Abstract
Background
Recent genome-wide association study showed rs10940346 locus near EMB gene was significantly associated with schizophrenia and suggested that EMB gene is one of the potentially causal genes for schizophrenia, but no causal variant has been identified. Our study aims to further verify EMB gene is a susceptibility gene for schizophrenia and to identify potentially causal variants in EMB gene that lead to schizophrenia.
Methods
Targeted sequencing for the un-translated region and all exons of EMB gene was performed among 1803 patients with schizophrenia and 997 healthy controls recruited from Chinese Han population.
Results
A total of 58 high-quality variants were identified in case and control groups. Seven of them are nonsynonymous rare variations, EMB: p.(Ala52Thr), p.(Glu66Gly), p.(Ser93Cys), p.(Ala118Val), p.(Ile131Met), p.(Gly163Arg) and p.(Arg238Tyr), but none of them reached statistical significance. Among them, p.(Ile131Met), p.(Gly163Arg) and p.(Arg238Tyr), were predicted to be deleterious variants. In addition, a common variant, rs3933097 located in 3′-UTR of EMB gene, achieved allelic and genotypic significance with schizophrenia (Pallele = 3.82 × 10− 6, Pgenotype = 3.18 × 10− 5).
Conclusions
Our research first presented a comprehensive mutation spectrum of exons and un-translated region in EMB gene for schizophrenia and provided additional evidence of EMB gene being a susceptibility gene for schizophrenia. However, further functional validations are necessary to reveal its role in the etiology of schizophrenia.
Keywords: Schizophrenia, EMB gene, Targeted next-generation sequencing
Background
Schizophrenia (MIM181500) is one of the most common types of severe mental disorders, and epidemiological studies show that its lifetime prevalence in the general population is about 1%. Schizophrenia is often chronic or subacute onset in young adults, and clinically, it often manifests itself as a variety of barriers involving perception, thinking, emotion and behavior, as well as a lack of coordination of mental activities. The course of schizophrenia is generally prolonged, recurrent, aggravated or deteriorated, and some patients eventually experience recession and mental disability.
It is generally accepted that the onset of schizophrenia is the result of genetic and environmental synergistic pathogenesis, with an estimated heritability of 70–85% [1]. The existing genome-wide association studies (GWAS) with schizophrenia have found more than 100 loci reached genome-wide significance [2]. The recent GWAS newly reported rs10940346 (P = 1.11 × 10− 8, odds ratio (OR) = 0.949, standard error (SE) = 0.009) locus near EMB gene is significantly associated with schizophrenia and EMB gene was reported to be the notable gene. The research also suggested that the EMB gene is one of the prioritized candidate genes, which had more than one line of supporting evidence, implicated in schizophrenia [3]. Moreover, the latest GWAS in schizophrenia also suggested that EMB gene is one of the potentially causal genes for schizophrenia [4].
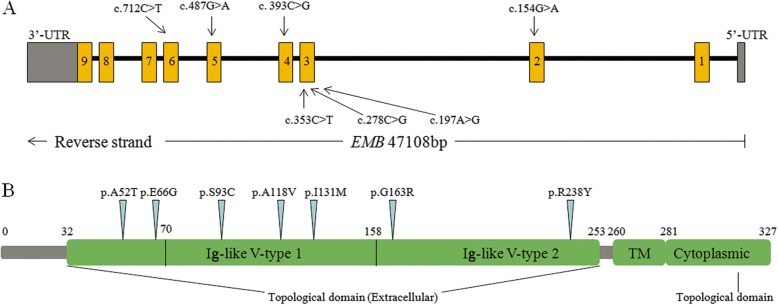
The EMB gene is located on a chromosome region of 5q11.1 and harbors 9 exons (Fig. 1a) which encodes a protein named embigin with a molecular weight of 30 kDa when unglycosylated [5]. Embigin, basigin and neuroplastin are in the same small subgroup of the immunoglobulin superfamily (IgSF) [6], while neuroplastin protein is coded by NPTN which was proven to be a susceptibility gene for schizophrenia [7] and played an important role in long-term potentiation, synaptic plasticity and neurite outgrowth, probably related to learning, memory and emotion [8]. Embigin is a highly glycosylated transmembrane protein consisting of an intracellular region containing 47 amino acids, a transmembrane region containing 29 amino acids, and an extracellular region containing two Ig domains [5](Fig. 1b). The previous study [9] shows that following muscle denervation, embigin promotes nerve terminal sprouting and the formation of additional acetylcholine receptor clusters at synaptic sites without affecting terminal Schwann cell number or morphology. It also delays the retraction of terminal sprouts following re-innervation of denervated endplates. So embigin plays an essential role in the growth of motor neurons and the formation of neuromuscular junctions.
Fig. 1.
Structure of EMB gene and embigin protein. a. EMB gene structure based on ENST00000303221, yellow boxes indicate the protein-coding exons. b. Embigin protein structure, TM is Transmembrane domain and the blue sites marked out represents the location of the mutation
Furthermore, embigin is an accessory protein of MCT2, participating in the ectopic process of MCT2 to the plasma membrane and maintaining the catalytic activity of MCT2 [10]. MCT2 acts as a monocarboxylic acid transporter, in combination with MCT1 and MCT4 synergistically transport lactic acid between glial cells and neurons [11]. Current research confirmed that lactic acid is not only an important energy substrate for energy metabolism in the brain, but also plays a vital role in the formation of long-term procedural memory [12]. Therefore, we infer that EMB gene may be related to brain energy metabolism and long-term memory formation.
Studies have shown that embigin is highly expressed in early embryos of mice, and subsequent expression is reduced [13], but it is still expressed in the heart, lung, brain and other tissues of adult rats [14]. Embigin was also reported to enhance integrin-mediated adhesion between cell matrices [15]. In summary, embigin is involved in early embryonic development, cell migration, and formation of neuromuscular junctions depending on neural cell adhesion molecules. The neurodevelopmental hypothesis of schizophrenia and the immune-inflammatory alterations have been widely supported. As a member of the IgSF, the EMB gene also plays an important role in embryonic development, which implies that there is a connection between the EMB gene and schizophrenia.
In recent years, next-generation sequencing technology has developed rapidly. Meanwhile its cost has been significantly reduced. Although EMB gene was suggested to be one of the potentially causal genes for schizophrenia, no real causal variant on EMB gene has been identified yet. Mutations in exons may lead to changes in amino acid sequence, which may alter the structure and function of the protein. The un-translated region (UTR) plays a key role in the regulation of gene expression. Therefore, the variants on the exons and UTR are more likely to be causal variants that cause changes in gene function and lead to disease. Sequencing the exons and UTR of EMB gene can help us to find out the causal variants of EMB gene for schizophrenia, further understand the role of EMB gene in schizophrenia, and lay a foundation for us to study the pathogenesis of schizophrenia. In order to scan pathogenic mutation sites related to schizophrenia in EMB gene, next-generation sequencing for the UTR and all exons of EMB gene in 1803 cases and 997 healthy controls were performed via the multiplex PCR technology and Illumina platform.
Methods
Subjects
The sample set includes 1803 unrelated patients with schizophrenia (1110 men and 693 women) and 997 unrelated healthy controls (437 men and 560 women). The mean age is 44.47 years (s.d. = 12.14) among patients with schizophrenia and 43.12 years (s.d. = 17.55) among healthy controls (Table 1).
Table 1.
Characteristics of the study sample set
| n | Age, years | ||||
|---|---|---|---|---|---|
| Men | Women | Total | Mean | s.d. | |
| Patients with SCZ | 1110 | 693 | 1803 | 44.47 | 12.14 |
| Healthy controls | 437 | 560 | 997 | 43.12 | 17.55 |
SCZ Schizophrenia
All the samples were recruited from Chinese Han population. All patients were interviewed by two independent psychiatrists from Wuxi Mental Health Center, Nanjing Medical University. Diagnoses were made strictly according to the DSM-IV criteria based on SCID-I (the Structured Clinical Interview for DSM-IV Axis I Disorders). Patients were excluded if they had suffered neurological illness, mood disorder, mental retardation, history of substance use and psychotic disorder due to general medical condition. Healthy controls were collected during the physical examination. All participants signed informed consent and the study was approved by the ethical committee.
DNA extraction, target region capture and next-generation sequencing
LifeFeng Genomic DNA Purification Kit (Lifefeng Biotech Co., Ltd., Shanghai, China) was used to extract genomic DNA from peripheral blood samples. DNA quality and concentration were examined by NanoDrop2000 (Thermo Scientific, United States). Thirty pairs of primers divided into two pools were designed covering all exons, UTRs and exon-intron boundary of EMB gene. All the primer sequences and target regions are shown in Table S1. The library construction method is a two-staged PCR process. The PCR reagents and protocol were provided by the Shanghai DYnastyGene Company. The size distribution of fragments was determined using 2100 Bioanalyzer and the High Sensitivity DNA kit (Agilent Technologies, United States). The final purified DNA libraries were sequenced on the Illumina HiSeq X Ten System (Illumina, United States) as PE 150 bp reads.
Variant identification and validation
The pipeline for germline short variant discovery of Genome Analysis Toolkit (GATK) Best Practices was run on every data set independently [16]. It mainly includes Burrows-Wheeler Aligner (BWA) [17] for aligning raw reads to the human reference genome (hg19), GATK haplotypercaller for variants (single nucleotide polymorphisms (SNPs), short insertions and deletions) calling and Annovar [18] for annotating variants. Multi-species alignments were performed using Clustal Omega online software [19]. Sanger sequencing was performed for the verification of the rare nonsynonymous variants.
Case-control study
A case-control study, including Hardy–Weinberg equilibrium (HWE), single locus association tests and pairwise linkage disequilibrium (LD) analysis were performed on the SHEsisPlus online software platform [20–22], which is a user-friendly platform designed for association studies. Pairwise LD analyses were performed in common variants and adjacent loci with D’ > 0.95 were classified in the same block. The Chi-square test or Fisher’s exact test for independence was used to infer whether the alleles and genotypes were associated with schizophrenia. All tests were two-tailed and statistical significance was set at P < 0.05. P-values were calibrated by the false discovery rate (FDR-BH).
Results
Variants identification
We performed Sanger sequencing for 21 samples containing missense mutations or frameshift indels detected by next-generation sequencing (Table S2). Among them, the mutations of 13 samples were verified to be true positives, and ten of these sites achieved sequencing depth with deeper than 15×. The other 8 samples were verified to be false positives, and their depths were all lower than 15×. Therefore, we only considered the sample sites whose depth were more than 15 × or the verified true functional variants to be successfully genotyped. Only loci with call rate greater than 85% or the verified loci were included in further statistical analysis.
A total of 58 high-quality variants, in which 51 are rare variants (SNVs) and 7 are common variants (SNPs), were identified in case and control groups, including 7 missense mutations, 4 synonymous mutations, 11 intron variants, 2 upstream variants and 34 UTR variants (Table S3). Rare mutations were filtered by three databases, the 1000 Genomes Project [23], the Exome Aggregation Consortium [24] and NHLBI Exome Sequencing Project [25]. Only loci with minor allele frequency (MAF) minor than 0.01 in each database were selected and in total 51 rare mutations were detected. On the whole, 3 rare variants in coding exons are newly reported (two are nonsynonymous and the other is synonymous). The information of common SNPs is shown in Table S4.
Analyses of rare variants in coding regions
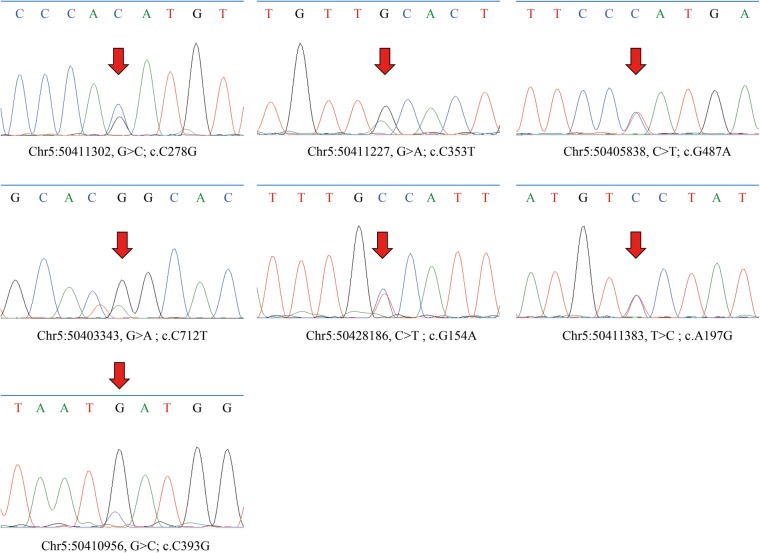
In total, 11 variants in coding exons including 4 synonymous mutations and 7 nonsynonymous mutations (Fig. 1) were identified. They are all heterozygous and rare mutations. Among the nonsynonymous variants, three single nucleotide variations (SNVs), EMB: p.(Ala118Val), p.(Ile131Met) and p.(Arg238Cys), occur only in schizophrenia subjects; EMB: p.(Glu66Gly) and p.(Ala52Thr) occur only in healthy controls and the others; p.(Gly163Arg) and p.(Ser93Cys) occur in both cases and healthy controls (Table 2). We performed Sanger sequencing to verify the nonsynonymous variants, EMB: c.712C > T, c.393C > G, c.353C > T, c.487G > A, c.197A > G, c.154G > A and c.278C > G. The results are shown in Fig. 2. All variants were verified to be heterozygous mutations. Moreover, the result of Sanger sequencing showed that the sample with c.712C > T variant has another mutation next to the mutation site: c.713G > A. Amino acid change, p.(Arg238Tyr), occurs in the case of simultaneous mutation of these two bases.
Table 2.
Detailed information of rare mutations detected in this study
| Variants | Variants status | SIFT | Polyphen-2 | MutationTaster | Novel or not | Individuals | Gender | group |
|---|---|---|---|---|---|---|---|---|
| c.712C > T/p.(Arg238Cys) | Heterozygous | D | D | D | rs749613196 | 55S0720 | Female | SCZ |
| c.393C > G/p.(Ile131Met) | Heterozygous | T | D | N | novel | QLS422 | Male | SCZ |
| c.353C > T/p.(Ala118Val) | Heterozygous | T | B | N | rs749459608 | QLS027 | Male | SCZ |
| c.487G > A/p.(Gly163Arg) | Heterozygous | D | D | N | rs553167245 | 55S0033 | Male | SCZ |
| DI128 | Female | |||||||
| Q17030 | Female | control | ||||||
| Q16488 | Male | |||||||
| c.278C > G/p.(Ser93Cys) | Heterozygous | T | B | N | rs148226429 | QLS295 | Male | SCZ |
| 55S0268、QLS255 | Female | |||||||
| Q16484 | Male | control | ||||||
| c.197A > G/p.(Glu66Gly) | Heterozygous | T | B | N | novel | Q17228 | Female | control |
| c.154G > A/p.(Ala52Thr) | Heterozygous | T | B | N | rs766190226 | Q16630 | Male | control |
SCZ Schizophrenia, D Deleterious for SIFT_pred,Probably damaging for Polyphen-2_pred, disease_causing for MutationTaster_pred; T Tolerated, B Benign
Fig. 2.
The results of Sanger sequence verifying the rare nonsynonymous variants. Arrows indicate the mutation sites
Annotations by Annovar were conducted to evaluate the pathogenicity of variants, which including PolyPhen-2 [26], MutationTaster [27] and SIFT [28] were used to predict the function of the variants. All the three applied prediction tools agreed that p.(Arg238Cys) was “deleterious”, while p.(Arg238Tyr) was predicted to be “deleterious” by SIFT, “possibly damaging” by PolyPhen-2 and “polymorphism” by MutationTaster. In addition, p.(Gly163Arg) was predicted to be “damaging” by SIFT and PolyPhen-2, and p.(Ile131Met) to be “damaging” only by PolyPhen-2. The other loci were predicted to be benign by all the three prediction tools.
Multiple alignments of embigin protein sequences of several available species show that EMB: p.(Gly163Arg) is relatively conserved across evolution, while p.(Ile131Met) and p.(Arg238Tyr) are respectively located in two Ig domains of embigin are not highly conserved (Fig. 3).
Fig. 3.
Multiple alignments of embigin protein sequences of various species. Three sites which were predicted to be deleterious variants by some prediction tools were box out
Association analysis in all variants
We performed a case-control study for all the variants except rs28528780, because it failed to reach Hardy–Weinberg equilibrium with the p-value being 1.72 × 10− 4 in the healthy controls. Significant associations of rare variants within EMB were not observed in schizophrenia in our sample. Then, we conducted a gene-based association study in which individuals carrying any rare nonsynonymous mutation were set as gene mutation carriers, but still, no statistically significant difference was detected (chi2 = 0.04, P > 0.05).
The statistical results of allele and genotype of common SNPs are shown in Table 3. Rs3933097 (Pallele = 3.820 × 10− 6, Pgenotype = 3.180 × 10− 5), achieved allelic and genotypic significance after FDR-BH correction. Rs35327819 (Pallele = 0.015, Pgenotype = 0.047) and rs199737787 (Pallele = 0.018, Pgenotype = 0.018) were significantly associated with schizophrenia both in allelic and genotypic distributions before FDR-BH correction. However, the significance disappeared after correction. Lower limits of odds ratio (OR) 95% confidence interval for these loci are both greater than 1.000 which suggests that the alternate alleles (“ins C” for rs35327819, “G” for rs3933097 and “A” for rs199737787) are both risk factor for schizophrenia. We also conducted association analyses in gender groups separately (Table S5 and Table S6). Only rs3933097 reached allelic and genotypic significance both in males and females after correction. Rs35327819 only achieved allelic significance in females.
Table 3.
Association results of 6 common variants
| SNP ID | Group | Allele frequency | Allelic P | Corrected Pa | OR | 95% CI | Genotype frequency | Genotypic P | Corrected Pa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs35327819 | - | C | -/- | -/C | C/C | |||||||
| chr5:50396796 | SCZ | 1690(0.494) | 1730(0.505) | 0.015* | 0.179 | 1.147 | [1.026~1.281] | 430(0.251) | 830(0.485) | 450(0.263) | 0.047* | 0.346 |
| Control | 1040(0.528) | 928(0.471) | 288(0.292) | 464(0.471) | 232(0.235) | |||||||
| rs3933097 | T | G | T/T | T/G | G/G | |||||||
| chr5:50398749 | SCZ | 1973(0.573) | 1467(0.426) | 1.98e-07** | 3.82e-06** | 1.354 | [1.208~1.518] | 571(0.331) | 831(0.483) | 318(0.184) | 1.64e-06** | 3.18e-05** |
| Control | 1268(0.645) | 696(0.354) | 413(0.420) | 442(0.450) | 127(0.129) | |||||||
| rs13172025 | G | A | G/G | G/A | A/A | |||||||
| chr5:50402346 | SCZ | 1535(0.480) | 1657(0.519) | 0.074 | 1.111 | [0.989~1.248] | 384(0.240) | 767(0.480) | 445(0.278) | 0.199 | ||
| Control | 895(0.507) | 869(0.492) | 239(0.270) | 417(0.472) | 226(0.256) | |||||||
| rs199737787 | C | A | C/C | C/A | ||||||||
| chr5:50403159 | SCZ | 3399(0.995) | 17(0.004) | 0.018* | 0.182 | 4.881 | [1.126~21.15] | 1691(0.990) | 17(0.009) | 0.018* | 0.216 | |
| Control | 1952(0.998) | 2(0.001) | 975(0.997) | 2(0.002) | ||||||||
| rs184038560 | A | G | A/A | G/A | ||||||||
| chr5:50403449 | SCZ | 3030(0.990) | 28(0.009) | 0.096 | 0.636 | [0.372~1.089] | 1501(0.981) | 28(0.018) | 0.095 | |||
| Control | 1792(0.985) | 26(0.014) | 883(0.971) | 26(0.028) | ||||||||
| rs72751723 | G | T | G/G | G/T | T/T | |||||||
| chr5:50441389 | SCZ | 3145(0.900) | 349(0.099) | 0.664 | 0.960 | [0.800~1.152] | 1417(0.811) | 311(0.178) | 19(0.010) | 0.843 | ||
| Control | 1757(0.896) | 203(0.103) | 787(0.803) | 183(0.186) | 10(0.010) | |||||||
SCZ Schizophrenia, SNP Single nucleotide polymorphism, OR Odds ratio, CI Confidence interval
aFDR correction
* P-values < 0.05; ** P-values < 0.01
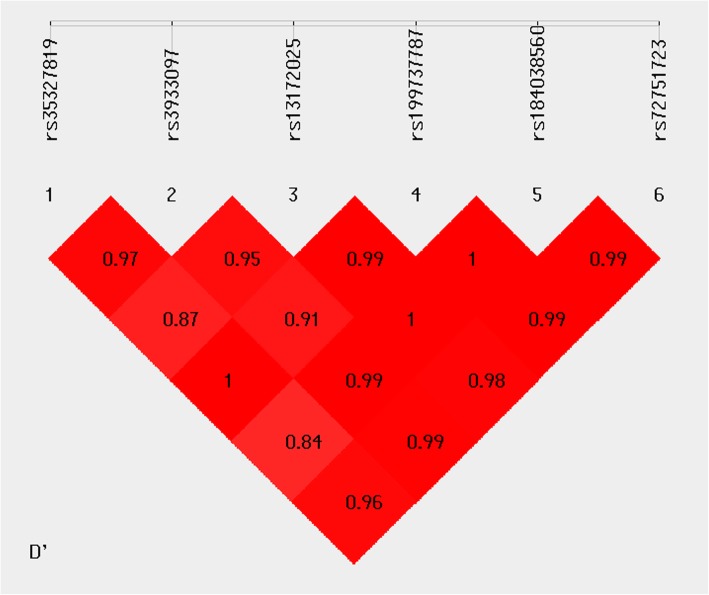
Pairwise LD analysis indicates that rs35327819-rs3933097 and rs13172025-rs199737787-rs184038560-rs72751723 form two haplotype blocks respectively (Fig. 4). Three haplotypes, −T/CG/CT, were identified for rs35327819-rs3933097 and the other three haplotypes, GCAG/ACAG/GCAT, for rs13172025-rs199737787-rs184038560-rs72751723. In the block rs35327819-rs3933097, haplotype “-T” and “CT” are implied to be significant protective factors for schizophrenia while “CG” is a significant risk factor for schizophrenia after FDR correction. The global Pearson’s P is 5.18 × 10− 10 after Bonferroni correction (Table 4). Haplotype “GCAG” for rs13172025-rs199737787-rs184038560-rs72751723 is also a significant protective factor while the block is not significantly associated with schizophrenia (global Pearson’s P = 0.114) (Table 4). We also performed haplotype analysis to evaluate whether the nonsynonymous SNVs we found are independent of the significant common SNP, rs3933097 in 3′-UTR of EMB gene discovered in our study. But because of the low frequency of the nonsynonymous SNVs, no significant haplotypes formed by any nonsynonymous SNVs and rs3933097 was found and the difference of odds radio could not be calculated between the haplotype with simultaneous mutation of both loci and the haplotype only with the mutation in rs3933097.
Fig. 4.
Pairwise linkage disequilibrium (LD) plot for the common variants in EMB gene. The pairwise D’ values are presented in the matrices, Deep red implicates relatively strong linkage disequilibrium, and vice versa
Table 4.
Results of Haplotype Analysis
| Haplotypea | Case frequency | Control frequency | Chi2 | P | Corrected Pb | OR [95% CI] | Global Chi2 | Global P | Corrected Pc | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs35327819-rs3933097 | -T | 1778 (0.493) | 1041 (0.522) | 4.319 | 0.037* | 0.037* | 0.890 [0.798~0.993] | 44.151 | 2.59e-10** | 5.18e-10** |
| CG | 1523 (0.422) | 691 (0.346) | 30.870 | 2.76e-08** | 5.52e-08** | 1.378 [1.230~1.544] | ||||
| CT | 294 (0.081) | 246 (0.123) | 25.796 | 3.79e-07** | 5.06e-07** | 0.630 [0.527~0.754] | ||||
| rs13172025-rs199737787- rs184038560-rs72751723 | GCAG | 1146 (0.381) | 734 (0.407) | 14.567 | 1.35e-04** | 2.03e-04** | 0.799 [0.712~0.897] | 4.341 | 0.114 | |
| ACAG | 1511 (0.502) | 849 (0.471) | 0.240 | 0.624 | 0.624 | 0.972 [0.870~1.086] | ||||
| GCAT | 306 (0.101) | 189 (0.105) | 1.569 | 0.210 | 0.252 | 0.885 [0.732~1.071] |
OR Odds ratio, CI Confidence interval
aHaplotypes with frequency < 0.03 were ignored; b FDR correction; c Bonferroni correction
* P-values < 0.05; ** P-values < 0.01
Discussion
In our results, seven rare missense single nucleotide variations in coding regions are identified, but there is no significant association between these loci and schizophrenia, which is probably caused by the low frequency of these mutations and the limit size of our sample set. Three mutations are only detected in the group of cases and c.393C > G is novel reported. The other two mutations, c.353C > T and c.712C > T, were reported before [29], and the alternative frequency of them in cases and healthy controls were as follow: c.712C > T: 6.19 × 10− 5 in cases and 5.14 × 10− 6 in controls, c.353C > T: 2.06 × 10− 5 in cases and 5.14 × 10− 6 in controls. Among the 7 mutations, three of them are predicted to be deleterious variants which may seriously affect embigin protein function. But the results of the multi-species analysis show that two of the variants in the IG domain are not completely conserved. Although the other variant is relatively conservative, it is detected both in cases and healthy controls and there is no significant difference detected between the two groups. Therefore, there is no sufficient evidence proving that the mutations at these three loci of EMB gene are associated with the pathogenesis of schizophrenia. In order to determine the relationship between EMB gene and schizophrenia, performing sequencing analysis for more schizophrenia samples and functional verification experiments are needed.
The SNP, rs3933097 located in 3′-UTR of EMB gene, is significantly associated with schizophrenia both in allelic and genotypic distributions in our study. It also achieves allelic and genotypic significance in both male and female subset. The SZDB2.0 eQTL data [30, 31] shows an association between rs3933097 and the expression of EMB gene (P = 4.90 × 10− 17 after FDR correction) [32]. In view of above-mentioned results, we propose that the association of EMB with schizophrenia partly depends on the affection of the mRNA expression level of EMB gene by rs3933097. From the data of the Psychiatry Genomics Consortium [33], we found that rs3933097 has been genotyped in a previous GWAS [2] and achieved nominal significance (P = 5.38 × 10− 5, odds ratio (OR) for allele “T” =0.956, standard error (SE) = 0.011). The results of latest GWAS in schizophrenia [4] showed that EMB gene is one of the potentially causal genes at 33 genome-wide significant loci (rs77853293, P = 1.77 × 10− 8, odds ratio (OR) for allele “C” =1.056, standard error (SE) = 0.010, gene tagged: EMB, PSMR = 1.12 × 10− 6). Our result of the direction effect for rs3933097 is consistent with the previous GWAS. Furthermore, our study confirms the previous GWAS result and suggests that the polymorphism of 3′-UTR sequence of EMB gene may be involved in the pathogenesis of schizophrenia. Recent GWAS indicated that, EMB gene with mRNA expression level in cis genetic linkage with rs10940346 which achieved genome-wide significance with schizophrenia [3]. Summary-data-based Mendelian Randomization (SMR) analysis was also applied in the latest GWAS [4] and rs77853293 was identified that might be causally linked through expression changes in EMB gene and achieved genome-wide significance (P < 1.17 × 10− 5, adjusted for 4276 probes). We also conducted Pairwise LD analysis among rs3933097, rs10940346 and rs77853293 using 1000 genomes data of Southern Han Chinese population [34]. The results show that the three SNPs are highly linked (between rs3933097and rs10940346, D’ = 0.99, R2 = 0.77; between rs3933097and rs77853293, D’ = 0.97, R2 = 0.77), which is also in line with our expectations. Our result reveals rs3933097 in 3′-UTR of EMB gene is significantly associated with schizophrenia by which the mRNA expression level of EMB gene might be regulated and affected. Overall, our results provide evidence that EMB gene is a susceptibility gene for schizophrenia which is consistent with the prediction of recent GWAS [3, 4].
3′-UTR plays a crucial role in the post-transcriptional regulation of gene expression. One such regulatory process is 3′-UTR facilitation of mRNA decay or translational repression of candidate genes through binding with micro RNAs (miRNAs) or RNA-binding proteins (RBPs) [35–37]. The previous study indicated that genetic changes in miRNAs potentially had an impact on psychiatry [38]. Several studies have revealed some SNPs located in the 3′-UTR affect gene expression by altering the binding of specific miRNAs to the 3′-UTRs, and thereby, affect the risk of schizophrenia. For example, rs1130354 within the 3′-UTR of human dopamine receptor D2 (DRD2) alters miR-326-mediated expression regulation [39], rs550067317, located in the 3′-UTR of ephrin B2 (EFNB2), affects miR-137-mediated repression of EFNB2 expression [40] as well as rs7219 within 3′-UTR of GRB2 alters the expression of GRB2 by affecting miR-1288-mediated inhibition [41], thereby affecting the risk or progression of schizophrenia. In our results, rs3933097 in 3′-UTR of EMB gene is significantly associated with schizophrenia and Several miRNAs, hsa-miR-508-3p, hsa-miR-182, hsa-miR-335 and hsa-miR-580, are predicted to bind with the seed region containing rs3933097 by SNPinfo Web Server [42]. The SNPs located in the gene of miR-182 were genotyped in the first research of genetic changes of miRNAs potentially having an impact in psychiatry [38]. Although in Spanish, potential associations (P < 0.05) were observed for rs2402961 (P = 0.01769) and rs73159662 (P = 0.02157), no association was observed in the overall sample set [38]. Previous research in vivo showed that overexpression of miR-182 within the lateral amygdala resulted in decreased expression of the protein and disrupted long-term but not short-term auditory fear memory [43]. However, more evidence is needed to confirm whether rs3933097 affects these miRNAs binding and thus regulates EMB expression.
Because RBPs act only as adaptors that connect 3′-UTRs to effector proteins and different effector proteins could be recruited, the biological functions of 3′-UTRs depending on the effector proteins could be various. 3′-UTRs also regulate mRNA localization [44] and protein-protein interactions [35]. Additionally, mRNA populations of BDNF transcripts are distinguished by the length of their 3′-UTR and different mRNA isoforms with different subcellular localization and functions in neurons [45]. All of these researches confirmed that 3′-UTRs of some genes involved in schizophrenia or other neuropsychiatric diseases have important regulatory functions. We discovered a SNP locus located in 3′-UTR of EMB gene is significantly associated with schizophrenia, but further functional validations are necessary for understanding the etiology correlated with 3′-UTR of EMB in schizophrenia.
The pivotal role of embigin in the growth of motor neurons and the formation of neuromuscular junctions has been confirmed. As an accessory protein of MCT2, embigin maintains the catalytic activity of MCT2 to synergistically transport lactic acid between glial cells and neurons [10], which plays an important role in brain energy metabolism and long-term procedural memory formation [11]. However, the research of EMB gene on the pathogenesis of schizophrenia is limited. Only recent GWAS found a locus, which was significantly associated with schizophrenia, was in cis genetic linkage with EMB gene mRNA levels [3]. Our results further validate a SNP located in the 3′-UTR of EMB gene is significantly associated with schizophrenia in Chinese Han population, but its influence to the regulation of EMB expression remains to be further studied, and the role of EMB gene in the pathogenesis of schizophrenia also needs to be confirmed by more research. In addition, the majority of the GWAS findings for complex traits are generally found in the regulatory regions of the genome. Therefore, the lack of sequencing of the non-exonic regions of EMB gene is a limitation in our study, which may cause some significant loci involved in the regulation of EMB gene expression were overlooked, and thus failed to reveal the comprehensive mutation spectrum of entire EMB gene in schizophrenia.
Conclusions
Our research first presented a comprehensive mutation spectrum of exons and UTR in EMB gene for schizophrenia. We identify two novel nonsynonymous mutations in EMB gene and first report the SNP, rs3933097, located in the 3′-UTR of EMB gene is significantly associated with schizophrenia. Our research provides additional evidence that EMB gene is a susceptibility gene for schizophrenia, but further functional validations are considered to be necessary for understanding EMB correlated with the etiology in schizophrenia.
Supplementary information
Additional file 1: Table S1. primers and target regions. Table S2. Variants validation results by Sanger Sequencing. Table S3 58 variants of EMB gene identified in this research. Table S4. Detailed information of the 7 common variants in the EMB Gene. Table S5. Association results of 6 common variants in male. Table S6. Association results of 6 common variants in female.
Acknowledgements
We thank all of the participants in the study. We are grateful for the help from psychiatrists during recruitment and diagnosis of schizophrenia patients, and we appreciate all patients and healthy controls who were involved in this study.
Abbreviations
- BWA
Burrows-Wheeler Aligner
- DRD2
Human dopamine receptor D2
- EFNB2
Ephrin B2
- FDR
False discovery rate
- GATK
Genome Analysis Toolkit
- GWAS
Genome-wide association study
- HWE
Hardy–Weinberg equilibrium
- IgSF
Immunoglobulin superfamily
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- OR
Odds ratio
- RBP
RNA-binding protein
- SE
Standard error
- SNP
Single nucleotide polymorphism
- SNV
Single nucleotide variation
- UTR
Un-translated region
Authors’ contributions
Y.S. designed and supervised the whole research process. J.Z. and C.M. carried out all experiments and managed the literature searches and analysis. H.Z. and X.L. conducted sample collection and verification. K.W. and Z.L. undertook the statistical analysis. J.C. was responsible for platform coordination and management. J.Z. wrote the first draft of the manuscript. J.C. and Z.L. proof read the article. All authors contributed to and approved the final manuscript.
Funding
This work was supported in part by Shanghai Municipal Science and Technology Major Project (Grant No. 2017SHZDZX01), the National Key R&D Program of China (2017YFC0908105, 2019YFA0905401 and 2016YFC1306900), the Natural Science Foundation of China (U1804284, 81421061, 81701321, 31571012, 91849104, 31770800, 81571329, 81501154 and 81871051), Shanghai Hospital Development Center (SHDC12016115), Shanghai Science and Technology Committee (17JC1402900 and 17490712200) and shanghai municipal health commission (ZK2015B01 and 201540114). The funding bodies did not participate in the design of the study, in collection, analysis and interpretation of data or in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
All participants signed informed consent and the study was approved by the ethical committee of Bio-X Institutes, Shanghai Jiao Tong University. The reference number is 20190918.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Juan Zhou and Chuanchuan Ma contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12888-020-02513-3.
References
- 1.Burmeister M, McInnis MG, Zöllner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–540. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- 2.Ripke S, Neale BM, Corvin A, Walters JTR, Farh KH, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49:1576. doi: 10.1038/ng.3973. [DOI] [PubMed] [Google Scholar]
- 4.Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozawa M, Huang RP, Furukawa T, Muramatsu T. A teratocarcinoma glycoprotein carrying a developmentally regulated carbohydrate marker is a member of the immunoglobulin gene superfamily. J Biol Chem. 1988;263(7):3059–3062. [PubMed] [Google Scholar]
- 6.Muramatsu T, Miyauchi T. Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol Histopathol. 2003;18(3):981–987. doi: 10.14670/HH-18.981. [DOI] [PubMed] [Google Scholar]
- 7.Paunio T, Tuulio-Henriksson A, Hiekkalinna T, Perola M, Varilo T, Partonen T, et al. Search for cognitive trait components of schizophrenia reveals a locus for verbal learning and memory on 4q and for visual working memory on 2q. Hum Mol Genet. 2004;13(16):1693–1702. doi: 10.1093/hmg/ddh184. [DOI] [PubMed] [Google Scholar]
- 8.Beesley P, Kraus M, Parolaro N. The neuroplastins: multifunctional neuronal adhesion molecules--involvement in behaviour and disease. Adv Neurobiol. 2014;8:61–89. doi: 10.1007/978-1-4614-8090-7_4. [DOI] [PubMed] [Google Scholar]
- 9.Lain E, Carnejac S, Escher P, Wilson MC, Lomo T, Gajendran N, et al. A novel role for embigin to promote sprouting of motor nerve terminals at the neuromuscular junction. J Biol Chem. 2009;284(13):8930–8939. doi: 10.1074/jbc.M809491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson MC, Meredith D, Fox JE, Manoharan C, Davies AJ, Halestrap AP. Basigin (CD147) is the target for organomercurial inhibition of monocarboxylate transporter isoforms 1 and 4: the ancillary protein for the insensitive MCT2 is EMBIGIN (gp70) J Biol Chem. 2005;280(29):27213–27221. doi: 10.1074/jbc.M411950200. [DOI] [PubMed] [Google Scholar]
- 11.Pérez-Escuredo J, Van Hée VF, Sboarina M, Falces J, Payen VL, Pellerin L, et al. Monocarboxylate transporters in the brain and in cancer. Biochim Biophys Acta. 2016;1863(10):2481–2497. doi: 10.1016/j.bbamcr.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki A, Stern Sarah A, Bozdagi O, Huntley George W, Walker Ruth H, Magistretti Pierre J, et al. Astrocyte-neuron lactate transport is required for Long-term memory formation. Cell. 2011;144(5):810–823. doi: 10.1016/j.cell.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang R-P, Ozawa M, Kadomatsu K, Muramatsu T. Developmentally regulated expression of embigin, a member of the immunoglobulin superfamily found in embryonal carcinoma cells. Differentiation. 1990;45(2):76–83. doi: 10.1111/j.1432-0436.1990.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 14.Guenette R, Sridhar S, Herley M, Mooibroek M, Wong P, Tenniswood M. Embigin, a developmentally expressed member of the immunoglobulin super family, is also expressed during regression of prostate and mammary gland. Dev Genet. 1997;21(4):268–278. doi: 10.1002/(SICI)1520-6408(1997)21:4<268::AID-DVG4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Huang R-P, Ozawa M, Kadomatsu K, Muramatsu T. Embigin, a member of the immunoglobulin superfamily expressed in embryonic cells, Enhances Cell-Substratum Adhesion. Dev Biol. 1993;155(2):307–314. doi: 10.1006/dbio.1993.1030. [DOI] [PubMed] [Google Scholar]
- 16.Genome Analysis Toolkit. https://software.broadinstitute.org/gatk. Accessed 13 August 2019.
- 17.Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clustal Omega. http://www.ebi.ac.uk/Tools/msa/clustalo/. Accessed 25 August 2019.
- 20.Shen J, Li Z, Chen J, Song Z, Zhou Z, Shi Y. SHEsisPlus, a toolset for genetic studies on polyploid species. Sci Rep. 2016;6:24095. doi: 10.1038/srep24095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision em algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn) Cell Res. 2009;19(4):519–523. doi: 10.1038/cr.2009.33. [DOI] [PubMed] [Google Scholar]
- 22.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 23.IGSR: The International Genome Sample Resource. http://www.1000genomes.org. Accessed 13 August 2019.
- 24.ExAC Browser (Beta) | Exome Aggregation Consortium. http://exac.broadinstitute.org. Accessed 13 August 2019.
- 25.NHLBI Exome sequencing project (ESP), Exome Variant Server http://evs.gs.washington.edu/EVS. Accessed 13 August 2019.
- 26.PolyPhen-2. http://genetics.bwh.harvard.edu/pph2/. Accessed 13 August 2019.
- 27.MutationTaster . http://www.mutationtaster.org. Accessed 13 August 2019.
- 28.SIFT .http://sift.jcvi.org/. Accessed 13 August 2019.
- 29.Exome meta-analysis results. http://schema.broadinstitute.org/results. Accessed 17 September 2019.
- 30.SZDB2.0 .http://www.szdb.org/. Accessed 21 November 2019.
- 31.Wu Y, Yao Y-G, Luo X-J. SZDB: a database for schizophrenia genetic research. Schizophr Bull. 2017;43(2):459–471. doi: 10.1093/schbul/sbw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Psychiatric Genomics Consortium. https://www.med.unc.edu/pgc/results-and-downloads/scz/. Accessed 17 September 2019.
- 34.1000genomes .http://phase3browser.1000genomes.org/Homo_sapiens/Info/Index. Accessed 20 November 2019.
- 35.Mayr C. Regulation by 3′-Untranslated regions. Annu Rev Genet. 2017;51:171–194. doi: 10.1146/annurev-genet-120116-024704. [DOI] [PubMed] [Google Scholar]
- 36.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 38.Toma C, Torrico B, Hervás A, Salgado M, Rueda I, Valdés-Mas R, et al. Common and rare variants of microRNA genes in autism spectrum disorders. World J Biol Psychiatry. 2015;16(6):376–386. doi: 10.3109/15622975.2015.1029518. [DOI] [PubMed] [Google Scholar]
- 39.Shi S, Leites C, He D, Schwartz D, Moy W, Shi J, et al. MicroRNA-9 and microRNA-326 regulate human dopamine D2 receptor expression, and the microRNA-mediated expression regulation is altered by a genetic variant. J Biol Chem. 2014;289(19):13434–13444. doi: 10.1074/jbc.M113.535203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S, Zhang R, Nie F, Wang X, Jiang C, Liu M, et al. MicroRNA-137 inhibits EFNB2 expression affected by a genetic variant and is expressed aberrantly in peripheral blood of schizophrenia patients. EBioMedicine. 2016;12:133–142. doi: 10.1016/j.ebiom.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Guo X, Zhu L, Huang J, Long J, Chen Q, et al. Rs7219 regulates the expression of GRB2 by affecting miR-1288-mediated inhibition and contributes to the risk of schizophrenia in the Chinese Han population. Cell Mol Neurobiol. 2019;39(1):137–147. doi: 10.1007/s10571-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SNPinfo Web Server. https://snpinfo.niehs.nih.gov/. Accessed 5 September 2019.
- 43.Griggs EM, Young EJ, Rumbaugh G, Miller CA. MicroRNA-182 regulates amygdala-dependent memory formation. J Neurosci. 2013;33(4):1734. doi: 10.1523/JNEUROSCI.2873-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 2009;19(9):465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 45.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134(1):175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. primers and target regions. Table S2. Variants validation results by Sanger Sequencing. Table S3 58 variants of EMB gene identified in this research. Table S4. Detailed information of the 7 common variants in the EMB Gene. Table S5. Association results of 6 common variants in male. Table S6. Association results of 6 common variants in female.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.