10.3389/fmicb.2018.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Foroozesh M, Qin Z. Transactivation of human endogenous retroviruses by tumor viruses and their functions in virus-associated malignancies. Oncogenesis. 2019;8:6. doi: 10.1038/s41389-018-0114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai L, Del Valle L, Miley W, et al. Transactivation of human endogenous retrovirus K (HERV-K) by KSHV promotes Kaposi's Sarcoma development. Oncogene. 2018;37:4534–4545. doi: 10.1038/s41388-018-0282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medstrand P, Mager DL. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrison KE, Jones RB, Meiklejohn DA, et al. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 2007;3:e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monde K, Terasawa H, Nakano Y, et al. Molecular mechanisms by which HERV-K Gag interferes with HIV-1 Gag assembly and particle infectivity. Retrovirology. 2017;14:27. doi: 10.1186/s12977-017-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blaise S, de Parseval N, Bénit L, et al. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imakawa K, Nakagawa S, Miyazawa T. Baton pass hypothesis: Successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution. Genes Cells. 2015;20:771–788. doi: 10.1111/gtc.12278. [DOI] [PubMed] [Google Scholar]
- 42.Kamp C, Hirschmann P, Voss H, et al. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- 43.Horie M, Honda T, Suzuki Y, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87. doi: 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katsura Y, Satta Y. Evolutionary history of the cancer immunity antigen MAGE gene family. PLoS One. 2011;6:e20365. doi: 10.1371/journal.pone.0020365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poiesz BJ, Ruscetti FW, Gazdar AF, et al. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalyanaraman VS, Sarngadharan MG, Robert-Guroff M, et al. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science. 1982;218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 47.Calattini S, Chevalier SA, Duprez R, et al. Human T-cell lymphotropic virus type 3: complete nucleotide sequence and characterization of the human Tax3 protein. J Virol. 2006;80:9876–9888. doi: 10.1128/JVI.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barré-Sinoussi F, Chermann JC, Rey F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 49.Kanki PJ, Barin F, M'Boup S, et al. New human T-lymphotropic retrovirus related to simian T-lymphotropic virus type III (STLV-IIIAGM) Science. 1986;232:238–243. doi: 10.1126/science.3006256. [DOI] [PubMed] [Google Scholar]
- 50.Khabbaz RF, Rowe T, Heneine WM, et al. Simian immunodeficiency virus needlestick accident in a laboratory worker. Lancet. 1992;340:271–273. doi: 10.1016/0140-6736(92)92358-m. [DOI] [PubMed] [Google Scholar]
- 51.Schweizer M, Falcone V, Gänge J, et al. Simian foamy virus isolatedfrom an accidentally infected human individual. J Virol. 1997;71:4821–4824. doi: 10.1128/jvi.71.6.4821-4824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brooks JI, Rud EW, Pilon RG, et al. Cross-species retroviral transmission from macaques to human beings. Lancet. 2002;360:387–388. doi: 10.1016/S0140-6736(02)09597-1. [DOI] [PubMed] [Google Scholar]
- 53.Achong BG, Mansell PW, Epstein MA. A new human virus in cultures from a nasopharyngeal carcinoma. J Pathol. 1971;103:P18. [PubMed] [Google Scholar]