Abstract
It has been shown that severe acute respiratory syndrome‐associated coronavirus (SARS‐CoV) 3a and 7a proteins, but not membrane (M) protein, induce apoptosis in mammalian cells. Upon expression of SARS‐CoV M protein using the baculovirus/insect cell expression system, however, we found that the expressed M protein triggered accelerated apoptosis in insect cells, as characterized by rapid cell death, elevated cytotoxicity, cell shrinkage, nuclear condensation and DNA fragmentation. Conversely, the M protein expressed in mammalian cells did not induce apoptosis. This is the first report describing the induction of apoptosis by SARS‐CoV M protein in animal cells and possible implications are discussed.
Keywords: Apoptosis, Baculovirus, Insect cell, Membrane protein, SARS-CoV
1. Introduction
Severe acute respiratory syndrome (SARS) is a newly described infectious disease and a novel SARS‐associated coronavirus (SARS‐CoV) has been identified as the causative agent. SARS‐CoV possesses a positive single‐stranded RNA genome (≈29 727 nucleotides) encoding at least five structural proteins: spike (S), membrane (M), envelope (E), nucleocapsid (N) proteins [1, 2, 3] and the newly identified ORF3a [4]. The S glycoprotein mediates virus binding to the target cells and subsequent membrane fusion [5]. The N protein binds to a defined packaging signal on the RNA, leading to formation of the helical nucleocapsids [6]. The E protein is a minor yet critical structural component in coronavirus assembly. The M glycoprotein is the most abundant structural protein on the viral envelope; it is a triple‐spanning membrane protein with a long carboxyl‐terminal tail in the virion interior which interacts with the E and S proteins during assembly [7], hence M protein has been implicated as a key player in coronavirus assembly and budding [7]. Besides, monoclonal antibodies to the M protein of mouse hepatitis virus (MHV), a mouse coronavirus, could neutralize virus infectivity in vitro and protect animals against lethal viral challenge in vivo [8]. Antiserum to M protein was found to contain neutralizing antibody towards SARS‐CoV infection in vivo [9]. SARS patients‐derived human recombinant antibodies to M protein also efficiently neutralize SARS‐CoV infectivity [10] and two immunodominant epitopes (a.a. 1–31 and a.a. 132–161) on the SARS‐CoV M protein have been recently identified [11].
Among the proteins SARS‐CoV encodes, we were interested in M protein due to its key role in the induction of neutralizing antibodies and virus assembly. To further characterize the structure and functions of SARS‐CoV M protein, large quantities of purified proteins are required. Therefore, we aimed at developing a method for mass production of SARS‐CoV M protein. Owing to the generally high production yield, ease of virus construction and safety, baculovirus/insect cell expression system was selected for the expression of recombinant M protein with or without histidine (His6) tags. Surprisingly, we found that the expression of SARS‐CoV M protein in insect cells triggered early induction of apoptosis, and led to exceedingly low expression level. This is the first report regarding the induction of apoptosis in animal cells by the expression of SARS‐CoV M protein.
2. Materials and methods
2.1. Cells and media
Sf‐9 cells were maintained in spinner flasks using TNM‐FH medium [12]. HuH‐7 (human hepatoma), Vero E6 (African green monkey kidney epithelial), 293 (human embryonic kidney), HeLa (human cervix carcinoma) and RD (human muscle rhabodomyosarcoma) cells were maintained in Dulbecco's modified Eagle's medium (DMEM). A549 (human lung carcinoma) and BEAS‐2B (human lung bronchus epithelial) cells were maintained in RPMI 1640 medium. All culture media were supplemented with 10% fetal bovine serum (Gibco BRL). The cell density was counted by a hemacytometer and the viability was determined by trypan blue dye exclusion.
2.2. Construction of recombinant plasmids and baculoviruses
The recombinant baculovirus expressing the histidine‐tagged (His6) N protein (rNH), vBacNH, was constructed previously [13]. To construct the recombinant baculovirus expressing M protein, the full‐length gene fragment of SARS‐CoV M protein (TW1 strain, GenBank Accession No. http://AY291451) was amplified by polymerase chain reaction (PCR) from the vector provided by Prof. Pei‐Jer Chen (College of Medicine, National Taiwan University) and subsequently subcloned into pFastBac HTb (Invitrogen). The subcloning resulted in the fusion of a His6 tag at the N‐terminus of M gene under the control of polyhedrin promoter. Alternatively, the M gene was PCR‐amplified and subcloned into pFastBac1 vector (Invitrogen). The resultant plasmids were designated pBac‐MH and pBac‐M, respectively (Fig. 1 ). For protein expression in mammalian cells, the polyhedrin and p10 promoters in the pFastBac DUAL vector (Invitrogen) was deleted, and replaced by cytomegalovirus immediate‐early promoter (CMV‐IE) which was PCR‐amplified from pcDNA3.0 plasmid (Invitrogen). The gene fragments encoding the M protein with or without His6 tag were PCR‐amplified from pBac‐MH and pBac‐M, respectively and subcloned into the corresponding site downstream of the CMV‐IE promoter. The resultant plasmids were designated pBacMam‐MH and pBacMam‐M (Fig. 1). Similarly, the gene encoding enhanced yellow fluorescent protein (EYFP) was cloned under the control of CMV‐IE promoter to obtain pBacMam‐EYFP. The recombinant baculoviruses were generated based on these plasmids using Bac‐to‐Bac® system (Invitrogen) and designated vBacMH, vBacM, vBacMam‐MH, vBacMam‐M and vBacMam‐EYFP, respectively. The storage, amplification and titration of the recombinant baculoviruses were performed as described [14].
Figure 1.
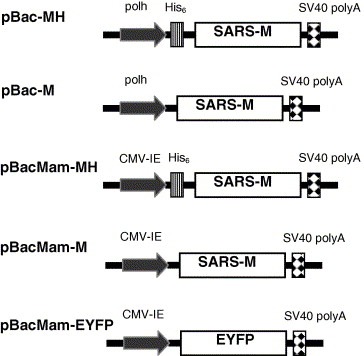
Schematic illustration of the recombinant baculovirus vectors, pBac‐MH, pBac‐M, pBacMam‐MH, pBacMam‐M and pBacMam‐EYFP. CMV‐IE, cytomegalovirus immediate‐early promoter; polh, polyhedrin promoter.
2.3. Western blot analysis
Sf‐9 cells were infected by recombinant baculovirus at a multiplicity of infection (MOI) of 10, and collected at three days post‐infection (dpi). The Western blot analysis of rNH protein was performed as described [13]. To detect rMH protein, the vBacMH‐infected cells (1 × 106 cells) were lysed in 100 μl RIPA buffer (20 mM Tris–HCl, 150 mM NaCl, 1% NP‐40, 0.5% sodium deoxycholate, 0.1% SDS, pH 7.5) for 30 min on ice. After centrifugation (13 000 rpm, 30 min), lysates and pellets were treated at 95 °C for 5 min or 37 °C for 30 min in the sample buffer. The samples were resolved in the 12% SDS–PAGE, and electrotransferred to nitrocellulose membranes for Western blot following standard procedures (Bio‐Rad Laboratories). The primary antibody was mouse anti‐His6 MAb (1:1500 dilution, Amersham Biosciences) while the secondary antibody was horseradish peroxidase‐conjugated goat anti‐mouse IgG (1:1000 dilution, Kirkegaard and Perry Laboratories). The membranes were developed with chemiluminescence reagent (ECL Western Blotting Substrate, Pierce).
2.4. MTT assay
The cytotoxicity resulting from protein expression in Sf‐9 cells was analyzed by MTT (3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5diphenyl tetrazolium bromide, Sigma) assay. The cells (5 × 105 cells/ml) were infected at MOI 10, seeded into 96‐well plates (100 μl/well) and cultured at 27 °C for three days. At the end of culture, 10 μl MTT (5 mg/ml) dissolved in phosphate buffer‐saline (PBS) was added. After 3 h of incubation at 27 °C, the solution was withdrawn, followed by the addition of 100 μl dimethyl sulfoxide (Sigma) to solublize the MTT reduction product, formazan. The color change was recorded by OD570 10 min later. The cytotoxicity was calculated by the formula: 1 − ODsample/ODcontrol, where the control was the infected cell at 0.5 dpi.
2.5. Terminal deoxynucleotidyl transferase‐mediated dUTP nick‐end labeling (TUNEL) assay
The DNA fragmentation was analyzed by flow cytometry coupled with a TUNEL assay. The cells were infected at MOI 10, collected at 2 dpi and treated with ApoAlert™ DNA Fragmentation Assay Kit according to the manufacturer's instructions (BD Biosciences). Approximately 10 000 cells from each sample were examined by flow cytometry (FACSCalibur, BD Biosciences) to measure fluorescin isothiocyanate (FITC) labeling. The mock‐infected cells were treated in the same way and set at 1% as TUNEL‐negative control.
2.6. DNA fragmentation by electrophoresis
The cells were seeded onto T‐75 flasks (1 × 107 cells/flask) and infected at MOI 10. Cells were collected at 3 dpi, washed with PBS, and lysed in 750 μl lysis buffer (20 mM Tris, 10 mM EDTA, 0.5% SDS and 0.5% RNase A, pH 8.0) at 37 °C for 1 h. After centrifugation (13 000 rpm for 15 min), 60 μl proteinase K (7 mg/ml) was added and the incubation continued at 55 °C for 2.5 h. At the end of incubation, genomic DNA in the mixture was extracted with phenol/chloroform/isoamyl alcohol (24:25:1) and chloroform, followed by precipitation with isopropanol. After drying, the DNA pellet was resuspended in 50 μl TE buffer and subject to 2% agarose gel electrophoresis.
2.7. Determination of sub‐G1 phase percentage in baculovirus‐transduced mammalian cells
Baculovirus transduction of mammalian cells was performed as described previously [15]. At 2 days post‐transduction (dpt), the cells were harvested and resuspended in PBS. After fixing with alcohol at −20 °C overnight, the cells were centrifuged (1500 rpm for 5 min), resuspended in PI (propidium iodide) staining solution (10 μg PI and 1 mg RNase A per milliliter PBS), and incubated at 37 °C for 45 min. Finally, the cells were analyzed by flow cytometer at FL‐2 filter.
2.8. Statistical analysis
Quantitative data were analyzed using Student's t‐tests and are expressed as means ± standard deviation (S.D.) or mean values of three or four independent experiments. P‐values less than 0.05 were considered significant.
3. Results
3.1. Expression of the rMH protein in Sf‐9 cells
To confirm the expression of His6‐tagged M protein, rMH, Sf‐9 cells were infected by vBacMH (MOI 10), harvested at 3 dpi and analyzed by Western blot. In parallel, cells were also infected by vBacNH and the expression of His6‐tagged N protein (rNH) was analyzed as a control. As shown in Fig. 2 (left panel), rNH protein (≈50 kDa) was expressed abundantly and readily extracted into the cell lysates (lys). In sharp contrast, nearly no His6‐tagged rMH was detectable using BCIP/NBT for color development (data not shown), indicating the low expression level. Consequently, chemiluminescence enhancement was employed for imaging. By incubating the vBacMH‐infected cell lysates (lys) and pellets (pel) at 95 °C for 5 min, however, rMH proteins were stuck at the interface of stacking and separating gels (right panel). Therefore, the samples were incubated at 37 °C for 30 min prior to electrophoresis. By modifying the incubation condition, rMH protein with a molecular mass of ≈29 kDa was detected in the pellets fraction, but was barely detectable in the lysates fraction, indicating the strong association of rMH protein with cell membranes.
Figure 2.
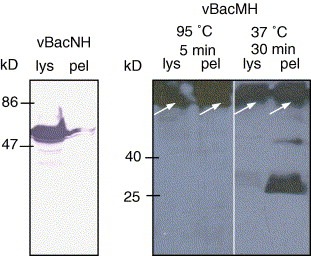
Western blot analysis of the lysates (lys) and pellets (pel) of Sf‐9 cells infected by vBacNH and vBacMH. The primary antibody was mouse anti‐His6 MAb while the secondary antibodies were either AP‐ (for rNH detection) or HRP‐conjugated (for rMH detection) goat anti‐mouse IgG. The membranes were developed with either BCIP/NBT color developing reagent or chemiluminescence reagent.
3.2. Cell death kinetics and cytotoxic effect
Upon vBacMH infection of cells, we noted that the cells died abnormally rapidly, thus Sf‐9 cells were infected by wild‐type AcMNPV (wt), vBacNH and vBacMH at MOI 10 and the death kinetics was monitored by trypan blue staining (Fig. 3 A). As shown, the viability of cells infected by wt AcMNPV and vBacNH declined slowly and remained higher than 55% at 3 dpi. Surprisingly, the viability of vBacMH‐infected cells started to decrease sharply at 1.5 dpi, and precipitously dropped to ≈20% at 3 dpi, suggesting strong adverse effects of rMH expression on Sf‐9 cells. To examine whether the dramatic decrease of cell viability resulted from the His6 tag fused to M protein, we constructed vBacM expressing rM devoid of the His6 tag for infection. Likewise, the vBacM‐infected cells exhibited rapid death kinetics and the viability dropped below 20% at 3 dpi.
Figure 3.
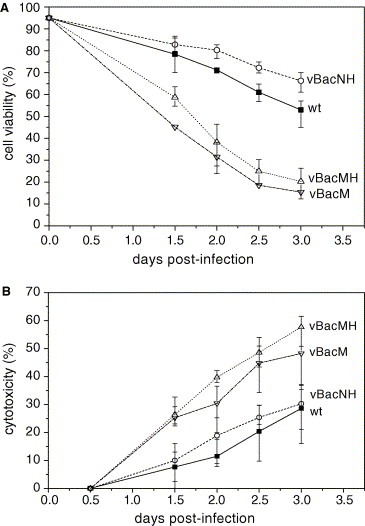
Time‐course profiles of viability (A) and cytotoxicity (B) of the cells infected by wt AcMNPV, vBacNH, vBacM and vBacMH at MOI 10. The data represent the means ± S.D. of four independent experiments.
To verify the distinction in cell death kinetics, the cytotoxicity of infected cells was analyzed at different time points by MTT assays (Fig. 3B). Compared to the wt AcMNPV infection, vBacNH infection resulted in similar cytotoxicity, whereas the vBacMH and vBacM infection led to significantly higher cytotoxicity (P < 0.05). Such elevated cytotoxicity upon expression of SARS CoV M protein was also observed in E. coli [16], and was attributed to budding of M protein, repression of certain host cell genes or alteration of cell membrane permeability.
3.3. Early induction of apoptosis in insect cells expressing rM and rMH
Loss of cell viability can occur following a toxic insult by triggering necrosis or apoptosis. Since SARS‐CoV infection causes cell apoptosis in patients [2] or cultured cells [17], we suspected that the dramatically lower viability for the vBacMH‐ and vBacM‐infected cells stemmed from the induction of apoptosis. To explore this hypothesis, Sf‐9 cells were infected as in Fig. 3. Fig. 4 A depicts that the cells infected by wt AcMNPV and vBacNH barely exhibited cytopathic effect (CPE) at 2 dpi, whereas cell shrinkage and nuclear condensation were apparently observed for the cells infected by vBacMH and vBacM. To further confirm the induction of apoptosis, the infected cells were harvested at 2 dpi for TUNEL analysis. Fig. 4B illustrates that very low percentages of cells infected by wt AcMNPV (5.2 ± 1.0%) and vBacNH (3.2 ± 1.3%) cells were TUNEL‐positive, which agreed with the high cell viabilities (Fig. 3A) and indicated the low degree of apoptosis in cells infected by wt AcMNPV and vBacNH at 2 dpi. In contrast, up to 34.3 ± 1.5% of vBacMH‐infected cells and 21.4 ± 3.6% of vBacM‐infected cells were TUNEL‐positive.
Figure 4.
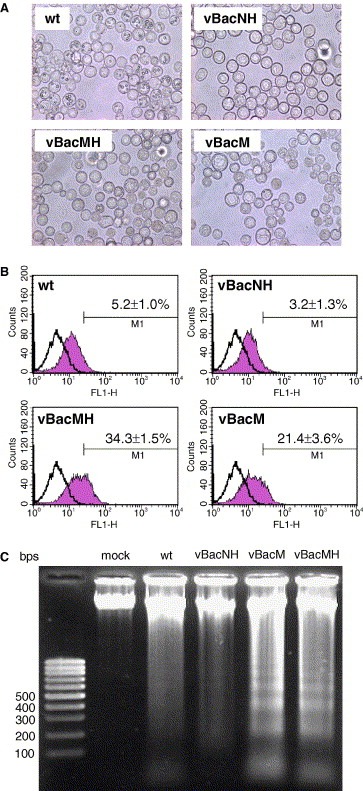
Induction of apoptosis by vBacMH and vBacM infection of Sf‐9 cells. (A) Photomicrographs showing the cell morphology of Sf‐9 cells at 2 dpi after infection with wt AcMNPV, vBacNH, vBacMH and vBacM at MOI 10. (B) TUNEL staining and flow cytometry analysis of Sf‐9 cells. Sf‐9 cells were mock‐infected or infected as in (A) and subject to TUNEL assay at 2 dpi. Samples were analyzed by flow cytometry and representative experiments are shown. The open and shaded histograms represent the fluorescence intensities of mock‐infected and baculovirus‐infected cells, respectively. The data shown in the figures represent the means ± S.D. of four independent experiments. (C) Cellular DNA fragmentation analysis. The figure is representative of four independent experiments.
To visualize the DNA fragmentation, a hallmark of apoptosis, induced by different baculoviruses, the genomic DNA of infected cells was extracted at 3 dpi and subject to gel electrophoresis. Fig. 4C reveals visible DNA fragmentation for wt AcMNPV‐ and vBacNH‐infected cells, but DNA laddering was not apparent. In contrast, low‐molecular‐weight DNA fragments appeared at 1.5 dpi (data not shown) and became clearly visible at 3 dpi for cells infected by vBacM and vBacMH. The TUNEL analyses and DNA fragmentation assays confirmed the early induction of apoptosis in insect cells by the expression of rM and rMH.
3.4. Absence of apoptosis in mammalian cells expressing rM and rMH
To examine whether rM and rMH also induced apoptosis in mammalian cells, we constructed vBacMam‐EYFP, vBacMam‐M and vBacMam‐MH for the expression of EYFP, rM and rMH in mammalian cells. These viruses were used to transduce mammalian cells of different origins, including A549, Vero E6, HeLa, BEAS‐2B, 293, HuH‐7 and RD cells, and the expression was confirmed by Western blot (data not shown). As a control, the cells were also mock‐transduced. The transduced and mock‐transduced cells were analyzed for the percentages of sub‐G1 phase cells. As shown in Fig. 5 , the transduction by vBacMam‐M and vBacMam‐MH resulted in sub‐G1 percentages below 10% in all cell types tested, which were statically similar to the percentages in the mock‐transduced or vBacMam‐EYFP‐transduced cells (P > 0.05) and indicated the absence of apoptosis induced by rM and rMH.
Figure 5.
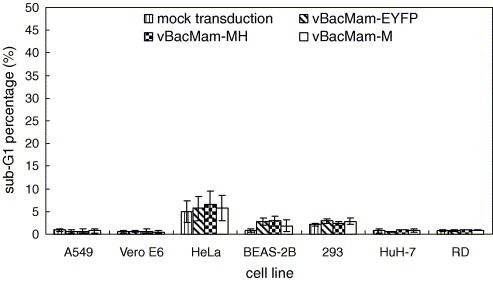
Determination of sub‐G1 phase percentage in mammalian cells that were mock‐transduced or transduced with vBacMam‐EYFP, vBacMam‐MH and vBacMam‐M at MOI 10. The data represent the means ± S.D. of three independent experiments.
4. Discussion
The importance of M protein in the life cycle of coronaviruses and elicitation of immunity makes it an attractive target for anti‐SARS drug research, vaccine development and the establishment of a serological detection assay. To obtain large quantities of recombinant SARS‐CoV M protein, the recombinant baculovirus vBacMH expressing rMH protein was constructed for expression in insect cells and subsequent purification. In contrast to the soluble rNH, rMH protein was strongly associated with membranes and formed protein aggregates if proteins were boiled at 95 °C prior to electrophoresis. Only treatment of samples under non‐denaturing conditions at 37 °C could partially alleviate the aggregation problem. Such thermal aggregation was also observed for SARS‐CoV M protein expressed in Vero E6 cells and the region (a.a. 51–170) responsible for the thermal aggregation was identified [18]. The molecular mass of rMH (≈29 kDa) was consistent with that of the glycosylated M protein. These attributes suggested the similarity between recombinant M protein expressed in insect and mammalian cells.
One unusual finding, however, was that expression of rMH in Sf‐9 cells resulted in accelerated cell death (Fig. 3A) and high cytotoxicity (Fig. 3B). These phenomena were concomitant with the early emergence of the hallmarks of apoptosis such as cell shrinkage, nuclear condensation (Fig. 4A) and DNA fragmentation (Fig. 4B and C) at 2 dpi, thus confirming the accelerated induction of apoptosis in insect cells by the expression of rMH. The apoptosis was also triggered early by the expression of rM, thus excluding the possibility that the additional His6 tag solely accounted for the accelerated induction. Normally, insect cells infected by wt or recombinant baculoviruses (e.g. vBacNH, Fig. 3A) do not exhibit obvious CPE until 3 dpi [19] because the baculovirus‐encoded p35 protein blocks baculovirus‐induced apoptosis in insect cells during the early stage of virus infection [20], which is an important strategy to prolong virus replication. The early induction of apoptosis, therefore, interfered with viral gene expression and virus replication and at least partly contributed to the exceedingly low expression level (Fig. 2). Intriguingly, when Sf‐9 cells were co‐infected with three recombinant baculoviruses individually expressing E, M and S proteins or singly infected with a recombinant baculovirus expressing all 3 SARS‐CoV proteins simultaneously, SARS‐CoV VLPs (virus‐like particles) comprising all three proteins were obtained [21, 22], but no apoptosis was noted. The M protein of other coronaviruses (e.g. MHV) accumulates in the Golgi complex as homomultimeric complexes when expressed alone [23]. However, in combination with the E protein, M protein is retained in the budding compartment and interacts with E protein to assemble into VLPs which are exported from the cell [24]. Similarly, SARS‐CoV M protein is a triple‐spanning membrane protein and predominantly localizes in the Golgi [25]; it is thus reasonable to hypothesize that when expressed alone, M protein accumulated in the Golgi and jammed the sorting of other cellular proteins, thereby triggering the apoptosis. But when co‐expressed with E and S proteins, M protein interacted with other proteins to assemble into VLPs that were released all over in the cytoplasm [22] or bud out of the cells [21], thus no apoptosis was observed.
It has been shown that in mammalian cells apoptosis can be induced by SARS‐CoV 3a [26] and 7a [27] proteins, and even N protein under restricted conditions [28], yet M protein has never been implicated in apoptosis. Our data again confirmed that M protein, with or without His6 tag, did not induce appreciable apoptosis in the mammalian cells (Fig. 5). However, we found, for the first time, that the expression SARS‐CoV M protein triggered early induction of apoptosis in insect cells and led to low expression levels. The discrepancy was probably due to the differences in cell type. Furthermore, the baculovirus‐expressed rM and rMH proteins were highly insoluble, difficult to extract from cell lysates and prone to thermal aggregation. These attributes created extra difficulties in the production of full‐length M protein using the baculovirus/insect cell expression system. One possible solution to this hurdle is the fusion of a signal peptide and removal of the transmembrane domain (a.a. 21–99) [29] while leaving the immunodominant epitopes [11] intact. This concept has been demonstrated feasible as high level expression of SARS‐CoV M protein lacking the transmembrane domain in Pichia pastoris was achieved and no apoptosis was reported [30]. However, the glycosylation pattern, a property critical to the M protein activity, necessitates further investigation. Alternatively, SARS‐CoV M protein may be co‐expressed with E protein and co‐purified in the form of VLP.
Acknowledgements
We thank Dr. Pei‐Jer Chen for providing vectors harboring the N and M genes. We are also grateful to the financial support from the National Science Council (NSC 92‐2751‐B‐007‐001‐Y) and National Health Research Institutes (NHRI‐EX95‐9412EI), Taiwan.
Lai Chia-Wei,Chan Zun-Ren,Yang Ding-Gang,Lo Wen-Hsin,Lai Yiu-Kay,Chang Margaret Dah-Tsyr and Hu Yu-Chen(2006), Accelerated induction of apoptosis in insect cells by baculovirus-expressed SARS-CoV membrane protein, FEBS Letters, 580, doi: 10.1016/j.febslet.2006.06.003
References
- 1. Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science, 300, (2003), 1394– 1399. [DOI] [PubMed] [Google Scholar]
- 2. Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet, 361, (2003), 1319– 1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med., 348, (2003), 1953– 1966. [DOI] [PubMed] [Google Scholar]
- 4. Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X.M., Lim S.G., Hong W., Tan Y.J., The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun., 330, (2005), 286– 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S., The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun., 312, (2003), 1159– 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J., The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology, 218, (1996), 52– 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rottier P.J.M., Siddell S.G. The Coronaviridae (1995), Plenum Press ; New York, NY: 115– 139. [Google Scholar]
- 8. Fleming J.O., Shubin R.A., Sussman M.A., Casteel N., Stohlman S.A., Monoclonal antibodies to the matrix (E1) glycoprotein of mouse hepatitis-virus protect mice from encephalitis. Virology, 168, (1989), 162– 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pang H., Liu Y., Han X., Xu Y., Jiang F., Wu D., Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J. Gen. Virol., 85, (2004), 3109– 3113. [DOI] [PubMed] [Google Scholar]
- 10. Liang M.F., Du R.L., Liu J.Z., Li C., Zhang Q.F., Han L.L., SARS patients-derived human recombinant antibodies to S and M proteins efficiently neutralize SARS-coronavirus infectivity. Biomed. Environ. Sci., 18, (2005), 363– 374. [PubMed] [Google Scholar]
- 11. He Y.X., Zhou Y.S., Siddiqui P., Niu J.K., Jiang S.B., Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol., 43, (2005), 3718– 3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Y.-C., Liu H.-J., Chung Y.-C., High level expression of the key antigenic protein, sC, from avian reovirus into insect cells and its purification by immobilized metal affinity chromatography. Biotechnol. Lett., 24, (2002), 1017– 1022. [Google Scholar]
- 13. Lai C.-W., Chung Y.-C., Lai Y.-K., Chang M.D.-T., Hu Y.-C., Expression and purification of N and E proteins from Severe Acute Respiratory Syndrome (SARS)-associated coronavirus: a comparative study. Biotechnol. Lett., 27, (2005), 883– 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang K.-C., Wu J.-C., Chung Y.-C., Ho Y.-C., Chang M.D., Hu Y.-C., Baculovirus as a highly efficient gene delivery vector for the expression of hepatitis delta virus antigens in mammalian cells. Biotechnol. Bioeng., 89, (2005), 464– 473. [DOI] [PubMed] [Google Scholar]
- 15. Hsu C.-S., Ho Y.-C., Wang K.-C., Hu Y.-C., Investigation of optimal transduction conditions for baculovirus-mediated gene delivery into mammalian cells. Biotechnol. Bioeng., 88, (2004), 42– 51. [DOI] [PubMed] [Google Scholar]
- 16. Zhang X.L., Wang J.R., Zhang Y., Chen M.L., Zhang W., Yang S., Jiang W.H., Expression, purification and identification of recombinant SARS coronavirus membrane protein. Acta. Biochim. Biophys. Sin., 35, (2003), 1140– 1144. [PubMed] [Google Scholar]
- 17. Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K., Zheng C., Gao G.F., SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol., 73, (2004), 323– 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee Y.N., Chen L.K., Ma H.C., Yang H.H., Li H.P., Lo S.Y., Thermal aggregation of SARS-CoV membrane protein. J. Virol. Methods, 129, (2005), 152– 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clem R.J., Fechheimer M., Miller L.K., Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science, 254, (1991), 1388– 1390. [DOI] [PubMed] [Google Scholar]
- 20. LaCount D.J., Friesen P.D., Role of early and late replication events in the induction of apoptosis by baculoviruses. J. Virol., 71, (1997), 1530– 1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mortola E., Roy P., Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett., 576, (2004), 174– 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C., Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun., 318, (2004), 833– 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J., Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol., 68, (1994), 6523– 6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J., Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J., 15, (1996), 2020– 2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nal B., Chan C.M., Kien F., Siu L., Tse J., Chu K., Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol., 86, (2005), 1423– 1434. [DOI] [PubMed] [Google Scholar]
- 26. Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K.S., The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol., 86, (2005), 1921– 1930. [DOI] [PubMed] [Google Scholar]
- 27. Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H.P., Lim S.G., Hong W.J., Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol., 78, (2004), 14043– 14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K., The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J., 383, (2004), 13– 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu W.L., Lu Y., Chen Y.H., Bioinformatics analysis of SARS-Cov M protein provides information for vaccine development. Prog. Nat. Sci., 13, (2003), 844– 847. [Google Scholar]
- 30. Han X.Q., Bartlam M., Jin Y.H., Liu X.T., He X.J., Cai X.P., Xie Q.Q., Rao Z.H., The expression of SARS-CoV M gene in P. pastoris and the diagnostic utility of the expression product. J. Virol. Methods, 122, (2004), 105– 111. [DOI] [PMC free article] [PubMed] [Google Scholar]


