Abstract
Introduction
Cervical cancer (CC) is a major health threat to women worldwide. Long non-coding RNA (lncRNA) has been reported to play crucial roles in regulating carcinogenesis, including CC.
Methods
In this work, levels of lncRNA forkhead box P4 antisense RNA 1 (FOXP4-AS1) in CC cell lines and normal cell lines were analyzed with quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) method. Effects of FOXP4-AS1 on CC cellular behaviors including proliferation, migration, and invasion were explored. Bioinformatic prediction tools and luciferase activity reporter assay were conducted to explore the downstream molecules for FOXP4-AS1.
Results
We found FOXP4-AS1 expression was significantly higher in CC cell lines than in normal cell line. Functionally, force FOXP4-AS1 expression increased CC cell proliferation, migration, and invasion, while FOXP4-AS1 knockdown caused opposite effects. Mechanistically, we found FOXP4-AS1 acts as competing endogenous RNA (ceRNA) for microRNA-136-5p (miR-136-5p) to regulate chromobox 4 (CBX4) expression.
Discussion
These findings indicated FOXP4-AS1 plays an oncogenic role in CC, which may provide novel therapeutic biomarker against CC.
Keywords: FOXP4-AS1, miR-136-5p, CBX4, cervical cancer
Introduction
Cervical cancer (CC) remains the second-leading reason for cancer-related deaths in females worldwide although its incidence was gradually reduced due to the wide usage of human papillomavirus vaccine.1 Carcinogenesis is accompanied with many cellular hallmarks including resistance to death, uncontrolled cell growth, activated cell metastasis and so on, which is mediated by the aberrantly expressed of protein-coding and non-coding genes.2–4 Hence, investigations on molecular mechanisms behind CC progression can help to identify novel biomarkers for cancer diagnosis or treatment.
Long non-coding RNA (lncRNA) is a type of non-coding RNA with the length of above 200 nucleotides.5 lncRNAs are reported to have limited protein or peptide coding ability.6 In recent years, lncRNAs are documented to have crucial roles in regulating cancer progression and cancer cell behaviors via sponging microRNA (miRNA).7 lncRNA Forkhead box P4 antisense RNA 1 (FOXP4-AS1) is capable to regulate numerous cellular processes including cell proliferation, migration, invasion, and apoptosis.8–10 For instance, FOXP4-AS1 was found upregulated expression in gastric cancer patients at late tumor stages and with poorer overall survival 8. FOXP4-AS1 knockdown inhibits gastric cancer cell proliferation, migration, and invasion via EZH2/LSD1 axis.8 Elevated FOXP4-AS1 expression was also found in colorectal cancer patients with advanced tumor stages and large tumor size.9 In addition, Wang et al constructed a FOXP4-AS1 centered regulatory network and revealed two potential target genes, IGFBP3 and PRC1, in hepatocellular carcinoma.10
To date, it is unclear whether FOXP4-AS1 has a role in CC carcinogenesis. To address this issue, our work was conducted to explore the potential connections of FOXP4-AS1/miR-136-5p/chromobox 4 (CBX4) triplets in regulating CC cell proliferation, migration, and invasion. Our work will provide novel insights into the mechanisms behind progression of CC.
Materials and Methods
Cell Lines
Normal cervical epithelium cell (HCvEpC) and CC cells (Caski and C33A) obtained at Chinese Academy of Science (Shanghai, China), a cell collection center that contained authentic cell lines, were incubated at RPMI-1640 (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) in supplement with 10% fetal bovine serum (FBS, Invitrogen) and kept in a moist 37°C incubator contains 5% CO2.
Cell Transfection
miR-136-5p mimic and corresponding negative control (NC-mimic) were obtained from RiboBio (Guangzhou, Guangdong, China). Small interfering RNA targeting FOXP4-AS1 (si-FOXP4-AS1) and corresponding negative control (NC-siR) were also designed by RiboBio. pcDNA3.1 contains sequence of FOXP4-AS1 (pFOXP4-AS1) and CBX4 (pCBX4) were purchased from GenScript (Nanjing, Jiangsu, China). Transfection of these molecules was conducted using Lipofectamine 2000 (Invitrogen).
Quantitative Real-Time PCR (RT-qPCR)
Total RNA of cells was extracted using Trizol reagent (Invitrogen). After RNA sample concentration quantification using NanoDrop-1000 (Thermo Fisher Scientific, Inc.), equal amount of RNA was reverse transcribed into complementary DNA with reverse transcription kit (Takara, Dalian, Liaoning, China) based on the recommended protocols. RT-qPCR was conducted at ABI QuantStudio 6 (Thermo Fisher Scientific, Inc.) using SYBR Green (Takara) using the following primers: FOXP4-AS1: F: 5ʹ-GTGAGCTTCTGGGTTCGACA-3ʹ, R: 5ʹ-ATTGAGGGTTAGGGCAGCAC-3ʹ; CBX4: F: 5ʹ-ACCGTGCCAAGCTGGATTT-3ʹ, R: 5ʹ-AGGTCGTACATTTTGGGGTCG-3ʹ; GAPDH: F: 5ʹ-GGTGAAGGTCGGAGTCAACGG-3ʹ, R: 5ʹ-CCAGAGTTAAAAGCAGCCCTGG-3ʹ; miR-136-5p: F: 5ʹ-ACACTCCAGCTGGGACTCCATTTGTTTT-3ʹ, R: 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ, U6 snRNA: F: 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ, R: 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ. Relative expression level was calculated using 2−ΔΔCt method. The procedure was as follows: 94°C for 30 s, 55°C for 30 s and 72°C for 90 s, for a total of 40 cycles. Experiments were repeated in triplicates.
Cell Proliferation Assays
5 × 103 cells were seeded into 96-well plates to detect cell proliferation with cell counting kit-8 (CCK-8, Beyotime, Haimen, Jiangsu, China). At indicated time (0, 24, 48, and 72 h), 10 μL CCK-8 reagent was added to the well and further incubated for 4 h. Then, microplate reader was used to detect the absorbance at 450 nm. Experiments were repeated in triplicates.
Wound-Healing Assay
Cells were pre-treated with trypsin and then put into 6-well plates to grow until 100% confluence. Scratch was generated at cell surface using a 200 μL pipette tip, washed with PBS to remove debris, and incubated in serum-free medium. At 0 and 48 h, images were captured and observed under microscope. Cell migration distance was measured using Image J software. Experiments were repeated in triplicates.
Transwell Invasion Assay
Transwell invasion assay was conducted to measure cell invasion ability using 24-well artificial inserts with 8 µm precoated Matrigel membrane, which allow single to invade. 1 × 105 cells in serum-free medium were put into the upper chamber, while the serum contained medium was filled into the lower chamber. After 48 h of incubation, the non-invasive cells were gently removed by cotton, while invasive cells were fixed with methanol and stained with crystal violet. After washed by PBS, numbers of invasive cells were counted under microscope. Experiments were repeated in triplicates.
Bioinformatics Analysis
DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=site%2Ftools) was used to search for miRNA target for FOXP4-AS1, while ENCORI database (http://starbase.sysu.edu.cn/) was used to predict targets for miRNA.
Luciferase Reporter Assay
The amplified wild-type FOXP4-AS1 or CBX4 sequences were inserted into pmirGLO vector (GeneChem, Shanghai, China) to generate FOXP4-AS1-wt or CBX4-wt. Site-direct mutagenesis kit (Takara) was used to generate mutant luciferase vectors and termed as FOXP4-AS1-mt or CBX4-mt. Cells were co-transfected with luciferase constructs and miR-136-5p mimic or NC-mimic using Lipofectamine 2000. Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to measure relative luciferase activity after 48 h of transfection using Renilla luciferase as internal control. Experiments were repeated in triplicates.
Statistical Analysis
Data collected from three repeated experiments were analyzed with GraphPad PRISM 6 (GraphPad, San Diego, CA, USA) and presented at the manner of mean ± standard deviation. Differences in groups were analyzed by Student’s t-test or one-way ANOVA and Tukey post-hoc test. P < 0.05 was believed to indicate significant difference.
Results
FOXP4-AS1 Expression Was Elevated in CC Cell Lines
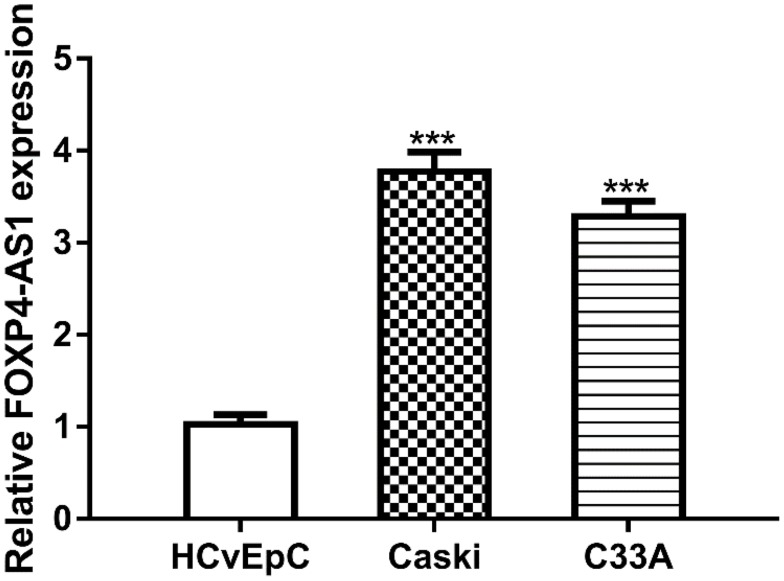
RT-qPCR assay was used to detect FOXP4-AS1 level in CC cell lines. As shown in Figure 1, expression of lncRNA FOXP4-AS1 in CC cells Caski (3.77 ± 0.21) and C33A (3.28 ± 0.17) was significantly higher than that in HCvEpC (1.03 ± 0.11).
Figure 1.
lncRNA FOXP4-AS1 was upregulated in human CC cells compared with normal cell.
Note: ***P < 0.001.
Abbreviations: lncRNA, long non-coding RNA; FOXP4-AS1, forkhead box P4 antisense RNA 1; CC, cervical cancer.
FOXP4-AS1 Overexpression Promotes CC Cell Proliferation, Migration, and Invasion
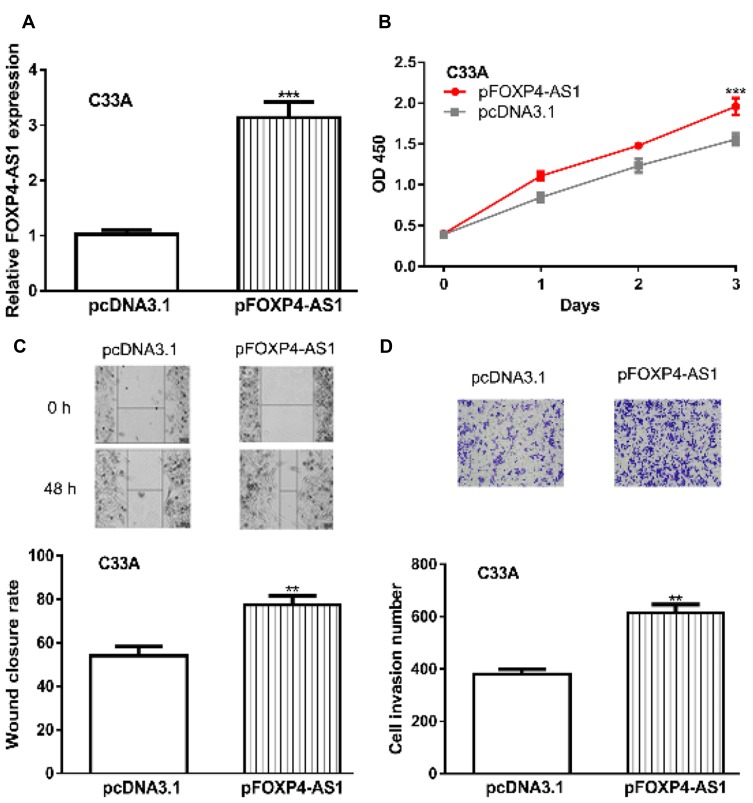
To investigate the functions of FOXP4-AS1 in CC, we upregulated FOXP4-AS1 levels in CC cells. RT-qPCR showed FOXP4-AS1 level was significantly elevated by pFOXP4-AS1 (3.14 ± 0.28 vs 1.03 ± 0.08) (Figure 2A). Cell proliferation analysis indicated cell viability was significantly stimulated by FOXP4-AS1 overexpression (1.96 ± 0.10 vs 1.56 ± 0.07) (Figure 2B). Wound-healing assay and transwell invasion assay revealed migration (77.33 ± 4.16 vs 54.03 ± 4.36) and invasion (615 ± 31 vs 381 ± 18) rates were significantly elevated in FOXP4-AS1 overexpressed groups compared with pcDNA3.1 groups (Figure 2C and D).
Figure 2.
Overexpression of lncRNA FOXP4-AS1 promotes CC cell proliferation, migration, and invasion in vitro.
Notes: (A) FOXP4-AS1 was overexpression in CC cells by transfection with pFOXP4-AS1. (B) Effects of FOXP4-AS1 overexpression on CC cell proliferation were measured by CCK-8 assay. (C) Roles of FOXP4-AS1 overexpression on CC cell migration were analyzed with wound-healing assay. (D) Effects of FOXP4-AS1 overexpression on CC cell invasion were measured by transwell invasion assay. **P < 0.01, *** P < 0.001.
Abbreviations: lncRNA, long non-coding RNA; FOXP4-AS1, forkhead box P4 antisense RNA 1; CC, cervical cancer; CCK-8, cell counting kit-8.
FOXP4-AS1 Knockdown Inhibits CC Cell Proliferation, Migration, and Invasion
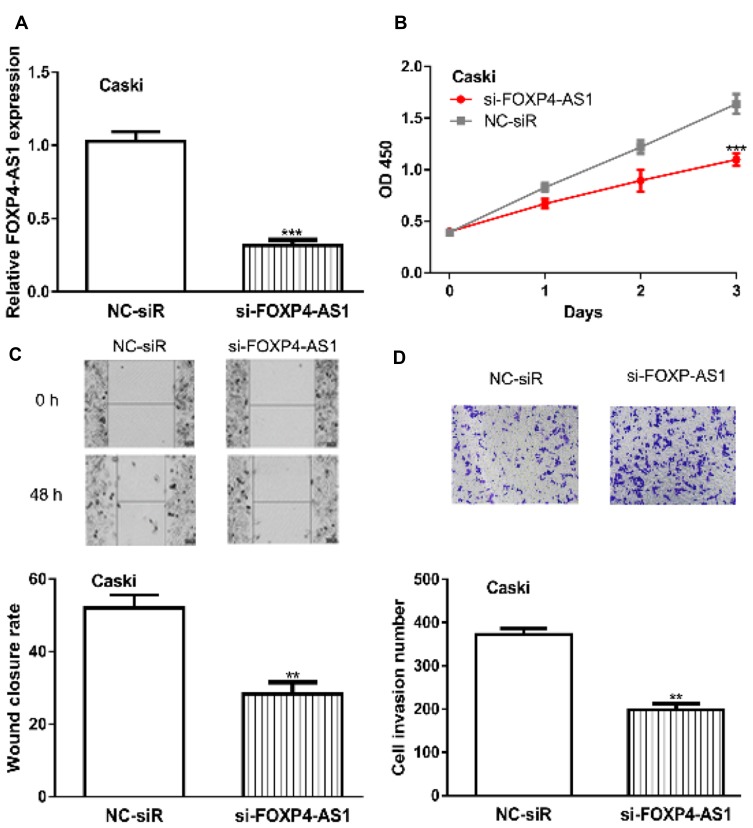
Furthermore, FOXP4-AS1 was silenced in CC cells by si-FOXP4-AS1 (0.32 ± 0.04 vs 1.03 ± 0.07) (Figure 3A). CCK-8 assay showed cell proliferation rate in cells transfected with si-FOXP4-AS1 was decreased compared those with NC-siR (1.10± 0.06 vs 1.64 ± 0.10) (Figure 3B). Likewise, the introduction of si-FOXP4-AS1 decreased cell migration (28.33 ± 3.21 vs 52.02 ± 3.61) and invasion (198 ± 14 vs 372 ± 13) abilities in comparison with NC-siR (Figure 3C and D).
Figure 3.
Knockdown of lncRNA FOXP4-AS1 inhibits CC cell proliferation, migration, and invasion in vitro.
Notes: (A) FOXP4-AS1 was knockdown in CC cells by transfection with si-pFOXP4-AS1. (B) Effects of FOXP4-AS1 knockdown on CC cell proliferation were measured by CCK-8 assay. (C) Roles of FOXP4-AS1 knockdown on CC cell migration were analyzed with wound-healing assay. (D) Effects of FOXP4-AS1 knockdown on CC cell invasion were measured by transwell invasion assay. **P < 0.01, *** P < 0.001.
Abbreviations: lncRNA, long non-coding RNA; FOXP4-AS1, forkhead box P4 antisense RNA 1; CC, cervical cancer; CCK-8, cell counting kit-8; si-FOXP4-AS1, small interfering RNA against FOXP4-AS1; NC-siR, negative control siRNA.
FOXP4-AS1 Acts as ceRNA by Binding with miR-136-5p
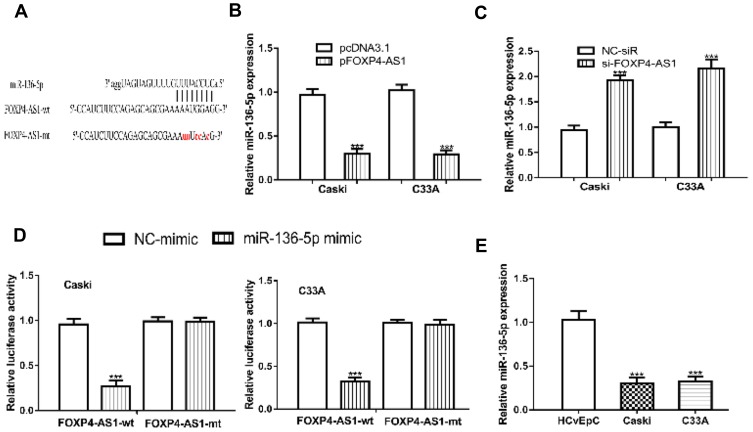
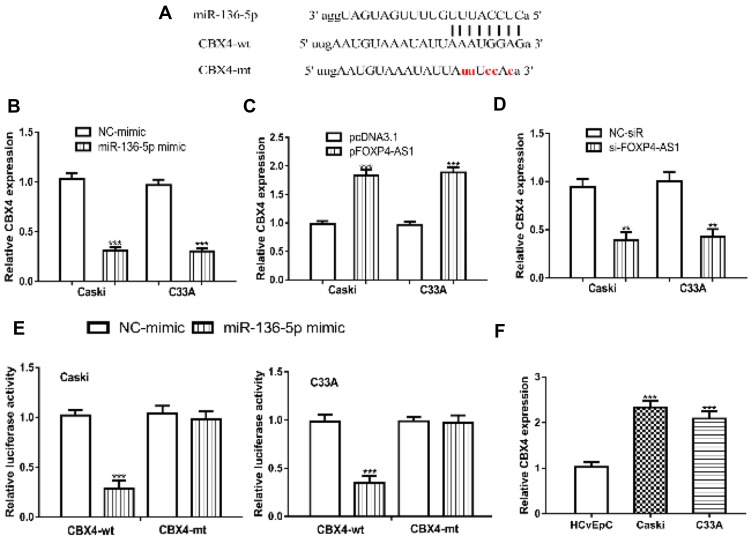
Through bioinformatic analysis, we found FOXP4-AS1 could complementarily bind with miR-136-5p (Figure 4A). Then, we detected the expression of miR-136-5p in CC cells after FOXP4-AS1 overexpress or silence. We found FOXP4-AS1 overexpression decreases (Caski: 0.97 ± 0.07 vs 0.30 ± 0.06, C33A: 1.02 ± 0.07 vs 0.29 ± 0.05), while FOXP4-AS1 knockdown increases (Caski: 0.94 ± 0.09 vs 1.92 ± 0.10, C33A: 1.00 ± 0.09 vs 2.15 ± 0.19) the levels of miR-136-5p levels in CC cells (Figure 4B and C). The luciferase activity reporter assay showed miR-136-5p overexpression could decrease the luciferase activities in cells transfected with FOXP4-AS1-wt (Caski: 0.96 ± 0.06 vs 0.27 ± 0.06, C33A: 1.01 ± 0.05 vs 0.32 ± 0.05) but not FOXP4-AS1-mt. Indicating the mutations on the predicted lncRNA-miRNA binding regions can disturb the interaction of FOXP4-AS1 and miR-136-5p (Figure 4D). In addition, we showed miR-136-5p expression was decreased in Caski (0.31 ± 0.06) and C33A (0.32 ± 0.06) comparted with HCvEpC (1.03 ± 0.11) (Figure 4E).
Figure 4.
lncRNA FOXP4-AS1 could directly bind with miR-136-5p.
Notes: (A) Binding site between FOXP4-AS1 and miR-136-5p. Effects of FOXP4-AS1 (B) overexpression, and (C) knockdown on the expression of miR-136-5p. (D) Relative luciferase activities in CC cells with luciferase activity vectors and synthesized miRNAs. (E) miR-136-5p was reduced expression in CC cells. ***P < 0.001.
Abbreviations: lncRNA, long non-coding RNA; FOXP4-AS1, forkhead box P4 antisense RNA 1; CC, cervical cancer; miR-136-5p, microRNA-136-5p; wt, wild-type; mt, mutant; NC-mimic, negative control miRNA for miR-136-5p mimic.
miR-136-5p Targets CBX4 and Regulate Its Expression
We further predicted targets of miR-136-5p using bioinformatic algorithm and found miR-136-5p could bind with 3ʹ-UTR of CBX4 (Figure 5A). The introduction of miR-136-5p mimic decreased the expression of CBX4 in Caski (1.03 ± 0.06 vs 0.31 ± 0.03) and C33A (0.97 ± 0.06 vs 0.30 ± 0.03) (Figure 5B). In addition, overexpression of FOXP4-AS1 increases (Caski: 0.98 ± 0.05 vs 1.84 ± 0.09, C33A: 0.97 ± 0.06 vs 1.88 ± 0.09), and knockdown of FOXP4-AS1 decreases CBX4 expression in CC cells (Caski: 0.94 ± 0.09 vs 0.39 ± 0.09, C33A: 1.00 ± 0.09 vs 0.43 ± 0.08) (Figure 5C and D). Notably, the transfection of miR-136-5p mimic decreased luciferase activity in cells transfected with CBX4-wt (Caski: 1.02 ± 0.05 vs 0.29 ± 0.08, C33A: 0.98 ± 0.08 vs 0.35 ± 0.07) (Figure 5E). Furthermore, the expression of CBX4 was revealed to be significantly elevated in Caski (2.33 ± 0.14) and C33A (2.09 ± 0.16) compared with HCvEpC (1.03 ± 0.11) (Figure 5F).
Figure 5.
miR-136-5p could directly bind with CBX4.
Notes: (A) Binding site between CBX4 and miR-136-5p. (B) Effects of miR-136-5p overexpression on the expression of CBX4. Effects of FOXP4-AS1 (C) overexpression, and (D) knockdown on the expression of CBX4. (E) Relative luciferase activities in CC cells with luciferase activity vectors and synthesized miRNAs. (F) CBX4 was increased expression in CC cells. **P < 0.01, ***P < 0.001.
Abbreviations: FOXP4-AS1, forkhead box P4 antisense RNA 1; CBX4, chromobox 4; CC, cervical cancer; miR-136-5p, microRNA-136-5p; wt, wild-type; mt, mutant; NC-mimic, negative control miRNA for miR-136-5p mimic.
FOXP4-AS1 Regulates CC Cell Behaviors via miR-136-5p/CBX4
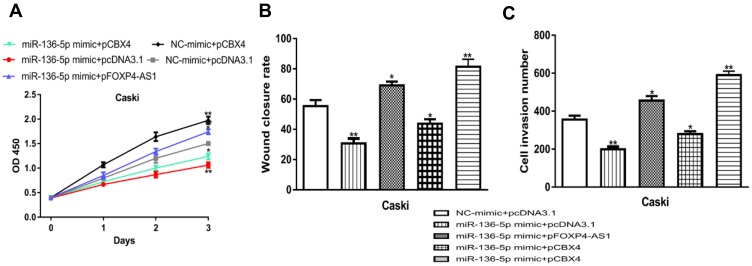
At length, we performed rescue experiments to investigate whether miR-136-5p/CBX4 involved in FOXP4-AS1 induced CC cell behaviors alteration. Functional assays revealed that cell proliferation, migration, and invasion abilities were increased by pCBX4 (proliferation: 1.98 ± 0.14 vs 1.50 ± 0.05, migration: 81.33 ± 5.03 vs 55.33 ± 4.04, invasion: 589 ± 22 vs 355 ± 21) and reduced by miR-136-5p mimic (proliferation: 1.06 ± 0.06 vs 1.50 ± 0.05, migration: 30.66 ± 3.21 vs 55.33 ± 4.04, invasion: 198 ± 14 vs 355 ± 21) (Figure 6A-C). Co-transfection of pFOXP4-AS1 and miR-136-5p mimic reversed the pFOXP4-AS1 induced stimulations on CC cell behaviors (proliferation: 1.74 ± 0.05, migration: 69.00 ± 2.65, invasion: 455 ± 24) (Figure 6A-C). Moreover, the co-transfection of pCBX4 and miR-136-5p mimic abolished the roles of miR-136-5p mimic on cell behaviors (proliferation: 1.24 ± 0.06 vs 1.06 ± 0.06, migration: 43.67 ± 3.06 vs 30.66 ± 3.21, invasion: 279 ± 15 vs 355 ± 21) (Figure 6A-C).
Figure 6.
lncRNA FOXP4-AS1 regulates CC cell proliferation, migration, and invasion via miR-136-5p/CBX4 axis.
Notes: (A) Effects of miR-136-5p or CBX4 on cell proliferation were analyzed with CCK-8 assay. (B) Roles of miR-136-5p or CBX4 on CC cell migration were analyzed with wound-healing assay. (C) Effects of miR-136-5p or CBX4 on CC cell invasion were measured by transwell invasion assay. *P < 0.05, **P < 0.01.
Abbreviations: lncRNA, long non-coding RNA; FOXP4-AS1, forkhead box P4 antisense RNA 1; miR-136-5p, microRNA-136-5p; CBX4, chromobox 4; CC, cervical cancer; CCK-8, cell counting kit-8; NC-mimic, negative control miRNA for miR-136-5p mimic.
Discussion
Survival of CC patients remains poor although the improvements in disease diagnosis or treatment methods.11 Emerging evidence indicated genomic mutations often occurred in regions that do not have the ability to encode proteins. Improvements in bioinformatic analyses methods and applications of high throughput sequencing technology have identified numerous aberrantly expressed lncRNAs in cancers.12,13 It should be noted lncRNAs play crucial roles in regulating CC carcinogenesis. Previous work found lncRNA SNHG7 was increased expression in CC tissues and correlated with poor overall survival.14 The knockdown of SNHG7 results in the inhibition of CC cell proliferation and invasion.14
Here, we reported lncRNA FOXP4-AS1 was elevated expression in Caski and C33A cells and to different content. The difference in expression level may be the different pathological and malignant status of cancer cells. Functional assays indicated that FOXP4-AS1 overexpression promoted CC cell proliferation, migration, and invasion. Moreover, we showed knockdown of FOXP4-AS1 inhibited CC cell behaviors. These results indicated an oncogenic role of FOXP4-AS1 in CC, which was consistent with its role in previous reported cancer types including gastric cancer, colorectal cancer, and hepatocellular carcinoma.8–10
Previous work found lncRNA can serve as sponge for miRNA as it contains miRNA binding sites.15 In this work, we found miR-136-5p was a potential target of FOXP4-AS1 using bioinformatic tools. miR-136-5p was reported to serve as a tumor-suppressive miRNA in renal cell carcinoma and has the potential to be used as biomarker for early detection and prognosis prediction.16 In addition, miR-136-5p was found decreased expression in lung squamous cell cancer and it may interact with UGT1A7 and ADH7 to regulate cellular metabolism processes.17 We validated the direct connection of FOXP4-AS1 and miR-136-5p using luciferase activity reporter assay. The previous works have identified multiplies miR-136-5p targets including UGT1A7, ADH7, and ROCK1 in cancers.17,18 Next, we found CBX4 as a target for miR-136-5p. CBX4 belongs to the Polycomb group (PcG) family and revealed to promote lung cancer proliferation and metastasis through recruiting BMI-1, indicating CBX4 may be a possible therapeutic target in cancer.19 Moreover, CBX4 was revealed to exert oncogenic roles in breast cancer progression via affecting the Notch1 signaling pathway.20 In our studies, we found lncRNA FOXP4-AS1 upregulates CBX4 expression via miR-136-5p. Rescue experiments by transfecting of miR-136-5p mimic, pFOXP4-AS1, and pCBX4 indicated lncRNA FOXP4-AS1 promotes CC cell proliferation, migration, and invasion through sponging miR-136-5p to regulate CBX4 expression as the introduction of pFOXP4-AS1 and pCBX4 could abolish the effects of miR-136-5p mimic on cellular behaviors.
Conclusion
Taken together, our results indicated that lncRNA FOXP4-AS1 promotes CC cell proliferation, migration, and invasion in vitro by targeting miR-136-5p and upregulating CBX4 expression, which helped us to understand the mechanisms of FOXP4-AS1 in CC development.
Funding Statement
This work was supported by the Scitech Program Foundation of Shaanxi Province (NO. 2017SF-016).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The author reports no conflicts of interest in this work.
References
- 1.Berman TA, Schiller JT. Human papillomavirus in cervical cancer and oropharyngeal cancer: one cause, two diseases. Cancer. 2017;123(12):2219–2229. doi: 10.1002/cncr.v123.12 [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 3.Wang B, Tang D, Zhang Z, Wang Z. Identification of aberrantly expressed lncRNA and the associated TF-mRNA network in hepatocellular carcinoma. J Cell Biochem. 2019. doi: 10.1002/jcb.29384 [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Tang D, Wang B, Wang Z, Liu M. Analysis of miRNA-mRNA regulatory network revealed key genes induced by aflatoxin B1 exposure in primary human hepatocytes. Mol Genet Genomic Med. 2019;7(11):e971. doi: 10.1002/mgg3.v7.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang JZ, Chen M, Chen D, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68(1):171–184. doi: 10.1016/j.molcel.2017.09.015 [DOI] [PubMed] [Google Scholar]
- 7.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 8.Chen RY, Ju Q, Feng LM, Yuan Q, Zhang L. The carcinogenic complex lncRNA FOXP4-AS1/EZH2/LSD1 accelerates proliferation, migration and invasion of gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23(19):8371–8376. doi: 10.26355/eurrev_201910_19148 [DOI] [PubMed] [Google Scholar]
- 9.Li J, Lian Y, Yan C, et al. Long non-coding RNA FOXP4-AS1 is an unfavourable prognostic factor and regulates proliferation and apoptosis in colorectal cancer. Cell Prolif. 2017;50(1):e12312. doi: 10.1111/cpr.2017.50.issue-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, Bai T, Chen G, et al. Upregulation of long non-coding RNA FOXP4-AS1 and its regulatory network in hepatocellular carcinoma. Onco Targets Ther. 2019;12:7025–7038. doi: 10.2147/OTT.S220923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 12.Zhao X, Tang DY, Zuo X, Zhang TD, Wang C. Identification of lncRNA-miRNA-mRNA regulatory network associated with epithelial ovarian cancer cisplatin-resistant. J Cell Physiol. 2019;234(11):19886–19894. doi: 10.1002/jcp.28587 [DOI] [PubMed] [Google Scholar]
- 13.Li L, Peng M, Xue W, et al. Integrated analysis of dysregulated long non-coding RNAs/microRNAs/mRNAs in metastasis of lung adenocarcinoma. J Transl Med. 2018;16(1):372. doi: 10.1186/s12967-018-1732-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng J, Ma YX, Liu ZH, Zeng YL. LncRNA SNHG7 contributes to cell proliferation, invasion and prognosis of cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23(21):9277–9285. doi: 10.26355/eurrev_201911_19420 [DOI] [PubMed] [Google Scholar]
- 15.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P, Zhao L, Pan X, et al. Tumor suppressor microRNA-136-5p regulates the cellular function of renal cell carcinoma. Oncol Lett. 2018;15(4):5995–6002. doi: 10.3892/ol.2018.8081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie ZC, Li TT, Gan BL, et al. Investigation of miR-136-5p key target genes and pathways in lung squamous cell cancer based on TCGA database and bioinformatics analysis. Pathol Res Pract. 2018;214(5):644–654. doi: 10.1016/j.prp.2018.03.028 [DOI] [PubMed] [Google Scholar]
- 18.Zhong Y, Yu C, Qin W. LncRNA SNHG14 promotes inflammatory response induced by cerebral ischemia/reperfusion injury through regulating miR-136-5p/ROCK1. Cancer Gene Ther. 2019;26(7–8):234–247. doi: 10.1038/s41417-018-0067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu C, Zhang Q, Tang Q, et al. CBX4 promotes the proliferation and metastasis via regulating BMI-1 in lung cancer. J Cell Mol Med. 2019. doi: 10.1111/jcmm.14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng JS, Zhang ZD, Pei L, et al. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. Int J Biochem Cell Biol. 2018;95:1–8. doi: 10.1016/j.biocel.2017.12.006 [DOI] [PubMed] [Google Scholar]