Abstract
Background
Bone defects are a common clinical condition that has gained an increasing amount of attention in recent years. Causes of bone defect include tumors, inflammation, and fractures. Bone tissue engineering is a novel treatment of bone defect, and human mesenchymal stem cells (hMSCs) are the ideal seed cells for bone tissue engineering due to their multi-lineage differentiation potential and immunogenicity. The laminin α2 (LAMA2) gene encodes the α2 subunit of laminins. Mutations in this gene have been reported to cause muscular dystrophy, but thus far no studies have elucidated the role of LAMA2 in the fate choices of MSCs. Here, we aimed to investigate the critical role of LAMA2 in the osteogenesis and adipogenesis of mesenchymal stem cells (MSCs).
Methods
We investigated LAMA2 function in osteogenic and adipogenic differentiation of MSCs in vitro and in vivo through loss- and gain-of-function experiments. In addition, molecular mechanism was clarified by Western blot and siRNA.
Results
Our results demonstrated that LAMA2 was a critical regulator for fate commitment of MSCs. Both in vitro and in vivo studies indicate that LAMA2 inhibits osteogenesis and promotes adipogenesis. Mechanistically, we found that LAMA2 regulated osteogenesis and adipogenesis of MSCs by modulating the hedgehog signaling pathway.
Conclusions
The present work confirms that LAMA2 is a new molecular target for MSC-based bone regeneration.
Keywords: LAMA2, Osteogenic differentiation, Human mesenchymal stem cells, Hedgehog signaling
Background
Many diseases can cause bone defects, and the presence of bone defects leads to a poor quality of life. Recent studies have illustrated bone tissue engineering as a prospective therapeutic approach for bone regeneration [1, 2]. Seed cells, growth factors, and biological scaffolds are the three main factors of tissue engineering. Mesenchymal stem cells (MSCs) are the current focus of researchers as they can self-regenerate, have multidirectional differentiation ability, and are easy to obtain; therefore, they are a good candidate for use as seed cells in bone tissue engineering and have promising clinical prospects.
Many studies have demonstrated that MSCs can differentiate into osteoblasts, chondrocytes, and adipocytes [3, 4]. These findings have significantly promoted research into tissue engineering [5–7]. The balance between osteoblast and adipocyte formation is related to metabolic homeostasis [8, 9]. Many studies have shown that adipogenesis and osteogenesis are antagonistic to each other [10–13], but the underlying mechanisms remain largely unknown. Once the balance between osteogenesis and adipogenesis is broken, it would lead to bone metabolic diseases and even bone defects [14, 15]. Therefore, investigations of the molecular mechanism of osteogenic and adipogenic differentiation of MSCs would help in developing MSC-based treatment strategies for bone loss.
Laminins are composed of three subunits, named α, β, and γ. They regulate cell growth, movement, and attachment and participate in the formation of the basement membrane and attach to other proteins in the muscle cell. The LAMA2 gene encodes the α2 subunit of laminins, and mutations in this gene have been shown to cause LAMA2-related muscular dystrophy [16, 17]. A study showed that laminin could regulate the osteogenic differentiation of dental follicle cells, and LAMA2 was found to be upregulated in dexamethasone-induced dental follicle cells [18]. However, it remains unclear how LAMA2 regulates the fate choices of MSCs. Therefore, we aimed to investigate the critical role of LAMA2 in the osteogenesis and adipogenesis of MSCs in this study. To this end, we studied the function of LAMA2 in osteogenic and adipogenic differentiation of MSCs in vitro and in vivo through loss- and gain-of-function experiments. The molecular mechanisms were studied by small interfering RNAs (siRNA) and Western blot analyses.
Methods
Culture, osteogenic induction, and adipogenic induction of MSCs
Primary human adipose-derived stem cells (hASCs) and human bone marrow mesenchymal stem cells (hBMMSCs) were obtained from ScienCell (San Diego, CA, USA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) or α-minimum essential medium (α-MEM). The proliferation medium (PM) contained 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) antibiotics. The osteogenic medium (OM) contained 10% (v/v) FBS, 1% (v/v) antibiotics, 10 nM dexamethasone, 200 μM ascorbic acid, and 10 mM β-glycerophosphate. The adipogenic medium (AM) contained 10% (v/v) FBS, 1% (v/v) antibiotics, 50 nM insulin, 100 nM dexamethasone, 500 μM 3-isobutyl-1-methylxanthine, and 200 μM indomecin.
Lentivirus infection
Lentiviruses targeting LAMA2 (shLAMA2-1 and shLAMA2-2), negative control vectors (NC), and lentiviruses that segmentally express the LAMA2 gene were purchased from GenePhama Co. (Suzhou, China). The sequences were as follows: shLAMA2-1, GCCTGAGATTTCAGAGGATCC, and shLAMA2-2, GCTCCCTATCTGGGAAACAAA. First, hASCs and hBMMSCs were cultured to 50% confluence and transfected in the presence of 5 μg/mL polybrene to promote transfection for 24 h. The lentivirus carried the plasmid vector containing fluorescence proteins and the puromycin resistance gene. Thus, stably transfected cells were detected by the addition of puromycin after incubation for 72 h.
RNA interference
siRNAs targeting GLI2 and NC were obtained from GenePharma, the sequences were presented in Additional file 1: Table S1. Cells were transfected with siRNAs using Lipofectamine 3000 (Invitrogen) and harvested for RNA and protein analyses after 48 h. For osteogenic and adipogenic differentiation, cells were transfected every 5 days in OM or AM and harvested after 7 or 21 days.
Alkaline phosphatase (ALP) staining and ALP activity, Alizarin Red S (ARS) staining and quantification
The cells were divided into PM and OM culture groups. ALP staining was performed using NBT/BCIP staining kit (CoWin Biotech, Beijing, China) on day 7 after osteogenic induction. ALP activity was quantified using ALP assay kit (Nanjing Jiancheng Bioengineering Institute). Absorbance was measured at 520 nm, and the ALP activity was calculated.
On day 14 after osteogenic induction, 2% Alizarin Red buffer (Sigma-Aldrich) was used to stain cells. To quantify mineral accumulation, 100 mM of cetylpyridine solution was added to the wells of a multiwell plate. Mineral accumulation was quantified by measuring the absorbance at 562 nm after completely dissolving the cells.
Oil Red O staining and quantification
Cells were inoculated and cultured in PM and AM separately, and Oil Red O staining and quantitative assessment were carried out on day 21 after adipogenic induction. The cells were fixed with 10% neutral formalin and then rinsed with 60% isopropanol. Cells were stained with Oil Red O working solution and observed under a microscope. For quantitative assessment, 100% isopropanol was added to each well containing the stained cells, after which absorbance was measured at 500 nm.
Real-time quantitative PCR (RT-qPCR)
Total cellular RNA was extracted from MSCs cultured in proliferation or differentiation medium for 7 and 14 days with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). PrimeScript RT Reagent Kit (Takara, Tokyo, Japan) was used to synthesize the cDNA. RT-qPCR was conducted with SYBR Green Master Mix on an ABI Prism 7500 real-time PCR System. Gene expression was normalized to the expression of GAPDH, which was used as the reference gene. The primer sequences of human GAPDH, RUNX2, BGLAP, PPARγ, C/EBPα, and LAMA2 and mouse Gapdh, Runx2, Pparγ, and Lama2 were presented in Additional file 1: Table S1.
Western blot analysis
For detection of proteins, the cells were lysed in lysis buffer containing 2% proteinase inhibitor. Proteins extracts were subjected to 5% SDS-PAGE and transferred to polyvinylidene fluoride membrane. The membrane was incubated with the primary antibodies overnight; then, it was incubated with peroxidase-conjugated secondary antibodies at room temperature. The visualized immunoreactive protein bands were detected using an enhanced chemiluminescence (ECL) kit (CWBIO, Beijing, China).
In vivo implantation of MSCs, ectopic bone, and ectopic adipose tissue formation
Lenti-NC-, Lenti-shLAMA2-1-, and Lenti-shLAMA2-2-transfected MSCs (P4) were cultured in PM for 1 week. The resultant cells were used for MSCs/β-TCP (RB-SK-005G) composite scaffold construction and implantation in nude mice. After trypsinization and resuspending, 1 × 106 cells from each of the three groups were inoculated into cryotubes containing β-TCP powder (approximately 3 × 2 × 2 mm3).
Lenti-NC-, Lenti-shLAMA2-1-, and Lenti-shLAMA2-2-transfected MSCs (P4) were cultured in AM for 1 week. 1 × 106 cells from each group were trypsinized and resuspended separately, mixed with the collagen membrane scaffold material (approximately 8 × 8 × 2 mm3 per tube), and placed in a cryotube.
Then, the mixtures (n = 10 per group) were placed in a shaker at 37 °C for 1 h and then centrifuged at 150g for 5 min to allow the cells to adhere to the β-TCP or collagen membrane scaffold. The mixtures were then implanted into female BALB/c nude mice. Eight weeks after implantation, the ectopic bone-like tissues were harvested and analyzed by hematoxylin and eosin (H&E), Masson’s trichrome, and immunohistochemical (IHC) staining. Adipose tissues were harvested after 6 weeks and the tissues were sectioned and analyzed by staining with H&E and Oil Red O.
Micro-computed tomography (micro-CT) analyses of mice
Eight-week-old mice (10 per group) underwent sham or ovarian surgery and were euthanized 3 months after the surgery. Femur samples were scanned using the Inveon MM System (Siemens) micro-CT. The scanning conditions were 60 kV, 500 μA, and precision 8.82 μm. Parametric analysis was performed using Inveon Research Workplace (Siemens) software. The analysis area was 0.5–1 mm proximal to the epiphysis. The parameters analyzed included bone volume/total volume (BV/TV), and trabecular thickness (Tb.Th), number of trabecular bone (Tb.N), and trabecular space (Tb.Sp).
Statistical analysis
SPSS 19.0 software was used for the statistical analysis. Data were expressed as means ± standard deviation. Differences between the two groups were analyzed by the independent two-tailed Student’s t tests. One-way ANOVA and Tukey’s post hoc test were used for comparison between groups. Values of p < 0.05 were considered statistically significant.
Results
LAMA2 is involved in MSCs cell fate determination
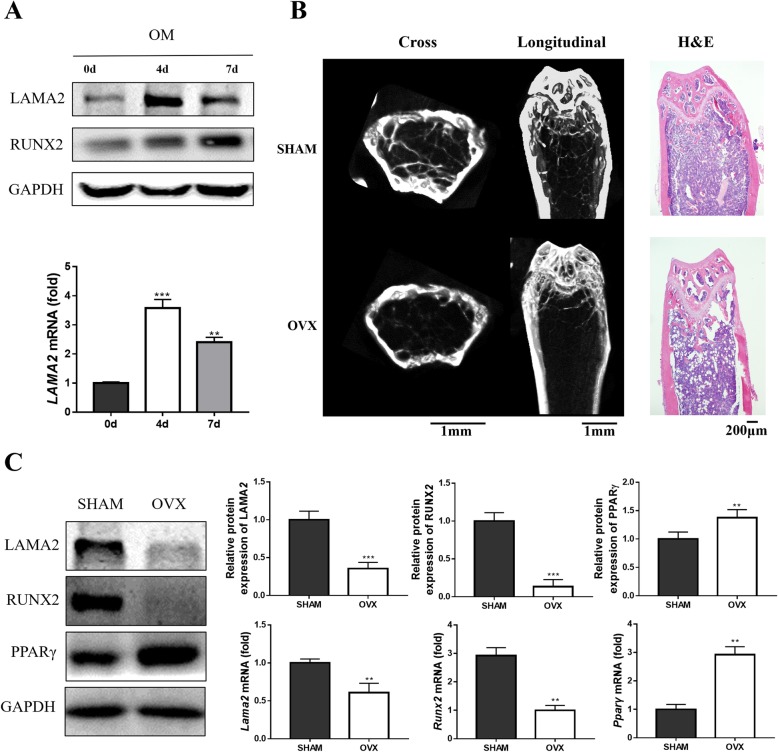
We studied the expression profile of LAMA2 to determine its involvement in osteogenic differentiation. LAMA2 expression initially increased during osteogenic differentiation and subsequently decreased (Fig. 1A). Then we investigate the status of LAMA2 in mice bone marrow mesenchymal stem cells. The results of the micro-CT scan and H&E staining showed a massive loss of trabecular bone in ovariectomy (OVX) mice (Fig. 1b and Additional file 2: Figure S1). Analysis of the LAMA2 mRNA and protein expression levels in BMMSCs revealed significantly decreased expression in OVX mice compared with sham mice (Fig. 1c).
Fig. 1.
LAMA2 is a potential target for osteoporosis treatment. a LAMA2 expression during the osteogenic differentiation determined by Western blot and RT-qPCR. b Representative micro-CT and H&E staining of bone loss in OVX mice. cLama2, Runx2, and Pparγ mRNA expression levels and LAMA2, RUNX2, and PPARγ protein levels in bone marrow-derived MSCs from OVX mice and sham mice. **p < 0.01, ***p < 0.001
LAMA2 inhibited osteogenic differentiation of MSCs in vitro
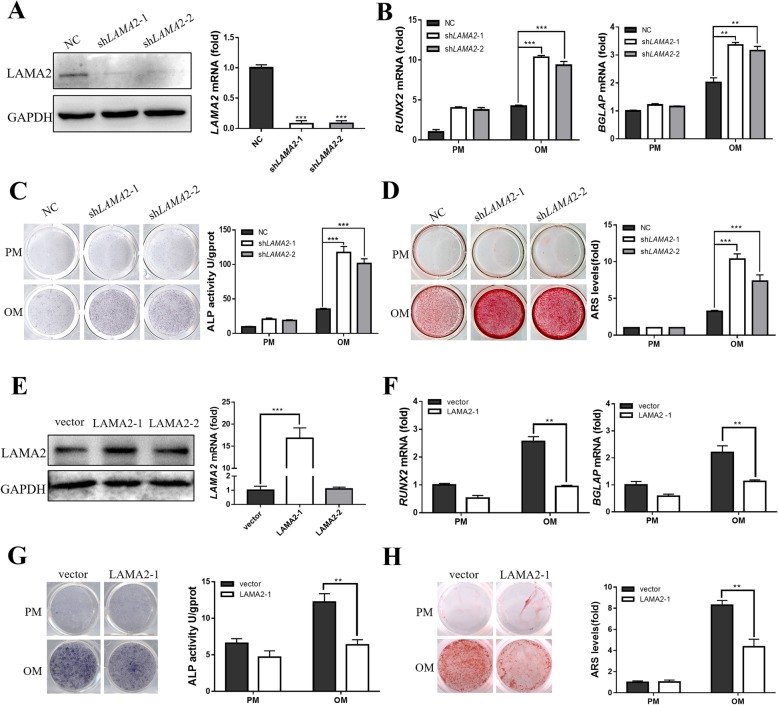
We established LAMA2 knockdown MSCs to further study the effects of LAMA2 on osteogenic differentiation. Transfection efficiency was confirmed by fluorescence microscopy (Additional file 3: Figure S2). We also analyzed the mRNA expression of LAMA2, and the LAMA2 knockdown groups (shLAMA2-1 and shLAMA2-2) exhibited a 90% decrease in expression compared with the NC group (Fig. 2a). Consistent with these findings, the results of the Western blot analysis also showed a significant decrease in the protein levels in the LAMA2 knockdown cells (Fig. 2a). Cells from the LAMA2 knockdown and control groups were incubated with either basal medium or osteogenic induction medium. RUNX2 and BGLAP expressions were detected on days 7 and 14, respectively. LAMA2 knockdown led to enhanced mRNA expression of RUNX2 and BGLAP (Fig. 2b). In addition, LAMA2 knockdown significantly enhanced osteogenesis, as indicated by ALP staining and quantification (Fig. 2c). Similar results were also observed following ARS staining and quantification (Fig. 2d).
Fig. 2.
LAMA2 inhibited osteogenic differentiation of MSCs in vitro. a LAMA2 expression detected by Western blot and RT-qPCR. b RT-qPCR results revealed that LAMA2 suppression enhanced the mRNA expression of RUNX2 and BGLAP. cLAMA2 knockdown enhanced osteogenesis, as indicated by ALP staining (left) and quantification (right) in MSCs. dLAMA2 knockdown enhanced ARS staining (left) and quantification (right). e LAMA2 expression detected by Western blot and RT-qPCR. fLAMA2 overexpression reduced the mRNA expression of RUNX2 and BGLAP detected by RT-qPCR. gLAMA2 overexpression reduced osteogenesis as indicated by ALP staining (left) and quantification (right) in MSCs. hLAMA2 overexpression reduced ARS staining (left) and quantification (right). **p < 0.01, ***p < 0.001
Next, we infected MSCs using fragments of the ectopic LAMA2 lentivirus (due to its long sequence and large molecular weight, we divided the LAMA2 protein into two fragments) and verified LAMA2 overexpression by RT-qPCR and Western blot analysis (Fig. 2e), the results showed that the cells infected by the first fragment could overexpress LAMA2 significantly. The results of the RT-qPCR analysis showed that RUNX2 and BGLAP expression levels were significantly decreased in LAMA2-overexpressing cells after osteogenic induction (Fig. 2f). Moreover, LAMA2 overexpression led to decreased staining with ALP (Fig. 2g), and ARS staining showed decreased mineralization in the MSCs overexpressing LAMA2 (Fig. 2h).
LAMA2 inhibited osteogenic differentiation of MSCs in vivo
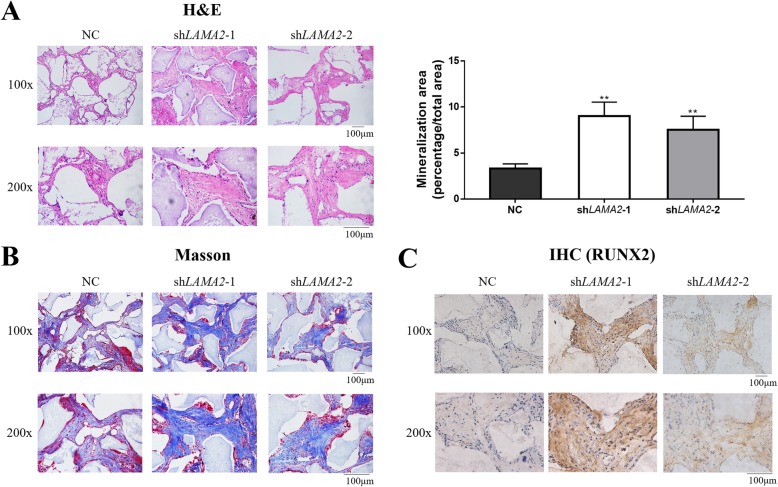
To further determine the role of LAMA2 in osteogenic differentiation, nude mice were implanted with MSCs stably expressing shLAMA2 and control cells mixed with β-TCP. The neo-generated tissues were collected for 8 weeks after implantation. Progressive tissue development was further characterized by H&E staining. The area of bone formation was larger in the shLAMA2 groups than in the NC group, as shown by quantitative measurements of bone-like tissues (Fig. 3a). In addition, LAMA2 knockdown groups had more bone tissue-like constructs as shown by Masson’s trichrome staining and IHC staining of RUNX2 (Fig. 3b, c).
Fig. 3.
LAMA2 inhibited osteogenic differentiation of MSCs in vivo. a The neo-generated tissues were characterized by H&E staining and showed newly formed bone-like tissue in the shLAMA2 groups. The area of bone formation of shLAMA2 groups were more than the NC group detected by quantitative measurements of bone-like tissues. b Masson’s staining of histological sections. c IHC staining of RUNX2. **p < 0.01
LAMA2 promoted adipogenic differentiation of MSCs
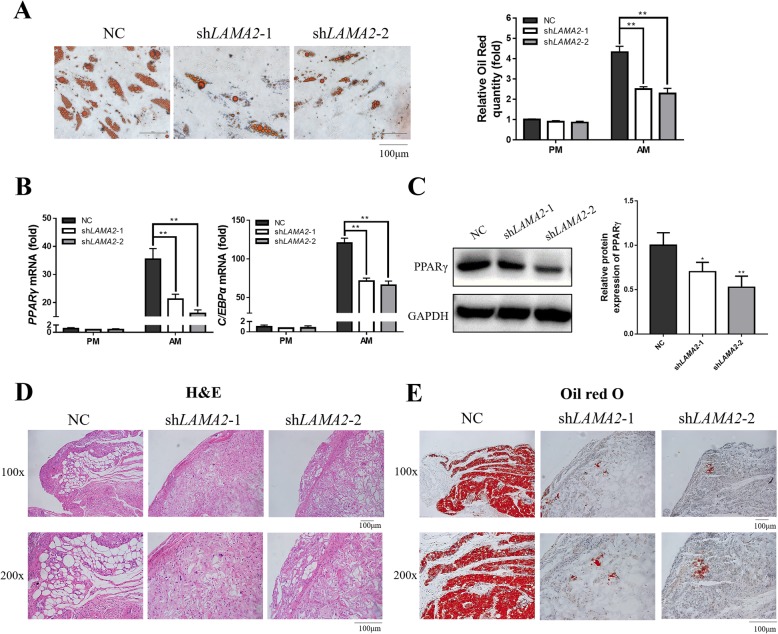
To further investigate the potential effect of LAMA2 during adipogenic differentiation of MSCs, LAMA2 knockdown MSCs were examined by Oil Red O staining after adipogenic induction. As shown in Fig. 4a, Oil Red O staining and quantification of LAMA2 knockdown cells showed significantly fewer lipid droplets compared with control cells. In addition, both PPARγ and C/EBPα expression levels in the control group were significantly higher than in the LAMA2 knockdown group (Fig. 4b). Moreover, Western blot analysis showed that PPARγ protein expression was downregulated during adipogenesis in LAMA2 knockdown MSCs (Fig. 4c). To further detect the effect of LAMA2 in adipogenic differentiation in vivo, we combined three MSCs groups (shLAMA2-1, shLAMA2-2, and NC) with a collagen sponge, and then implanted them into nude mice. The mixtures were collected after implantation for 6 weeks. Both H&E and Oil Red O staining results revealed fewer adipose tissue-like structures in the LAMA2 knockdown groups (Fig. 4d, e).
Fig. 4.
LAMA2 promoted adipogenic differentiation of MSCs in vitro and in vivo. a Knockdown of LAMA2 inhibited adipogenesis as indicated by Oil Red O staining. b Western blot (left) showed that the protein expression of PPARγ was upregulated in the LAMA2 knockdown group compared with the control group (NC) after 21 days of adipogenic induction. Western blot quantification (right) showed the same result. c Suppressing the expression of LAMA2 inhibited the mRNA expression of PPARγ and C/EBPα, as detected by RT-qPCR. d H&E staining illustrated that LAMA2 knockdown groups had fewer adipose tissue-like constructs in vivo. e Oil Red O staining illustrated that LAMA2 knockdown groups had fewer adipose tissue-like constructs in vivo. *p < 0.05, **p < 0.01
LAMA2 regulated the osteogenic and adipogenic differentiation of MSCs via hedgehog signaling pathway
To clarify the mechanism of LAMA2 regulating osteogenic and adipogenic differentiation, we screened key factors related to osteogenic and adipogenic differentiation and detected that hedgehog signaling was involved in the LAMA2-regulated osteogenic differentiation of MSCs.
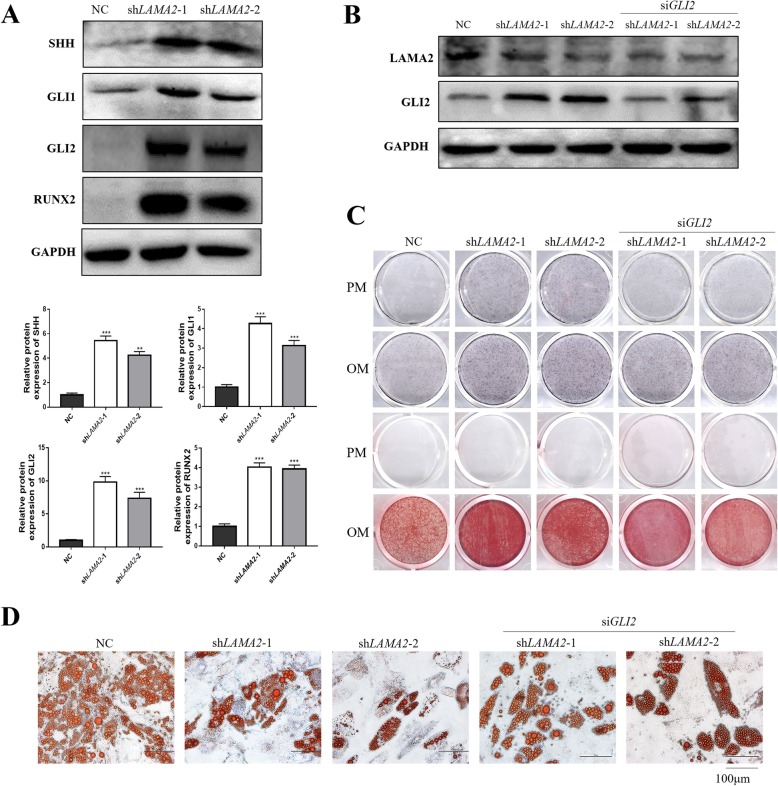
It was reported that the enhancement of the hedgehog pathway increased osteogenesis and inhibited adipogenesis [19–21]. We detected a dramatic increase in the expression of SHH, GLI1, and GLI2 of the hedgehog signaling pathway in LAMA2 knockdown cells (Fig. 5a). To further clarify LAMA2 regulation of osteogenesis and adipogenesis via the hedgehog pathway, we produced siGLI2 in MSCs to block the hedgehog pathway, and RT-qPCR and Western blot analysis were used to verify the efficiency of GLI2 silencing (Additional file 4: Figure S3). Then we established LAMA2 and GLI2 double knockdown cells and used Western blot analysis to verify the efficiency of double knockdown of LAMA2 and GLI2 (Fig. 5b). ALP and ARS staining showed that the double knockdown of LAMA2 and GLI2 inhibited the promoting effect of LAMA2 knockdown on osteogenesis (Fig. 5c). Moreover, the depressed adipogenic differentiation by LAMA2 knockdown was rescued in these double knockdown cells (Fig. 5d). Based on these results, our data suggest that LAMA2 regulates osteogenesis and adipogenesis by regulating the hedgehog signaling pathway.
Fig. 5.
LAMA2 regulated osteogenic differentiation of MSCs via hedgehog signaling pathway. aLAMA2 knockdown enhanced the expression of SHH, GLI1, GLI2, and RUNX2. b Double knockdown of LAMA2 and GLI2 validated by Western blot analysis. c ALP and ARS staining in the control, LAMA2 knockdown, and LAMA2/GLI2 knockdown groups. d Oil Red O staining in the control, LAMA2 knockdown, and LAMA2/GLI2 knockdown groups. **p < 0.01, ***p < 0.001
Discussion
In this study, we detected the crucial role of LAMA2 in the fate choices of MSCs. LAMA2 inhibition was found to promote osteogenic differentiation and inhibit adipogenic differentiation both in vitro and in vivo. Furthermore, LAMA2 overexpression could effectively suppress the osteogenic differentiation of MSCs.
Bone homeostasis depends on the resorption and formation of bones. Osteoporosis is caused by enhanced bone resorption and suppressed bone formation. Osteoporosis animal model is commonly used to study bone metabolism disorders and bone regeneration model, and OVX mice are a common osteoporosis model [22–24]. During the development of osteoporosis, BMMSCs have been shown to exhibit an increased number of adipogenic cells and a decline in the number of osteoblastic cells [25, 26]. It was worth noting that LAMA2 expression was reduced in OVX mice, but it increased first and then decreased during osteogenic differentiation. It was reasonable to assume that LAMA2 was degraded to prevent over-differentiation after early promotion of osteogenic differentiation.
Laminins are major basement membrane proteins consisting of alpha, beta, and gamma chains, and the LAMA1–5 genes encode laminin alpha 1–5 chains, respectively. Diseases associated with LAMA1 include the Poretti-Boltshauser syndrome and cerebellar dysplasia with cysts [27, 28]. The LAMA2 gene encodes the laminin alpha 2 chain, and mutations in this gene are thought to be responsible for merosin-deficient congenital muscular dystrophy [29]. LAMA3-related diseases include laryngeal cartilage skin syndrome and epidermolysis bullosa [30, 31], LAMA4-related diseases include cardiomyopathy [32], while LAMA5-related diseases include peripheral retinal degeneration and nephrotic syndrome [33, 34]. Several studies have shown that LAMA4 is closely related to adipogenesis. Vaicik et al. found that LAMA4 could affect adipose tissue expansion and function [35, 36], and Yamashita et al. found that laminin α4 chain fragment inhibited adipogenesis [37]. LAMA4 is located on chromosome 6q21 while LAMA2 is located on 6q22-q23 [38], so we want to further study whether LAMA2 plays a similar role as LAMA4 in adipogenesis. Some studies found an antagonistic relationship between the osteogenic and adipogenic differentiation of MSCs [39–41]. There is a balance between osteogenesis and adipogenesis, promoting the osteogenic differentiation and inhibiting the adipogenic differentiation of MSCs are the key directions in which bone regeneration progresses in bone tissue engineering. However, no studies have reported the effects of LAMA2 on osteogenesis and adipogenesis.
With an increase in age, the amount of adipose tissue increases while the amount of cancellous bone decreases. This is related to changes in the relative signaling pathways, which increases the differentiation of MSCs into adipocytes and decreases the differentiation of osteoblasts [8, 9, 42]. In this study, we found that LAMA2 regulates osteogenesis and adipogenesis of MSCs via modulating the hedgehog signaling pathway. The hedgehog signaling pathway is involved in bone formation and in osteogenic and adipogenic differentiation of MSCs which indicates that targeted regulation of hedgehog signaling is a potential target for the treatment of bone-related diseases such as osteoporosis and fracture healing [19, 20, 43–47].
Collectively, our research has some limitations. First, we did not determine the effects of LAMA2 overexpression on osteogenesis and adipogenesis in vivo. Second, we did not use a knockout mouse model in this study. Further studies on LAMA2 in a knockout mouse model investigating the effects on osteogenesis and adipogenesis should be performed.
Conclusions
This is the first study to demonstrate that LAMA2 is a negative regulator of osteogenesis and a positive modulator of adipogenesis both in vitro and in vivo via hedgehog signaling. This work pointed out a new important function of LAMA2 and clarified its underlying molecular mechanism of a directional way that could be used as a novel molecular target in bone tissue engineering.
Supplementary information
Additional file 1: Table S1. Sequences of RNA and DNA oligonucleotides.
Additional file 2: Figure S1. Bone volume, trabecular number, trabecular spacing, and trabecular thickness detected in Sham and OVX mice.
Additional file 3: Figure S2. Microscopic images of green fluorescence protein (GFP)-positive MSCs under ordinary and fluorescent light.
Additional file 4: Figure S3. RT-qPCR and Western blot analysis were used to verify the efficiency of GLI2 silencing in MSCs.
Acknowledgements
The authors are grateful to Dr. Xiao Zhao for the help in animal operations.
Abbreviations
- AM
Adipogenic medium
- ARS
Alizarin Red S
- BGLAP
Bone gamma-carboxyglutamic acid-containing protein
- BMMSCs
Bone marrow mesenchymal stem cells
- BV/TV
Trabecular bone volume/tissue volume
- C/EBPα
CCAAT/enhancer-binding protein α
- FBS
Fetal bovine serum
- GAPDH
Glyceraldehyde3-phosphate dehydrogenase
- GLI1
Glioma-associated oncogene family zinc finger 1
- GLI2
Glioma-associated oncogene family zinc finger 2
- H&E
Hematoxylin and eosin
- hASCs
Human adipose-derived stem cells
- hBMMSCs
Human bone marrow mesenchymal stem cells
- hMSCs
Human mesenchymal stem cells
- IHC
Immunohistochemical
- LAMA2
Laminin α2
- LP
Alkaline phosphatase
- Micro-CT
Micro-computed tomography
- MSCs
Mesenchymal stem cells
- OM
Osteogenic medium
- OVX
Ovariectomy
- PM
Proliferation medium
- PPARγ
Peroxisome proliferator-activated receptor γ
- RT-qPCR
Real-time quantitative PCR
- RUNX2
Runt-related transcription factor 2
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SHH
Sonic hedgehog signaling molecule
- siRNA
Small interfering RNA
- Tb.N
Trabecular number
- Tb.Sp
Trabecular spacing
- Tb.Th
Trabecular thickness
- β-TCP
β-Tricalcium phosphate
Authors’ contributions
YZ and XZ were responsible for the conception and design, collection and/or assembly of data, data analyses and interpretation, and manuscript writing. RG, XL, and SW were responsible for the collection and/or assembly of data and data analyses and interpretation in the animal experiments. DX, ZL, and XL were responsible for the collection and/or assembly of data and data analyses in the molecular biology experiments. PZ and YL were responsible for the conception and design, financial support, and manuscript writing. YZ was responsible for the conception and design and manuscript writing. The authors read and approved the final manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 81771039 and 81970908 to YL), the National Key Research and Development Program of China (No. 2016YFC1102900 to YL), Peking University Medicine Seed Fund for Interdisciplinary Research (No. BMU2018ME005 to YL), and the Beijing Nova Program (No. Z181100006218037 to PZ).
Availability of data and materials
The authors confirm that all data underlying the findings are fully available.
Ethics approval and consent to participate
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the Peking University Health Science Center (approval no. LA2016305). All surgeries were performed under anesthesia, and all efforts were made to minimize animal suffering.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuan Zhu and Xiao Zhang contributed equally to this work.
Contributor Information
Yuan Zhu, Email: pkukqzy@163.com.
Xiao Zhang, Email: zx8213163@163.com.
Ranli Gu, Email: guranli@163.com.
Xuenan Liu, Email: pkukqlxn@163.com.
Siyi Wang, Email: clara1970444525@163.com.
Dandan Xia, Email: dandan66x@126.com.
Zheng Li, Email: 704940647@qq.com.
Xiaomin Lian, Email: lianxiaomin@pku.edu.cn.
Ping Zhang, Email: zhangping332@bjmu.edu.cn.
Yunsong Liu, Email: liuyunsong@hsc.pku.edu.cn.
Yongsheng Zhou, Email: kqzhouysh@hsc.pku.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13287-020-01631-9.
References
- 1.Fröhlich M, Grayson WL, Wan LQ, Marolt D, Drobnic M, Vunjak-Novakovic G. Tissue engineered bone grafts: biological requirements, tissue culture and clinical relevance. Curr Stem Cell Res Therapy. 2008;3(4):254–264. doi: 10.2174/157488808786733962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marolt D, Knezevic M, Novakovic GV. Bone tissue engineering with human stem cells. Stem Cell Res Ther. 2010;1(2):10. doi: 10.1186/scrt10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres-Torrillas M, Rubio M, Damia E, Cuervo B, del Romero A, Peláez P, et al. Adipose-derived mesenchymal stem cells: a promising tool in the treatment of musculoskeletal diseases. Int J Molecular Sci. 2019;20(12):3105. [DOI] [PMC free article] [PubMed]
- 4.Ciuffi S, Zonefrati R, Brandi ML. Adipose stem cells for bone tissue repair. Clin Cases Mineral Bone Metabolism. 2017;14(2):217–226. doi: 10.11138/ccmbm/2017.14.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tae SK, Lee SH, Park JS, Im GI. Mesenchymal stem cells for tissue engineering and regenerative medicine. Biomed Materials (Bristol, England) 2006;1(2):63–71. doi: 10.1088/1748-6041/1/2/003. [DOI] [PubMed] [Google Scholar]
- 6.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 7.Seong JM, Kim BC, Park JH, Kwon IK, Mantalaris A, Hwang YS. Stem cells in bone tissue engineering. Biomed Materials (Bristol, England) 2010;5(6):062001. doi: 10.1088/1748-6041/5/6/062001. [DOI] [PubMed] [Google Scholar]
- 8.Muruganandan S, Roman AA, Sinal CJ. Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: cross talk with the osteoblastogenic program. Cell Molecular Life Sci. 2009;66(2):236–253. doi: 10.1007/s00018-008-8429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takada I, Kouzmenko AP, Kato S. Molecular switching of osteoblastogenesis versus adipogenesis: implications for targeted therapies. Expert Opin Ther Targets. 2009;13(5):593–603. doi: 10.1517/14728220902915310. [DOI] [PubMed] [Google Scholar]
- 10.Qiu W, Andersen TE, Bollerslev J, Mandrup S, Abdallah BM, Kassem M. Patients with high bone mass phenotype exhibit enhanced osteoblast differentiation and inhibition of adipogenesis of human mesenchymal stem cells. J Bone Miner Res. 2007;22(11):1720–1731. doi: 10.1359/jbmr.070721. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Z, Li Q, Luo S, Liu Z, Luo D, Zhang B, et al. PPARgamma and Wnt signaling in adipogenic and osteogenic differentiation of mesenchymal stem cells. Current Stem Cell Res Therapy. 2016;11(3):216–225. doi: 10.2174/1574888x10666150519093429. [DOI] [PubMed] [Google Scholar]
- 12.Lin Z, He H, Wang M, Liang J. MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Proliferation. 2019;52(6):e12688. [DOI] [PMC free article] [PubMed]
- 13.Yu B, Huo L, Liu Y, Deng P, Szymanski J, Li J, et al. PGC-1alpha controls skeletal stem cell fate and bone-fat balance in osteoporosis and skeletal aging by inducing TAZ. Cell Stem Cell. 2018;23(2):193–209. doi: 10.1016/j.stem.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Justesen J, Stenderup K, Ebbesen EN, Mosekilde L, Steiniche T, Kassem M. Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology. 2001;2(3):165–171. doi: 10.1023/a:1011513223894. [DOI] [PubMed] [Google Scholar]
- 15.Berendsen AD, Olsen BR. Osteoblast-adipocyte lineage plasticity in tissue development, maintenance and pathology. Cell Mole Life Sci. 2014;71(3):493–497. doi: 10.1007/s00018-013-1440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colognato H, Yurchenco PD. Form and function: the laminin family of heterotrimers. Dev Dynamics. 2000;218(2):213–234. doi: 10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira J, Gruber A, Cardoso M, Taipa R, Fineza I, Goncalves A, et al. LAMA2 gene mutation update: toward a more comprehensive picture of the laminin-alpha2 variome and its related phenotypes. Hum Mutat. 2018;39(10):1314–1337. doi: 10.1002/humu.23599. [DOI] [PubMed] [Google Scholar]
- 18.Viale-Bouroncle S, Gosau M, Morsczeck C. Laminin regulates the osteogenic differentiation of dental follicle cells via integrin-alpha2/-beta1 and the activation of the FAK/ERK signaling pathway. Cell Tissue Res. 2014;357(1):345–354. doi: 10.1007/s00441-014-1869-x. [DOI] [PubMed] [Google Scholar]
- 19.Spinella-Jaegle S, Rawadi G, Kawai S, Gallea S, Faucheu C, Mollat P, et al. Sonic hedgehog increases the commitment of pluripotent mesenchymal cells into the osteoblastic lineage and abolishes adipocytic differentiation. J Cell Sci. 2001;114(Pt 11):2085–2094. doi: 10.1242/jcs.114.11.2085. [DOI] [PubMed] [Google Scholar]
- 20.James AW, Leucht P, Levi B, Carre AL, Xu Y, Helms JA, et al. Sonic hedgehog influences the balance of osteogenesis and adipogenesis in mouse adipose-derived stromal cells. Tissue Eng Part A. 2010;16(8):2605–2616. doi: 10.1089/ten.tea.2010.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontaine C, Cousin W, Plaisant M, Dani C, Peraldi P. Hedgehog signaling alters adipocyte maturation of human mesenchymal stem cells. Stem Cells. 2008;26(4):1037–1046. doi: 10.1634/stemcells.2007-0974. [DOI] [PubMed] [Google Scholar]
- 22.Kalu DN. The ovariectomized rat model of postmenopausal bone loss. Bone Mineral. 1991;15(3):175–191. doi: 10.1016/0169-6009(91)90124-i. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Li W, Liu Y, Zhang X, Zhou Y. Co-administration of aspirin and allogeneic adipose-derived stromal cells attenuates bone loss in ovariectomized rats through the anti-inflammatory and chemotactic abilities of aspirin. Stem Cell Res Ther. 2015;6:200. doi: 10.1186/s13287-015-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Aijima R, Hoshino Y, Yamaza H, Tomoda E, Tanaka Y, et al. Transplantation of mesenchymal stem cells ameliorates secondary osteoporosis through interleukin-17-impaired functions of recipient bone marrow mesenchymal stem cells in MRL/lpr mice. Stem Cell Res Ther. 2015;6(1):104. doi: 10.1186/s13287-015-0091-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Zoelen EJ, Duarte I, Hendriks JM, van der Woning SP. TGFβ-induced switch from adipogenic to osteogenic differentiation of human mesenchymal stem cells: identification of drug targets for prevention of fat cell differentiation. Stem Cell Res Ther. 2016;7(1):123. doi: 10.1186/s13287-016-0375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambrosi TH, Scialdone A, Graja A, Gohlke S, Jank AM, Bocian C, et al. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell. 2017;20(6):771–784. doi: 10.1016/j.stem.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marlow E, Chan RVP, Oltra E, Rusu I, Gupta MP. Retinal avascularity and neovascularization associated with LAMA1 (laminin1) mutation in Poretti-Boltshauser syndrome. JAMA Ophthalmol. 2018;136(1):96–97. doi: 10.1001/jamaophthalmol.2017.5060. [DOI] [PubMed] [Google Scholar]
- 28.Aldinger KA, Mosca SJ, Tetreault M, Dempsey JC, Ishak GE, Hartley T, et al. Mutations in LAMA1 cause cerebellar dysplasia and cysts with and without retinal dystrophy. Am J Hum Genet. 2014;95(2):227–234. doi: 10.1016/j.ajhg.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yurchenco PD, McKee KK, Reinhard JR, Rüegg MA. Laminin-deficient muscular dystrophy: Molecular pathogenesis and structural repair strategies. Matrix Biol. 2018;71–72:174–187. doi: 10.1016/j.matbio.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLean WH, Irvine AD, Hamill KJ, Whittock NV, Coleman-Campbell CM, Mellerio JE, et al. An unusual N-terminal deletion of the laminin alpha3a isoform leads to the chronic granulation tissue disorder laryngo-onycho-cutaneous syndrome. Hum Mol Genet. 2003;12(18):2395–2409. doi: 10.1093/hmg/ddg234. [DOI] [PubMed] [Google Scholar]
- 31.Reimer A, Schwieger-Briel A, He Y, Leppert J, Schauer F, Kiritsi D, et al. Natural history and clinical outcome of junctional epidermolysis bullosa generalized intermediate due to a LAMA3 mutation. Br J Dermatol. 2018;178(4):973–975. doi: 10.1111/bjd.16088. [DOI] [PubMed] [Google Scholar]
- 32.Knoll R, Postel R, Wang J, Kratzner R, Hennecke G, Vacaru AM, et al. Laminin-alpha4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116(5):515–525. doi: 10.1161/CIRCULATIONAHA.107.689984. [DOI] [PubMed] [Google Scholar]
- 33.Napolitano F, Di Iorio V, Di Iorio G, Melone MAB, Gianfrancesco F, Simonelli F, et al. Early posterior vitreous detachment is associated with LAMA5 dominant mutation. Ophthalmic Genet. 2019;40(1):39–42. doi: 10.1080/13816810.2018.1558261. [DOI] [PubMed] [Google Scholar]
- 34.Braun DA, Warejko JK, Ashraf S, Tan W, Daga A, Schneider R, et al. Genetic variants in the LAMA5 gene in pediatric nephrotic syndrome. Nephrol Dialysis Trans. 2019;34(3):485–493. doi: 10.1093/ndt/gfy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaicik MK, Thyboll Kortesmaa J, Moverare-Skrtic S, Kortesmaa J, Soininen R, Bergstrom G, et al. Laminin alpha4 deficient mice exhibit decreased capacity for adipose tissue expansion and weight gain. PLoS One. 2014;9(10):e109854. doi: 10.1371/journal.pone.0109854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaicik MK, Blagajcevic A, Ye H, Morse MC, Yang F, Goddi A, et al. The absence of laminin α4 in male mice results in enhanced energy expenditure and increased beige subcutaneous adipose tissue. Endocrinology. 2018;159(1):356–367. doi: 10.1210/en.2017-00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita H, Goto C, Tajima R, Koparal AT, Kobori M, Ohki Y, et al. Cryptic fragment alpha4 LG4-5 derived from laminin alpha4 chain inhibits de novo adipogenesis by modulating the effect of fibroblast growth factor-2. Develop Growth Differ. 2008;50(2):97–107. doi: 10.1111/j.1440-169X.2007.00979.x. [DOI] [PubMed] [Google Scholar]
- 38.Aumailley M. The laminin family. Cell Adhes Migr. 2013;7(1):48–55. doi: 10.4161/cam.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawai M. Adipose tissue and bone: role of PPARgamma in adipogenesis and osteogenesis. Horm Mol Biol Clin Invest. 2013;15(3):105–113. doi: 10.1515/hmbci-2013-0036. [DOI] [PubMed] [Google Scholar]
- 40.Lee M-J, Chen H-T, Ho M-L, Chen C-H, Chuang S-C, Huang S-C, et al. PPARγ silencing enhances osteogenic differentiation of human adipose-derived mesenchymal stem cells. J Cell Mol Med. 2013;17(9):1188–1193. doi: 10.1111/jcmm.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marciano DP, Kuruvilla DS, Boregowda SV, Asteian A, Hughes TS, Garcia-Ordonez R, et al. Pharmacological repression of PPARgamma promotes osteogenesis. Nat Commun. 2015;6:7443. doi: 10.1038/ncomms8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veronesi F, Torricelli P, Borsari V, Tschon M, Rimondini L, Fini M. Mesenchymal stem cells in the aging and osteoporotic population. Crit Rev Eukaryot Gene Expr. 2011;21(4):363–377. doi: 10.1615/critreveukargeneexpr.v21.i4.60. [DOI] [PubMed] [Google Scholar]
- 43.Lv W-T, Du D-H, Gao R-J, Yu C-W, Jia Y, Jia Z-F, et al. Regulation of hedgehog signaling offers a novel perspective for bone homeostasis disorder treatment. Int J Mol Sci. 2019;20(16):3981. doi: 10.3390/ijms20163981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenzie JA, Maschhoff C, Liu X, Migotsky N, Silva MJ, Gardner MJ. Activation of hedgehog signaling by systemic agonist improves fracture healing in aged mice. J Orthop Res. 2019;37(1):51–59. doi: 10.1002/jor.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cousin W, Fontaine C, Dani C, Peraldi P. Hedgehog and adipogenesis: fat and fiction. Biochimie. 2007;89(12):1447–1453. doi: 10.1016/j.biochi.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Bao C, Kim JT, Cho JS, Qiu S, Lee HJ. Sulforaphene inhibition of adipogenesis via hedgehog signaling in 3T3-L1 adipocytes. J Agric Food Chem. 2018;66(45):11926–11934. doi: 10.1021/acs.jafc.8b04330. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y, Long F. Hedgehog signaling via Gli2 prevents obesity induced by high-fat diet in adult mice. eLife. 2017;6:e31649. doi: 10.7554/eLife.31649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sequences of RNA and DNA oligonucleotides.
Additional file 2: Figure S1. Bone volume, trabecular number, trabecular spacing, and trabecular thickness detected in Sham and OVX mice.
Additional file 3: Figure S2. Microscopic images of green fluorescence protein (GFP)-positive MSCs under ordinary and fluorescent light.
Additional file 4: Figure S3. RT-qPCR and Western blot analysis were used to verify the efficiency of GLI2 silencing in MSCs.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available.