Abstract
Knowledge of immunodominant regions in major viral antigens is important for rational design of effective vaccines and diagnostic tests. Although there have been many reports of such work done for SARS–CoV, these were mainly focused on the immune responses of humans and mice. In this study, we aim to search for and compare immunodominant regions of the spike (S) and nucleocapsid (N) proteins which are recognized by sera from different animal species, including mouse, rat, rabbit, civet, pig and horse. Twelve overlapping recombinant protein fragments were produced in Escherichia coli, six each for the S and N proteins, which covered the entire coding region of the two proteins. Using a membrane-strip based Western blot approach, the reactivity of each antigen fragment against a panel of animal sera was determined. Immunodominant regions containing linear epitopes, which reacted with sera from all the species tested, were identified for both proteins. The S3 fragment (aa 402–622) and the N4 fragment (aa 220–336) were the most immunodominant among the six S and N fragments, respectively. Antibodies raised against the S3 fragment were able to block the binding of a panel of S-specific monoclonal antibodies (mAb) to SARS–CoV in ELISA, further demonstrating the immunodominance of this region. Based on these findings, one-step competition ELISAs were established which were able to detect SARS–CoV antibodies from human and at least seven different animal species. Considering that a large number of animal species are known to be susceptible to SARS–CoV, these assays will be a useful tool to trace the origin and transmission of SARS–CoV and to minimise the risk of animal-to-human transmission.
Abbreviations: CPE, cytopathic effect; CoV, coronavirus; mAb, monoclonal antibody; oscELISA, one-step-competition ELISA; SARS, severe acute respiratory syndrome
Keywords: SARS, Coronavirus, Antibody, Spike protein, Nucleocapsid protein, ELISA
1. Introduction
In late 2002 to middle 2003, outbreaks of severe acute respiratory syndrome (SARS), also known as atypical pneumonia, spread through more than 30 countries and caused more than 8000 cases with close to 800 human deaths (Peiris et al., 2004). The aetiological agent was a previously unknown coronavirus, now named SARS associated coronavirus or SARS–CoV, which seems to be evolutionally distant to any of the existing coronaviruses known at the time (Rota et al., 2003, Marra et al.). Initial epidemiology studies indicated an animal origin (Xu et al., 2004), which was further confirmed by the detection and isolation of almost identical viruses from wild life animals in live animal markets (Guan et al., 2003). More recently, SARS–like-CoVs were identified among different species of horseshoe bats (Li et al., 2005, Lau et al., 2005), implicating bats as the potential reservoir of these novel coronaviruses (Wang et al., 2006).
Coronaviruses are enveloped positive-stranded RNA viruses containing several structural proteins including the spike glycoprotein (S), nucleocapsid protein (N), membrane protein (M), and envelope protein (E). Receptor-binding and membrane fusion between the virus and host cell is mediated by the S protein, which is also the main target for neutralizing antibodies (Lai et al., 2007). The S protein is a large class I transmembrane glycoprotein consisting of two domains: the S1 domain at the N terminus responsible for receptor-binding and the C-terminal S2 domain functioning in fusion. For SARS–CoV, the minimal receptor-binding domain has been defined as a 193-aa fragment (aa 318–510) which binds to the receptor molecule, ACE2, in vitro (Li et al., 2003, Wong et al., 2004). The N protein is thought to participate in the replication and transcription of viral RNA and interfere with cell cycle processes of host cells. In addition, the N protein of many coronaviruses is highly immunogenic and abundantly expressed during infection, making it an attractive target for detection of virus-specific antibodies (Lai et al., 2007).
There are several diagnostic approaches developed for the detection of SARS–CoV infection, including PCR-based assays, antigen detection and antibody detection (Shi et al., 2003, He et al., 2005, Houng et al., 2004, Manopo et al., 2005, Wang et al., 2005b, Chou et al., 2005). Most, if not all, of these detection methods were developed for human applications. There is a lack of attention to the study of immune responses in different animal species and a lack of proper diagnostic tools for wide range surveillance of animal populations. Yet, it is well documented that SARS–CoVs and SARS-like-CoVs can infect a wide range of animal species (Wang et al., 2006). In most cases, there is no obvious clinical sign associated with the infection, making early detection and diagnosis even more difficult. Knowing that SARS–CoV originated from an animal reservoir and animals played a key role in the adaptation and transmission to the human population (Holmes, 2005, Li et al., 2006), accurate diagnosis of SARS–CoV infection among different animals species should be an important and integral part of an effective prevention strategy for reducing animal-to-human transmission.
In this study we identified and characterized the major immunodominant domains of the SARS–CoV N and S proteins recognized by different animal species, and then developed competition ELISAs based on these findings. By conjugating the detection antibodies with horseradish peroxidase, we developed one-step-competition ELISAs (or oscELISA) which are simple to use and can detect SARS-specific antibodies from different species. With further improvement and validation, we are hopeful that this will become a powerful tool for early detection of SARS–CoV infections in animals, which will in turn help to reduce the risk of future SARS outbreaks in humans.
2. Materials and methods
2.1. Virus and cell culture
The HKU-39849 strain of SARS–CoV was used in this study and all live virus work was carried out in the Biosafety Level 4 (BSL-4) laboratory under protocol approved by the Institutional Biosafety Committee (IBC). Culturing of virus in Vero E6 cells was conducted as previously reported (Tu et al., 2004). For use outside the BSL-4 laboratory, virus preparation was inactivated by gamma-irradiation at a dose of 50 kGy.
2.2. Antisera and monoclonal antibodies
Anti-SARS–CoV sera from rabbits, rats and mice were generated in the BSL-4 animal facility at the Australian Animal Health Laboratory (AAHL). To test the susceptibility and to generate antibodies from live virus, 100,000 TCID50 SARS–CoV was administered oronasally to rabbits (New Zealand white, n = 2), mice (Balb/c, n = 6) and rats (Sprague–Dawley, n = 4). Daily monitoring for clinical signs did not detect disease and after three weeks the animals were euthanized and blood samples were taken for serology. Anti-SARS–CoV antibody in serum was detected using immunofluorescence antibody test (IFAT) and Western blot assays, and confirmed by virus neutralization test (VNT). In addition, a collection of human and animal antisera was also used in this study. They included human sera from SARS patients (Liang et al., 2004), hyperimmune horse, pig and rabbit sera raised against formalin inactivated SARS–CoV (C. Tu, unpublished results), pig sera from experimental infection studies (Weingartl et al., 2004), sera from naturally infected civets (Tu et al., 2004) and bats (Li et al., 2005), and a panel of mouse monoclonal antibodies (Berry et al., 2004).
2.3. Recombinant antigens
For cDNA preparation, SARS–CoV RNA was extracted from the inactivated virus pellet using the QIAamp Viral RNA Mini Kit (Qiagen), followed by cDNA synthesis using the Omniscript RT Kit (Qiagen) and random hexamer primers. PCR primers were designed using published sequence to amplify fragments coding overlapping regions of the S and N proteins, respectively. All forward primers contain the MluI site and the reverse primers contain the NotI site (primer sequences will be provided on request) to facilitate cloning into the modified expression vectors pHAN and pBAN, which were modified, respectively, from the pRSET vector (Invitrogen) for expression of His6-tagged fusion proteins and the pDW363 vector (Tsao et al., 1996) for expression of fusion proteins with a biotin tag at the N terminus. Both vectors have in-frame AscI and NotI sites in their multiple cloning sites.
E. coli BL21 (DE3) (Novagen) harbouring appropriate recombinant plasmid was grown in 2 ml of LB broth containing ampicillin (50 μg/ml) at 37 °C with shaking overnight. The culture was diluted 1:100 and incubated at 37 °C with shaking until OD600 reached 0.5. Protein expression was induced by addition of Isopropyl-beta-d-thiogalactoside (IPTG) at a final concentration of 1 mM. After 4 h, bacterial cells were harvested by centrifugation. The cell pellet was resuspended in lysis buffer (500 mM NaCl, 50 mM phosphate buffer, pH7.8) and lysed by sonication. The insoluble protein fraction was collected by centrifugation and solubilised in 8 M urea (in the same lysis buffer). The recombinant proteins were then purified by preparative sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (Tachedjian et al., 2006).
2.4. Production, purification and conjugation of chicken mono-specific antibodies
Four 16-week-old chickens were immunized with recombinant proteins for antibody production, two with His6-S3 and two with biotin-Nc (see Fig. 1 ). For primary immunization, 50 μg of recombinant protein was mixed with the CSIRO Triple Adjuvant (Tachedjian et al., 2006) before injection. Three weeks later, the same injection was repeated for boost immunization. The final boost was carried out by injection of 50 μg protein without adjuvant at three weeks after the second injection. Chickens were bled before each injection and SARS–CoV reactive antibodies were determined by ELISA using inactivated SARS–CoV antigen (see below).
Fig. 1.
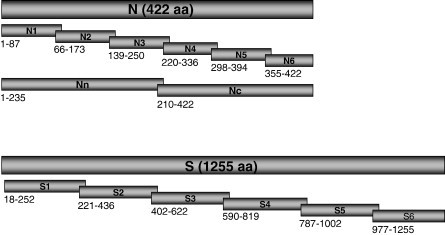
Schematic diagram of the S and N protein fragments generated in this study. The numbers shown below indicate the region (aa residue positions) of each fragment expressed.
Eggs were collected for a period of five months and egg yolk was pooled for purification of IgY antibodies. The IgY Extraction and Purification Kit (Pierce) was used for purification as follows: one part of egg yolk (approximately 10 ml) was mixed with 4 parts of phosphate buffered saline (PBS), pH 7.5, followed by the addition of PEG 6000 to a final concentration of 3.5% (w/v). The mixture was stirred for 20 min at room temperature and centrifuged for 25 min at 5000 ×g. The supernatant was then filtered through cotton wool in a funnel, and PEG 6000 was added to the clear supernatant to a final concentration of 12% (w/v). The mixture was stirred for 10 min at room temperature and centrifuged as above. After dissolving the precipitate in 25 ml of PBS, the same procedure was repeated once by addition of PEG 6000 to 12%, mixing and centrifugation. The final precipitate was slowly dissolved in 2.5 ml of PBS and dialysed against PBS overnight at 4 °C. Aliquots of IgY antibodies were stored at − 20 °C for future use.
Conjugation of IgY antibodies to horseradish peroxidase (HRP) was carried out following the protocol provided by the EZ-Link Plus Activated Peroxidase Kit (Pierce). Briefly, 1 ml of IgY antibodies at approximately 1 mg/ml in conjugation buffer was added to the lyophilized EZ-Link Plus Activated Peroxidase. The mixture was incubated for 1 h at room temperature, followed by the addition of 10 μl Reductant Solution and a further incubation of 15 min. The reaction was stopped by adding 20 μl of Quench Buffer, followed by incubation at room temperature for 15 min. The conjugate was dialysed against PBS buffer overnight at 4 °C and aliquots were stored at − 20 °C in the presence of 50% glycerol.
2.5. Detection of specific antibodies by immunofluorescent antibody test, Western blot and virus neutralization test
All immunofluorescent antibody tests were carried out following the standard protocols previously described by our group (Tu et al., 2004, Zhang et al., 2006). Western blot analysis was carried out as described (Wang et al., 1996). To produce membrane strips for testing reactivity of the same antigen with a panel of different sera, 20 μg of purified protein was loaded into a wide comb, separated by SDS-PAGE and transferred to nitrocellulose membrane. The membrane was then sliced into 5 mm-wide strips and each strip tested with a different serum sample. Alkaline phosphatase (AP)-conjugated secondary antibodies and AP-streptavidin were obtained from Chemicon Australia. For virus neutralization, serial two-fold dilutions of test sera and control sera were prepared in duplicate in a 96-well tissue culture plate in 50 μl cell media (Minimal Essential Medium containing Earle's salts and supplemented with 2 mM glutamine, 500 µg/ml fungizone, 100 units/ml penicillin, 100 µg/ml streptomycin and 10% fetal calf serum). Under BSL4 conditions, an equal volume of SARS–CoV working stock (HKU-39849) containing 200 TCID50 was added and the virus-sera mix incubated for 30 min at 37 °C in a humidified 5% CO2 incubator. 100 μl of Vero cell suspension containing 2 × 105 cells/ml was added and the plate incubated at 37 °C in a humidified 5% CO2 incubator. After three days, the plate was observed for CPE.
2.6. ELISAs
All ELISA assays were carried out using standard procedures established in our group (Yu et al., 2005, Zhang et al., 2006). All incubations were conducted at 37 °C for 1 h with gentle shaking, and all washes (five times between each step) were carried out at room temperature using PBS containing 0.05% Tween 20 (PBST). Antigens were coated in 0.1 M carbonate buffer (pH 9.3) and antibody dilutions were done in blocking solution (2% skim milk in PBST). PVC 96-well plate (Dynatech) was used for all ELISA analysis. For indirect ELISAs, viral or recombinant antigens were coated at a concentration of 50–150 ng/well, and bound antibodies were detected using HRP-conjugated species-specific secondary antibodies (Chemicon) or HRP-conjugated protein A/G (Zymed). Color development was conducted using the substrate 3,3,5,5, tetramethylbenzidine (TMB) and the absorbance at 450 nm was determined. For competition ELISA, detecting antibody (mAb or chicken mono-specific antibody) was added directly after the antigen-coated plate was pre-incubated with test sera without wash steps. The percentage inhibition of detecting antibody binding was calculated using the following formula:
3. Results
3.1. Production of SARS–CoV antisera in different experimental animals
To test for susceptibility and to generate antisera in experimental small animals, rabbits, rats and mice were inoculated with 100,000 TCID50 SARS–CoV oronasally and monitored daily for clinical signs. None of the animals showed any signs of disease. Three weeks post-infection, all animals were euthanized and serum samples were taken for analysis using three different assays, i.e., immunofluorescent antibody test (IFAT), Western blot and virus neutralization test (VNT). The results are shown in Table 1 . Seroconversion was observed in all three species although not in all animals. From the limited number of animals used in this study, it appeared that rats were the most susceptible and displayed the highest VNT titres in their sera varying from 1:80 (n = 3) to 1:160 (n = 1). Four of the six mice produced antibodies detected by IFAT and Western blot, but only two of them were positive in VNT. One of the two rabbits also seroconverted, with a low VNT titre of 1:20.
Table 1.
Serologic analysis of sera from animals inoculated oronasally with live SARS–CoV
| Animal Species | Animal # | VNT Titera | IFATb | Western Blotc |
|---|---|---|---|---|
| Mouse | 1 | −ve | +++ | ++ |
| 2 | 1:20 | ++++ | ++ | |
| 3 | 1:10 | +++ | ++ | |
| 4 | −ve | −ve | ++ | |
| 5 | −ve | ++ | ++ | |
| 6 | −ve | ++ | ++ | |
| Rat | 1 | 1:80 | ++ | ++ |
| 2 | 1:80 | ++ | ++ | |
| 3 | 1:80 | ++ | ++ | |
| 4 | 1:160 | ++ | ++ | |
| Rabbit | 1 | −ve | −ve | −ve |
| 2 | 1:20 | ++++ | ++++ |
Titre of antibody that neutralized infectivity of 200 TCID50 of SARS–CoV.
Immunofluorescent antibody test was done using a serum dilution of 1:50 and the intensity of staining in SARS–CoV-infected cells, but not in mock infected cells, was indicated by the sign + with ++++ representing the strongest signal observed.
Western blot was carried out using a serum dilution of 1:200 and the intensity of the reactivity against a recombinant full-length N protein was indicated by the sign + with ++++ representing the strongest reactivity.
3.2. Expression and purification of recombinant proteins
Recombinant proteins, full-length or serially overlapping fragments, were produced in E. coli for determination of immunodominant regions in the S and N proteins. PCR fragments were cloned into two expression systems, the pRSET-derived pHAN vector for production of His6-tagged proteins and the pDW363-derived pBAN vector for biotin-tagged proteins. All of the recombinant proteins generated in this study are summarised in the diagram in Fig. 1 with their aa positions indicated. For each construct, two positive clones were confirmed by DNA sequencing for correct insert sequence and in-frame fusion with the vector-encoded tagging sequences. As expected, the level of expression varied with the different constructs, and some expressed better in the His6-tagging system and other vice versa (data not shown). Overall, the expression level varied from 0.5–1 mg/l to approximately 10–20 mg/l. Most of the expressed proteins were present largely in the insoluble fractions when analysed by Western blot using anti-His6 antibody (for His6-tagged proteins) or AP-conjugated streptavidin (for biotin-tagged proteins). This observation prompted us to purify the recombinant proteins using preparative SDS-PAGE under denaturing conditions. A purity of 90% or greater was achieved for all recombinant proteins when analysed by direct staining with Commassie blue.
3.3. Determination of the immunodominant regions recognized by sera from different animal species
For mapping of immunodominant regions of the S and N proteins, Western blot using membrane strips was conducted. Each of the 12 recombinant protein fragments, His6-tagged S1 to S6 and biotin-tagged N1 to N6, was tested against a panel of animal sera from six different animal species. The results are compiled in Table 2 together with the reactivity of each serum sample in VNT. The hyperimmune horse serum was known to contain a very high titre of antibodies and was used as a positive control in this study. As shown in Table 2, the horse serum reacted with all of the S and N protein fragments tested. Overall, the hyperimmune sera (especially those from horse and rabbit) displayed more reactivity towards different fragments than sera obtained from experimental or natural infection (civet, rabbit, mouse and rat). However, the results in Table 2 clearly indicate that S3 and N4 represented the most cross-reactive regions of the S and N proteins, respectively. In addition, the data also reveal that the C-terminal half of the N protein is more immunogenic than the N-terminal half, at least for the epitopes which were recognizable under denaturing conditions, i.e., in Western blot.
Table 2.
Summary of western blot profile for different animal sera against the N and S-protein fragments
| Animal Seraa | VNT titre | Western blot: N fragmentsb |
Western blot: S fragmentsb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | N3 | N4 | N5 | N6 | S1 | S2 | S3 | S4 | S5 | S6 | ||
| Horse/IM | 1:1280 | + | + | + | + | + | + | + | + | + | + | + | + |
| Civet/IF | 1:240 | + | + | + | + | + | + | + | |||||
| Mouse/IF | 1:20 | + | + | + | + | + | |||||||
| Pig/IM | 1:40 | + | + | + | + | + | |||||||
| Rat/IF | 1:180 | + | + | + | + | + | + | + | + | ||||
| Rabbit/IM | 1:80 | + | + | + | + | + | + | + | + | + | + | ||
| Rabbit/IF | 1:20 | + | + | + | + | + | |||||||
For each animal species, a representative positive serum with the best reactivity in VNT and IFAT was used for this study. Abbreviations: IF, infected sera; IM, immunized sera;
Positive reactivity in Western blot is indicated by the + sign.
3.4. Generation and characterization of chicken anti-Nc and anti-S3 antibodies
Based on the results obtained above, mono-specific antibodies were raised in chickens against the S3 and Nc fragments. Two chickens each were used for production of anti-S3 and anti-Nc antibodies. After three immunizations, the peak antibody titres were determined by ELISA using whole virus antigen. For both S3 and Nc, a final titre of 1:100,000 to 200,000 was obtained (data not shown). The specificity of the antibody was further confirmed using Western blot against viral antigen (data not shown) and IFAT using SARS–CoV infected Vero cells. As shown in Fig. 2 , both antibodies reacted with SARS–CoV infected Vero cells whereas the pre-bleed chicken sera did not. When tested in VNT, neither of the anti-S3 or anti-Nc antibodies showed any evidence of neutralization activity (data not shown).
Fig. 2.
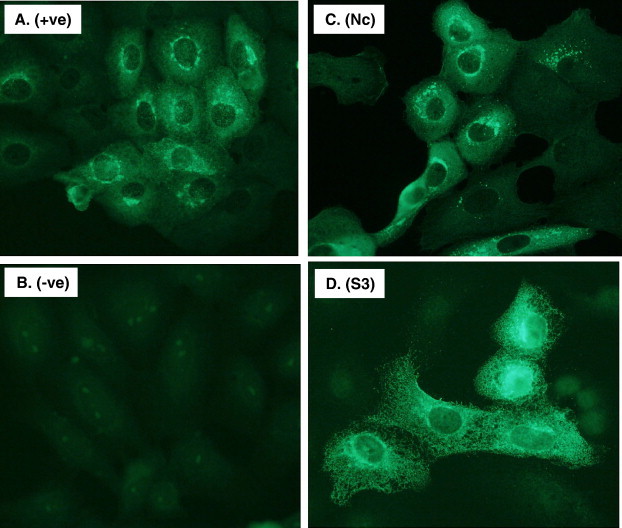
Test of mono-specific chicken antibodies by IFAT. Vero E6 cells infected with SARS–CoV was tested with the following antibodies (dilution of antibodies given in parenthesis): A. Rabbit anti-SARS–CoV serum (1:500); B. Pre-bleed chicken serum (1:200); C. Chicken anti-Nc serum (1:200); and D. Chicken anti-S3 serum (1:200).
For conjugation of HRP to the chicken antibodies, crude IgY antibodies were purified from egg yolks using the PEG precipitation method and protein concentration and purity were determined by SDS-PAGE in comparison with serially diluted BSA standards run on the same gel. The protein concentration of anti-S3 and anti-Nc IgY was at 2 mg/ml and 5 mg/ml, respectively. The purity for both antibodies was approximately 85–90%. The antibody titres of the partially purified anti-S3 and anti-Nc IgY were 1:3200 and 1:6400, respectively, tested using the same ELISA as for the serum antibodies above. An aliquot (approximately 1 mg) of each IgY antibody was then conjugated to HRP using the Pierce EZ-Link Plus Activated Peroxidase Kit. The final titres of the HRP-anti-S3 and HRP-anti-Nc antibodies were very similar, 1:200 and1:100, respectively.
3.5. Mapping binding sites of S-specific monoclonal antibodies
Among a panel of mAbs generated using inactivated SARS–CoV, five were directly shown to be S-specific by Western blot analysis (Berry et al., 2004). In addition, four other mAbs were shown to neutralize SARS–CoV although failed to react with the S protein in Western blot. In this study, competition ELISA (cELISA) was carried out between chicken anti-S3 antibodies and these mAbs, using chicken anti-Nc as a control. The data shown in Fig. 3A indicated that the binding of three known S-specific mAbs and four neutralizing mAbs to SARS–CoV antigen was blocked by the anti-S3 antibodies, but not by the anti-Nc antibodies. Among these mAbs, F26G8 displayed the most efficient blocking by the chicken anti-S3 antibodies. Western blot analysis of the six S-protein fragments indicated that F26G8 was able to bind both the S3 and S4 fragments (Fig. 3B), suggesting that its epitope is located in the overlapping region (aa 590–622) of the two protein fragments.
Fig. 3.
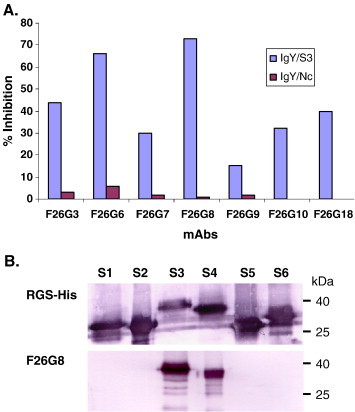
Mapping of mAb binding regions by cELISA and Western blot. A. Competition ELISA using whole virus as antigen. The IgY antibodies were used at 1:10 for both Nc and S3. The percentage inhibition was calculated using the formula given in Materials and methods. B. Western blot analysis of all six S fragments using mAbs RGS-His and F26G8.
3.6. Competition ELISAs for detection of SARS–CoV antibodies from different species
One of the main aims of this study was to assess the feasibility of developing a competition ELISA for detection of SARS–CoV antibodies from different animal species. Based on the results above, it was evident to us that a cELISA based on either mAb F26G8 or the chicken mono-specific antibodies might be ideal for such an application. To confirm this, three separate cELISAs were developed and tested against a panel of negative and positive SARS–CoV sera from different species. Firstly, a cELISA based on mAb F26G8 was tested against a panel of known positive and negative sera from different species, and the results in Fig. 4 suggested that each serum was able to inhibit F26G8 binding, with the civet sera displaying the strongest inhibition.
Fig. 4.
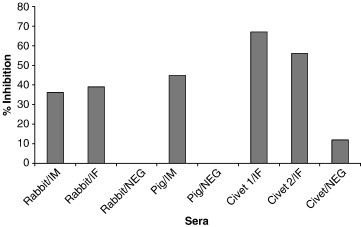
Inhibition of F26G8 binding to SARS–CoV by sera from different species. All animal sera were used at a dilution of 1:10. The percentage inhibition was calculated using the formula given in Materials and methods. Abbreviations: IF, infected sera; IM, immunized sera; NEG, negative control sera.
We then tested the feasibility of using the HRP-conjugated chicken mono-specific antibodies as detecting antibody in a cELISA. Since the ELISA only requires one-step incubation with both detection and testing antibodies, we termed the assay one-step-competition ELISA or oscELISA. As shown in Fig. 5 , although different sera showed slightly different inhibition patterns against the HRP-IgY/S3 or HRP-IgY/Nc antibodies, both assays, especially the oscELISA using HRP-IgY/S3 antibody, were able to detect positive sera from a wide range of animal sera. It is worth noting that the sera from bats infected with SARS-like-CoVs inhibited HRP-IgY/Nc antibodies more effectively than HRP-IgY/S3 antibodies (see more in Discussion).
Fig. 5.
Inhibition of binding of mono-specific chicken antibodies to SARS–CoV by sera from different species. All animal sera were used at 1:10 for this study. Since animal sera of different species gave rise to different level of background inhibition, the percentage inhibition was calculated using the following formula:% inhibition = 100 − (ODN − ODT / ODB) × 100, in which ODN = OD obtained in the presence of known negative sera from the same species; ODT = OD obtained in the presence of test sera; and ODB = OD obtained with buffer only. Abbreviations: IF, infected sera; IM, immunized sera; NEG, negative control sera.
4. Discussion
Compared with the great success in rapid identification of the SARS causative agent involved in and the control of the SARS outbreaks, our understanding of the origin and mechanism of animal-to-animal and animal-to-human transmission of SARS–CoV is less satisfactory. With recent identification of SARS-like-CoVs in bats (Li et al., 2005, Lau et al., 2005) and the key role played by civets in direct transmission of SARS–CoV to humans (Liang et al., 2004, Wang et al., 2005a, Wang et al., 2006), it is evident that further research is required before we are able to get a better understanding of what was the true origin of the SARS–CoVs responsible for the 2002–2003 outbreaks and whether such events can happen again in the future.
There are two major difficulties associated with the study of roles played by animals leading to potential SARS outbreaks. First, there are more than a dozen animal species known to be susceptible to SARS–CoV by either natural or experimental infection (Wang et al., 2006), and it is likely that more animal species could be susceptible too. It is important to differentiate between those which play a key role in the origin and transmission of SARS–CoV and those that are infected, but play no significant role in disease spread and spill over to the human population. Secondly, there is a lack of an ideal detection approach to monitor infection status of different animal species. The most commonly used tool is direct PCR analysis for the presence of viral material in animal specimens (Guan et al., 2003, Poon et al., 2005, Kan et al., 2005, Woo et al., 2006, Tang et al., 2006). While this is effective to detect active infection, it is not very sensitive to monitor prior exposure in a given animal population. We believe that a serological test which detects SARS–CoV antibodies will be a more powerful tool for investigation of SARS–CoV prevalence among different animal populations.
To develop an antibody test which can be applied to different animal species, it is essential to first have a panel of different animal sera which are known to be positive to SARS–CoV. This was achieved by a broad international collaboration among groups who have access to such sera from naturally infected wild life animals, experimentally infected animals or animals immunized with inactivated virus.
For production of SARS–CoV antisera in small animals at AAHL, we chose to use live virus for two reasons. Firstly, this allowed us to test the susceptibility of these animals to SARS–CoV (the work began in 2003, before any experimental animal infection studies were published). Secondly, if the animals were susceptible, this would provide us better quality reagent than that obtained from immunization with inactivated virus and adjuvant. The seroconversion data in this study indicate that rats, mice and rabbits are susceptible to infection of SARS–CoV by the oronasal route. When the same amount of virus was administered subcutaneously and intra-peritoneally, none of the animals seroconverted (data not shown), suggesting that the seroconversion is due to the replication of SARS–CoV rather than immunization and that the route of administration is important for the infection to occur.
To determine the immunodominant regions of the N and S proteins recognized by different animal species, a series of overlapping truncated recombinant fragments were produced in E. coli. Reactivity of different animal sera against each of the 12 protein fragments was conducted by Western blot. While it is known that Western blot is not ideal for detecting conformational epitopes, it is the most feasible and effective way to unequivocally locate at least the linear or continuous immunodominant epitopes. The two most immunodominant regions, S3 and N4, were identified for the S and N proteins, respectively. For the N protein, it seems that the C-terminal region is more immunogenic than the N-terminal region. For the S protein, the location of S3 (aa 402–622) is consistent with previous studies conducted with SARS patient sera, which revealed that the immunodominant regions are located in the following regions of the S protein, aa 528–635 (He et al., 2004), 540–559 (Lu et al., 2005) and 441–700 (Manopo et al., 2005). Combined with our data, these results suggest that the common immunodominant regions are shared among humans and animals.
Based on the results above, mono-specific antibodies targeting the S3 and Nc regions were produced in chickens. The chicken was chosen for several reasons. It was known that avian species were not susceptible to SARS–CoV (Weingartl et al., 2004, Swayne et al., 2004), and antibodies produced in chicken can be used as detecting antibodies in a competition ELISA for SARS–CoV sera from any mammalian species without the concern of potential cross reactivity of anti-species secondary antibodies. Collection and purification of IgY antibodies from egg yolk have proven to be a very effective and economical way of producing large quantities of antibodies (Tachedjian et al., 2006). By collecting the lymphocytes (bursa or spleen) of antigen-sensitized chickens, the immune gene repertoire can be stored for later production of recombinant chicken single chain antibodies, which is technically less challenging than the production of rabbit or mouse single chain antibodies (Foord et al., 2007). The successful production of S3- and Nc-specific antibodies was confirmed by several assays, including IFAT, ELISA and Western blot. The results presented in Fig. 2 demonstrated not only the specific staining of viral antigens in SARS–CoV infected cells, but also the different staining patterns for the two antibodies. This was expected considering that S3 antibodies should react to the extracellular domain of the S proteins whereas the N proteins are known to be located intra-cellularly (Lai et al., 2007).
In addition to the main application of these antibodies in the development of cELISA, these mono-specific antibodies also proved useful in determining the specificity of several mAbs produced in a previous study (Berry et al., 2004). The data presented in Fig. 3 indicated that all four conformational neutralizing mAbs (F26G3, F26G7, F26G9 and F26G10) are S-specific and bind to a domain overlapping with the S3 region.
In comparison with an indirect ELISA, the cELISA has the advantage in that it can be used for detection of antibodies from different species without having to use species-specific secondary antibodies. Most cELISAs employ mAbs as the detecting antibodies to achieve maximal specificity. However, for viruses like SARS–CoV, which are known to have many variants and are rapidly mutating, a mAb-based cELISA may potentially give false negative diagnosis due to subtle changes of protein sequence close to or in the epitope region. For this reason, we have developed in parallel cELISAs using mono-specific antibodies as detecting antibodies. Also, by directly conjugating the chicken antibodies with HRP, we were able to develop oscELISAs for detection of SARS–CoV antibodies from any species, from human, civets, to bats.
It is important to point out that due to the limited availability of positive sera from different animal species, the cELISAs developed in this study need more optimization and validation before they can be used as robust diagnostic assays. It is also important to note that depending on the purpose of application, one may wish to use different assays under different circumstances. Although sequence analysis would suggest low probability of interference by cross-reactive antibodies to other animal coronaviruses, this is yet to be confirmed experimentally for the HRP-IgY/NC oscELISA. However, we are certain that this will not be an issue in the S-protein-based cELISAs. This was deduced from the preliminary data obtained with bat SL-CoV antisera in this study. It was clear that not all bat SL-CoV sera were detectable in the HRP-IgY/S3 oscELISA. The failure to detect some positive bat sera using the HRP-IgY/S3 oscELISA can be explained by the significant sequence difference present in the N-terminal half of the S proteins of SARS–CoVs and SARS-like-CoVs (Li et al., 2005). The development of three cELISAs was intended to provide choices for different applications. If the most specific antibody test is needed to detect SARS–CoV infection, the mAb-based cELISA may be the best choice. On the other hand, if one wishes to detect antibodies to all of the related SARS–CoVs and SARS-like-CoVs, the anti-Nc-based cELISA will be more appropriate. The anti-S3-based cELISA will be most useful to detect antibodies for SARS–CoVs only, but allow for sequence variations in the S proteins of different SARS–CoV strains.
Acknowledgements
The authors would like to thank D. Middleton, J. Bingham, K. Halpin, and R. Fogarty for help with animal infection experiments; M. Tachidjian and S. Wilson for assistance in production of chicken antibodies; K. Selleck, and J. Brangwyn for technical support; D. Magoffin and K. Halpin for critical reading of the manuscript; and T. Pye and E. Hansson for DNA sequencing service. The major component of the study described in this paper was funded by a grant (Project 1.007R) from the Australian Biosecurity Cooperative Research Centre for Emerging Infectious Diseases to L.-F.W.
References
- Berry J.D., Jones S., Drebot M.A., Andonov A., Sabara M., Yuan X.Y., Weingartl H., Fernando L., Marszal P., Gren J., Nicolas B., Andonova M., Ranada F., Gubbins M.J., Ball T.B., Kitching P., Li Y., Kabani A., Plummer F. Development and characterisation of neutralising monoclonal antibody to the SARS–coronavirus. J Virol. Meth. 2004;120:87. doi: 10.1016/j.jviromet.2004.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.-F., Shen S., Tan Y.-J., Fielding B.C., Tan T.H.P., Fu J., Xu Q., Lim S.G., Hong W. A novel cell-based binding assay system reconstituting interaction between SARS–CoV S protein and its cellular receptor. J. Virol. Meth. 2005;123:41. doi: 10.1016/j.jviromet.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foord A.J., Muller J.D., Yu M., Wang L.-F., Heine H.G. Production and application of recombinant antibodies to foot-and-mouth disease virus non-structural protein 3ABC. J. Immunol. Meth. 2007;321:142. doi: 10.1016/j.jim.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- He Q., Du Q., Lau S., Manopo I., Lu L., Chan S.-W., Fenner B.J., Kwang J. Characterization of monoclonal antibody against SARS coronavirus nucleocapsid antigen and development of an antigen capture ELISA. J. Virol. Meth. 2005;127:46–53. doi: 10.1016/j.jviromet.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Wu H., Luo B., Chen J., Li W., Jiang S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173:4050. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- Holmes K.V. Adaptation of SARS coronavirus to humans. Science. 2005;309:1822. doi: 10.1126/science.1118817. [DOI] [PubMed] [Google Scholar]
- Houng H.-S.H., Norwood D., Ludwig G.V., Sun W., Lin M., Vaughn D.W. Development and evaluation of an efficient 3'-noncoding region based SARS coronavirus (SARS–CoV) RT-PCR assay for detection of SARS–CoV infection. J. Virol. Meth. 2004;120:33. doi: 10.1016/j.jviromet.2004.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Qi X., Chen K., Du L., Gao K., Zhao Y.T., Zou X.Z., Feng Y.J., Gao Y.F., Hai R., Yu D., Guan Y., Xu J. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Perlman S., Anderson L.J. Coronaviridae. In: Knipe D.M., Griffin D.E., Lamb R.A., Straus S.E., Howley P.M., Martin M.A., Roizman B., editors. Fields Virology. 5th Ed. Lippincott Williams & Wilkins; Philadelphia: 2007. p. 1587. [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wong S.-K., Li F., Kuhn J.H., Huang I.-C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Viol. 2006;80:4211. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.-F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Liang G., Chen Q., Xu J., Liu Y., Lim W., Peiris J.S., Anderson L.J., Ruan L., Li H., Kan B., Di B., Cheng P., Chan K.H., Erdman D.D., Gu S., Yan X., Liang W., Zhou D., Haynes L., Duan S., Zhang X., Zheng H., Gao Y., Tong S., Li D., Fang L., Qin P., Xu W., SARS Diagnosis Working Group Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg. Infect. Dis. 2004;10:1774. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Wu X.-D., Shi M.D., Yang R.F., He Y.Y., Bian C., Shi T.L., Yang S., Zhu X.-L., Jiang W.-H., Li Y.X., Yan L.-C., Ji Y.Y., Lin Y., Lin G.-M., Tian L., Wang J., Wang H.X., Xie Y.H., Pei G., Wu J.R., Sun B. Synthetic peptides derived from SARS coronavirus S protein with diagnostic and therapeutic potential. FEBS Lett. 2005;579:2130. doi: 10.1016/j.febslet.2005.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manopo I., Lu L., He Q., Chee L.L., Chan S.-W., Kwang J. Evaluation of a safe and sensitive Spike protein-based immunofluorescence assay for the detection of antibody responses to SARS–CoV. J. Immunol. Meth. 2005;296:37. doi: 10.1016/j.jim.2004.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Jones, S.J., Astell, C.R., Holt, R.A., Brooks-Wilson, A., Butterfield, Y.S., Khattra, J., Asano, J.K., Barber, S.A., Chan, S.Y., Cloutier, A., Coughlin, S.M., Freeman, D., Girn, N., Griffith, O.L., Leach, S.R., Mayo, M., McDonald, H., Montgomery, S.B., Pandoh, P.K., Petrescu, A.S., Robertson, A.G., Schein, J.E., Siddiqui, A., Smailus, D.E., Stott, J.M., Yang, G.S., Plummer, F., Andonov, A., Artsob, H., Bastien, N., Bernard, K., Booth, T.F., Bowness, D., Czub, M., Drebot, M., Fernando, L., Flick, R., Garbutt, M., Gray, M., Grolla, A., Jones, S., Feldmann, H., Meyers, A., Kabani, A., Li, Y., Normand, S., Stroher, U., Tipples, G.A., Tyler, S., Vogrig, R., Ward, D., Watson, B., Brunham, R.C., Krajden, M., Petric, M., Skowronski, D.M., Upton, C., Roper, R.L., The Genome Sequence of the SARS-Associated Coronavirus. Science 300, 1399. [DOI] [PubMed]
- Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., Ma Q., Cao C. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapside antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2003;41:5781. doi: 10.1128/JCM.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D.E., Suarez L., Spackman E., Tumpey T.M., Beck J.R., Erdman D., Rollin P.E., Ksiazek T.G. Domestic poultry and SARS coronavirus, southern China. Emerg. Infect. Dis. 2004;10:914. doi: 10.3201/eid1005.030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachedjian M., Yu M., Lew A.M., Rockman S., Boyle J.S., Andrew M.E., Wang L. Molecular cloning and characterization of pig, cow and sheep MAdCAM-1 cDNA and the demonstration of cross-reactive epitopes amongst mammalian homologues. Tissue Antigens. 2006;67:419. doi: 10.1111/j.1399-0039.2006.00587.x. [DOI] [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L., Xu K.M., Song W.J., Vijaykrishna D., Poon L.L., Peiris J.S., Smith G.J., Chen H., Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao K.-L., DeBarbieri B., Michel H., Waugh D.S. A versatile plasmid expression vector for the production of biotinylated proteins by site-specific, enzymatic modification in Escherichia coli. Gene, 1996;169:59. doi: 10.1016/0378-1119(95)00762-8. [DOI] [PubMed] [Google Scholar]
- Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M., Xiang H., Xia X., Liu S., Ren T., Yu Y., Eaton B.T., Xuan H., Wang L.-F. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 2004;10:2244. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-F., Shi Z., Zhang S., Field H., Daszak P., Eaton B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006;12:1834. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-F., Yu M., White J.R., Eaton B.T. BTag: a novel six-residue epitope tag for surveillance and purification of recombinant proteins. Gene. 1996;169:53. doi: 10.1016/0378-1119(95)00795-4. [DOI] [PubMed] [Google Scholar]
- Wang M., Yan M., Xu H., Liang W., Kan B., Zheng B., Chen H., Zheng H., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Liu Y.F., Guo R.T., Liu X.N., Zhan L.H., Zhou D.H., Zhao A., Hai R., Yu D., Guan Y., Xu J. SARS–CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 2005;11:1860. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sakhastskyy P., Chou T.-H.W., Lu S. Assays for the assessment of neutralizing antibody activities against severe acute respiratory (SARS) associated coronavirus (SCV) J. Immunol. Meth. 2005;301:21. doi: 10.1016/j.jim.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H.M., Copps J., Drebot M.A., Marszal P., Smith G., Gren J., Andova M., Pasick J., Kitching P., Czub M. Susceptibility of pigs and chickens to SARS coronavirus. Emerg. Infect. Dis. 2004;10:179. doi: 10.3201/eid1002.030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li M., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molecular diversity of coronaviruses in bats. Virology. 2006;350:180. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.H., He J.F., Evans M.R., Peng G.W., Field H.E., Yu D.W., Lee C.K., Luo H.M., Lin W.S., Lin P., Li L.H., Liang W.J., Lin J.Y., Schnur A. Epidemiologic clues to SARS origin in China. Emerg. Infect. Dis. 2004;10:1030. doi: 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Zeng W., Pagnon J., Walker J., Shosh S., Wang L.-F., Jackson D. Identification of dominant epitopes of synthetic immunocontraceptive that induce antibodies in dogs. Vaccine. 2005;23:4589. doi: 10.1016/j.vaccine.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Zhang F., Yu M., Weiland E., Morrissy C., Zhang N., Westbury H., Wang L.-F. Characterization of epitopes for neutralizing monoclonal antibodies to classical swine fever virus E2 and Erns using phage-displayed random peptide library. Arch. Virol. 2006;151:37. doi: 10.1007/s00705-005-0623-9. [DOI] [PubMed] [Google Scholar]


