Abstract
Background and aims
Inflammation plays a key role in atherosclerosis. The complement system is involved in atherogenesis, and the complement receptor 1 (CR1) plays a role facilitating the clearance of immune complexes from the circulation. Limited evidence suggests that CR1 may be involved in cardiovascular disease. We investigated the relationship between CR1 gene polymorphisms and cardiovascular risk.
Methods
Single nucleotide polymorphisms (SNPs) within the CR1 region (n = 73) on chromosome 1 were assessed in 5244 participants in PROSPER (PROspective Study of Pravastatin in the Elderly at Risk) (mean age 75.3 years), who had been randomized to pravastatin 40 mg/day or placebo and followed for a mean of 3.2 years. Logistic regression, adjusted for gender, age, country and use of pravastatin, was used to assess the association between the SNPs and cardiovascular disease.
Results
All 73 SNPs within the genomic region of the CR1 gene on chromosome 1 were extracted. In this region, strong LD was present leading to the occurrence of two haploblocks. Twelve of the 73 investigated CR1 SNPs were significantly associated with the risk of fatal or nonfatal myocardial infarction (all p < 0.05). Moreover, most of the associated SNPs were also associated with levels of serum C-reactive protein (CRP). The global p-value for the tail strength method to control for multiple testing was 0.0489, implying that the null hypothesis of no associated SNPs can be rejected.
Conclusions
These data indicate that genetic variation within the CR1 gene is associated with inflammation and the risk of incident coronary artery disease.
Keywords: Apolipoprotein B, Atherosclerosis, C-reactive protein, Inflammation, Myocardial infarction
Highlights
-
•
The complement receptor 1 (CR1) may be involved in atherosclerosis.
-
•
12 SNPs within the CR1 region were associated with myocardial infarction.
-
•
7 SNPs were also associated with levels of C-reactive protein.
-
•
These results imply that CR1 may be involved in atherogenesis.
1. Introduction
Inflammation plays a key role in the development of atherosclerosis. Lipoproteins can migrate into the subendothelial space, where they can induce inflammation, foam cell formation and atherosclerotic plaque development [1], [2]. Several inflammatory markers, such as C-reactive protein (CRP), interleukin-6 and leukocyte count have been associated with the risk of cardiovascular disease [3], [4], and after myocardial infarction, levels of interleukin-6, CRP and leukocyte count increase [5]. Interleukin-6 is the main stimulant of the hepatic synthesis of acute phase proteins, such as CRP [6].
The complement system is involved in this inflammatory condition leading to atherogenesis. The terminal complement complex, C5b-9, colocalizes with CRP in human atherosclerotic lesions [7]. Furthermore, elevated levels of serum complement component 3 (C3) have been associated with the presence of myocardial infarction, and predict the risk of future coronary events [8], [9].
The complement receptor 1 (CR1) is found on the membranes of many cell types, including erythrocytes, granulocytes, monocytes and macrophages [10]. CR1 is a receptor for the complement proteins C3b and C4b [10]. Via this receptor, erythrocytes carry immune complexes from the circulation to the spleen and liver, where the immune complexes are transferred to phagocytic cells [11]. Recently, it has been postulated that CR1 on erythrocytes may also be involved in the clearance of atherogenic lipoproteins [12], [13], [14]. Besides a role in the clearance of immune complexes and lipoproteins, CR1 can also inhibit complement activation, by acting as a co-factor for the factor I-mediated breakdown of C3b into iC3b [10], [15].
A large intra-individual variation in the number of CR1 molecules per erythrocyte has been described, with values ranging between less than 100 to 1200 molecules per cell [16], [17]. Erythrocytes loose CR1 molecules during their aging process in the circulation [18]. Accelerated and sometimes reversible loss of erythrocyte-CR1 has been described in patients with several types of inflammatory and non-inflammatory diseases, such as severe acute respiratory syndrome [19], tuberculosis [20], insulin-dependent diabetes mellitus [21] and systemic lupus erythematosus [22]. In addition to these conditions, several polymorphisms in the CR1 gene have been related to erythrocyte CR1 expression, including the Pro1827Arg (C5507G, rs3811381) SNP in exon 33, the His1208Arg (A3650G, rs2274567) SNP in exon 22 and the HindIII restriction fragment length polymorphism (RFLP, T520C, rs11118133) in intron 27 [17], [23], [24]. These three polymorphisms are in strong linkage disequilibrium (LD) [25].
The role of the CR1 gene in cardiovascular disease and atherosclerosis remains unclear. The goal of the present study was to investigate the relationship between CR1 polymorphisms and cardiovascular disease.
2. Materials and methods
2.1. Study population
All data come from the PROspective Study of Pravastatin in the Elderly at Risk (PROSPER). A detailed description of the study has been published elsewhere [26], [27]. In short, elderly subjects (aged 70–82 years) with a history of vascular disease, or increased vascular risk, were enrolled in Scotland, Ireland and the Netherlands. The primary study endpoint was death from coronary heart disease, non-fatal myocardial infarction (MI), and fatal and non-fatal stroke. Secondary endpoints were the separate coronary and cerebrovascular components of the primary endpoint. The study protocol was approved by the medical ethics committees of each participating institution. All study subjects gave written informed consent. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
2.2. Genotyping
In the PHASE project, whole genome wide screening has been performed, as has been described in detail previously [28]. From this GWAS study, we selected all single nucleotide polymorphisms within the CR1 region on chromosome 1 (n = 73) with PLINK software. Taking a relatively stringent R2 threshold (>0.8), using LDlink (https://analysistools.nci.nih.gov/LDlink/) [29], we observed three sets of SNPs. The R2-matrix for these 12 SNPs is shown in Supplementary Table 1.
2.3. Laboratory measurements
All measurements were performed on samples stored at −80 °C. CRP was measured by automated particle-enhanced immunoturbidimetric assay (Roche, UK). This method has an inter- and intra-assay coefficient of variation of 3%. IL-6 was determined using a high-sensitivity enzyme-linked immunosorbent assay (R&D Systems, Abingdon, UK) with inter- and intra-assay coefficients of variation of <6% and sensitivity of 0.16 pg/mL. White blood cell count (WBC) was measured by a fully automated system Sysmex XE-2100 (TOA Medical Electronics, Kobe, Japan).
2.4. Statistical analysis
Allele frequencies were estimated and pairwise LD between the investigated SNPs was estimated and plotted with the program Haploview. Associations between the CR1 SNPs and laboratory measurements were assessed with linear regression adjusted for sex, age, and country. Logistic regression was used to associate the CR1 SNPs with cardiovascular outcomes adjusted for sex, age, country, and pravastatin treatment. All statistical analyses were performed with PLINK statistical software (http://pngu.mgh.harvard.edu/∼purcell/plink/download.shtml#download). To control for multiple testing, we calculated global p-values using the tail strength method [30]. In short, the tail strength measures how much p-values in a set differ from the expected uniform distribution under the null hypothesis and sums up these differences into single-test statistics. The tail strength is powerful when many small effects exist in the data [30]. Since SNPs are not independent, empirical p-values were computed using permutations. SNPs were permuted as a block, keeping intact the relationship between covariates and outcome. Individual tests were based on a Cox-model (coxph) and 2 × 104 permutations were used. Computations were parallelized using package parallelize.dynamic [31]. Global p-values were computed using R version 3.2.
3. Results
Table 1 shows the baseline characteristics of the 5244 participants of the PROSPER Study. Supplementary Table 1 shows the characteristics of subjects who developed fatal or nonfatal myocardial infarction versus those without myocardial infarction. The mean age of participants was 75.3 years and approximately 50% were female. Due to the inclusion criteria of PROSPER, almost 50% of the participants had a history of vascular disease. Mean follow-up of study subjects was 3.2 years (range 2.8–4.0).
Table 1.
Baseline characteristics of the 5,244 subjects of the PROSPER study.
| Participants (n = 5244) | |
|---|---|
| Demographics | |
| Female, n (%) | 2720 (51.9) |
| Age, years | 75.3 ± 3.4 |
| Current smoker, n (%) | 1392 (26.5) |
| Body Mass Index, kg/m2 | 26.8 ± 4.2 |
| History of diabetes, n (%) | 544 (10.4) |
| History of hypertension, n (%) | 3257 (62.1) |
| History of myocardial infarction, n (%) | 708 (13.5) |
| History of stroke or TIA, n (%) | 586 (11.2) |
| History of vascular diseasea, n (%) | 2336 (44.5) |
| Hyperlipidemiab, n (%) | 1424 (27.2) |
| Laboratory measurements | |
| White blood cell count, x109/L | 6.43 ± 1.62 |
| C-reactive protein, LN transformed, mg/L | 1.13 ± 1.13 |
| Interleukin-6, LN transformed, ng/L | 0.97 ± 0.66 |
| LDL-cholesterol, mmol/L | 3.79 ± 0.80 |
| HDL-cholesterol, mmol/L | 1.28 ± 0.35 |
Data are presented as number (percentage) or mean ± SD.
Any of: stable angina, intermittent claudication, stroke, transient ischemic attack, myocardial infarction, peripheral artery disease surgery, or amputation for vascular disease more than 6 months before study entry.
Hyperlipidemia was defined according to the NCEP criteria as total cholesterol >6.21 mmol/L (240 mg/dL) or triglycerides > 5.5 mmol/L (400 mg/dL).
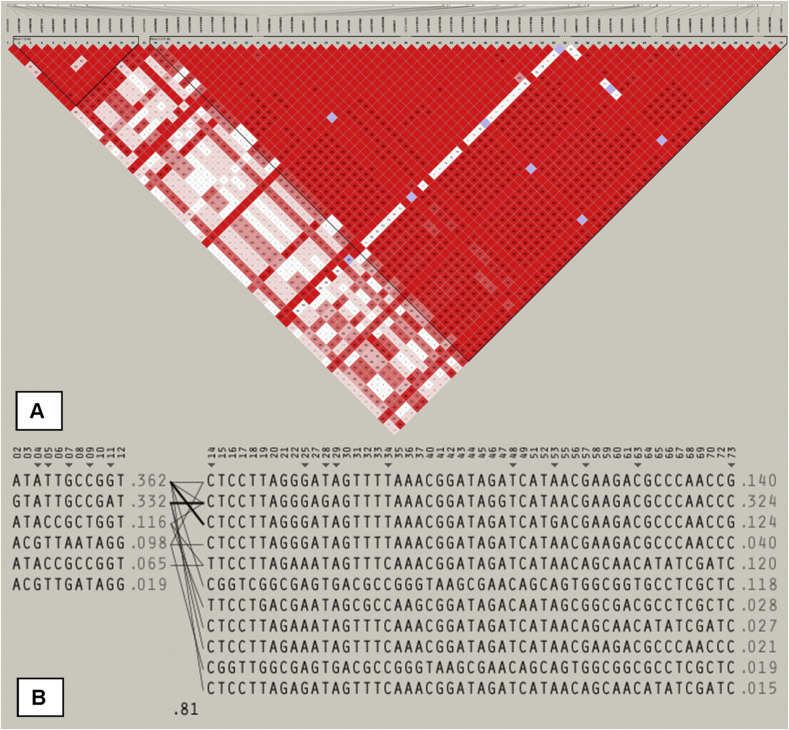
From the GWAS database including 2.5 million SNPs, we extracted all SNPs within the genomic region of the CR1 gene on chromosome 1 (n = 73). Fig. 1 shows the CR1 gene structure with the location of the investigated SNPS. Strong LD is present within the genomic region of CR1 leading to the occurrence of two haploblocks as shown in Fig. 1. All SNPs were in Hardy Weinberg equilibrium (p > 0.05).
Fig. 1.
Details of the genomic region of the CR1 gene. The linkage disequilibrium (LD) between (A) the single nucleotide polymorphisms examined, and (B) the existing haplotypes with corresponding frequencies. Two haploblocks are shown with strong LD (red blocks present LD > 95%). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
During follow-up, 12.7% of the patients developed a fatal or nonfatal MI, 4.8% experienced fatal or nonfatal stroke, 5.3% died from cardiovascular disease and the overall mortality was 11.7%.
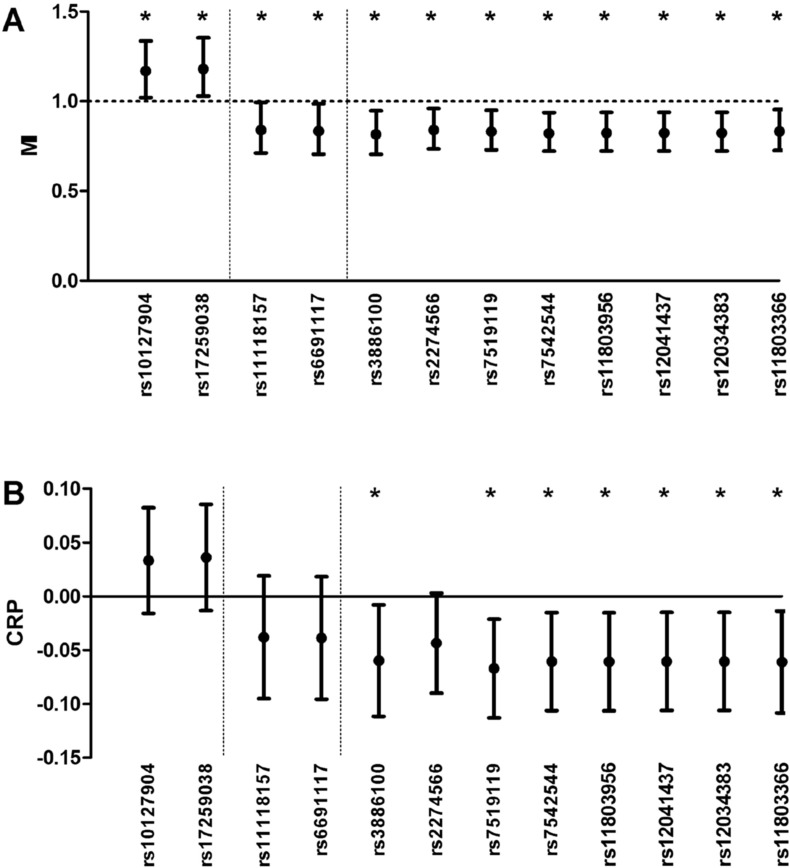
Twelve of the 73 investigated CR1 SNPs were significantly associated with the risk of fatal or nonfatal MI. These SNPs, and their corresponding odds ratio for MI with 95% confidence intervals, are depicted in Fig. 2 A. The minor allele frequency of the CR1 SNPs rs3886100, rs2274566, rs11118157, rs6691117, rs7519119, rs7542544, rs11803956, rs12041437, rs12034383 and rs11803366 was associated with a decreased risk of coronary artery disease, while the minor allele frequency of SNPs rs10127904 and rs17259038 was associated with increased risk. Seven out of 10 SNPs associated with decreased risk of MI, were also associated with lower levels of CRP. Hence, the two SNPs associated with an increased risk of MI showed an association with higher CRP levels, although not statistically significant. The 12 SNPs were not associated with leukocyte count or levels of interleukin-6 (data not shown). The global p-value for the tail strength method to control for multiple testing was 0.0489, implying that the null hypothesis of no associated SNPs can be rejected.
Fig. 2.
CR1 SNPs in relation to risk of MI and CRP. (A) The relationship of all investigated SNPs with risk of future fatal or nonfatal myocardial infarction (odds ratio with 95% confidence interval). (B) The relationship of these SNPs with serum C-reactive protein levels (beta-coefficient with 95% confidence interval). There were three sets of SNPs, which are separated by the dotted lines. *p < 0.05.
Supplementary Fig. 1 shows the association of the investigated SNPs with vascular and all-cause mortality and with fatal and nonfatal stroke. In line with the results for risk of MI, SNPs rs3886100 and rs2274566 were associated with decreased risk of vascular and all-cause mortality, while SNPs rs10127904 and rs17259038 were associated with increased vascular and all-cause mortality. SNPs rs7542544, rs11803956, rs12041437 and rs12034383 were associated with decreased vascular mortality, but not with all-cause mortality. None of the investigated SNPs was associated with risk of stroke.
Further adjustments for smoking, diabetes, hypertension, average LDL cholesterol during follow-up, history of MI, history of stroke, and log-transformed CRP did not materially change the results (data not shown).
4. Discussion
We investigated the relationship between CR1 polymorphisms and cardiovascular risk in PROSPER, a prospective randomized trial in which elderly subjects received pravastatin or placebo. After a mean follow-up of 3.2 years, 12 SNPs in the CR1 gene were associated with risk of fatal and nonfatal MI. In addition, several SNPs associated with decreased risk of MI were also associated with lower levels of CRP. Furthermore, many of the investigated SNPs were associated with the risk of vascular and all-cause mortality.
To date, two previous studies have investigated the relationship between CR1 polymorphisms and cardiovascular risk. Buraczynska et al. found that the GG phenotype of the Pro1827Arg polymorphism, corresponding to decreased erythrocyte-CR1 expression [23], [32], was more prevalent in end-stage renal disease patients with a history of cardiovascular disease than in those without cardiovascular disease [33]. In contrast, Boiocchi et al. described a lower prevalence of the GG variant in hypercholesterolemic patients with a history of coronary artery disease than in healthy controls [34]. However, in that study, the total number of patients with the GG variant was only 20. Unfortunately, this polymorphism was not present in the database of our GWAS.
No previous studies have assessed the relationship between the 12 presently described CR1 SNPs and cardiovascular disease. However, the minor alleles of the rs12034383 and rs6691117 SNPs have been associated with lower erythrocyte sedimentation rate [35]. The lower risk of MI, which we observed in carriers of the minor allele rs12034383 and rs6691117, and lower levels of CRP in rs12034383, are in line with the previously reported lower inflammation in these subjects [35]. Several markers of inflammation, including CRP [3], IL-6 [3] and white blood cell count [4], have been associated with increased cardiovascular risk. Several CR1 SNPs that were significantly associated with cardiovascular risk in the present study, were also associated with levels of CRP, but not with IL-6 or white blood cell count. CRP is synthesized in the liver, primarily in response to IL-6 [36]. When CRP binds to the phosphocholine groups on the surface of, for instance, bacteria, this activates the complement system and induces an inflammatory cascade to destroy the ligand [37]. Complexes of CRP with soluble ligands may bind to CR1 on the erythrocyte surface and thus be cleared from the circulation [38]. This direct interaction between CRP and CR1 may explain the observed association with the CR1 SNPs. In the present study, we found no evidence for a direct effect of CR1 gene polymorphisms on levels of IL-6 or leukocyte count.
We hypothesize that the mechanism behind the observed relationship between CR1 polymorphisms and the risk of coronary artery disease involves the level of CR1 on circulating erythrocytes. This may affect cardiovascular risk in several ways. Firstly, polymorphisms leading to lower expression of CR1 on these cells may result in reduced clearance of immune complexes from the circulation, resulting in a pro-inflammatory and therefore, a pro-atherogenic situation. Secondly, lower erythrocyte-CR1 expression may be pro-atherogenic due to less binding of atherogenic lipoproteins to erythrocytes. We have previously demonstrated in vivo that circulating human erythrocytes are able to bind atherogenic apolipoprotein B-containing lipoproteins [12], [39]. The binding of these lipoproteins by circulating blood cells was associated with a reduced prevalence of atherosclerosis [12], [39]. Possibly, this binding of atherogenic particles by blood cells prevents their interaction with the endothelium, and we have speculated that erythrocytes contribute to removal of the lipoproteins from the circulation [40]. In vitro and ex vivo work from our group indicates that CR1 is a likely candidate receptor for the binding of lipoproteins to circulating blood cells [13].
A limitation of the present study is that we did not quantify erythrocyte-bound CR1 in our patients. We do not know whether these SNPs lead to functional changes in the CR1 protein, although the observed relationship with CRP levels and the previously reported relationship with erythrocyte sedimentation rate suggest functionality. Another limitation is that we did not correct for multiple testing with the more common Bonferroni correction. Due to the large number of SNPs analyzed, the Bonferroni adjusted p-value would have been 6.8 × 10−6 (0.05/73). However, we noticed by checking the haplotype structure within the gene, that there were only two major haploblocks with very strong LD. This indicates that the 73 investigated SNPs were not all independent of each other and that using the Bonferroni correction would have been too conservative. Instead, we used the tail strength method to control for multiple testing, since this test is powerful for studies with strong LD (dependency) and with small effect sizes. Since we found a significant p-value with this test as well, we consider our results to be valid and not false-positive. Another limitation is that these data were obtained in a single cohort, and these findings need to be confirmed in an independent cohort. In addition, we investigated a group of subjects aged 70 years and above. While the impact of CR1 gene polymorphisms, and their influence on inflammation and atherosclerosis, may be most pronounced in a group of elderly individuals, if CR1 gene polymorphisms lead to premature death before the age of 70, we may underestimate the true effect of these polymorphisms. Furthermore, future studies investigating the functionality of these CR1 SNPs are necessary.
In conclusion, genetic variation within the CR1 gene is associated with inflammation and risk of incident coronary artery disease. These data further strengthen the evidence for the role of the complement system in atherosclerosis.
Conflict of interest
The authors declared they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
Financial support
The PROSPER study was supported by an investigator initiated grant obtained from Bristol-Myers Squibb. Prof. Dr. J. W. Jukema is an Established Clinical Investigator of the Netherlands Heart Foundation (grant 2001 D 032). Support for genotyping was provided by the seventh framework program of the European commission (grant 223004) and by the Netherlands Genomics Initiative (Netherlands Consortium for Healthy Aging grant 050-060-810).
Author contributions
ST and JWJ designed the study. ST and JWJ performed the study. MAV, ST and SPM performed the statistical analysis. MAV, ST and MCC drafted the manuscript. MAV, ST, SPM, MCC and JWJ critically revised the manuscript.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.atherosclerosis.2016.12.017.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Libby P. Inflammation in atherosclerosis. Arter. Thromb. Vasc. Biol. 2012;32:2045–2051. doi: 10.1161/ATVBAHA.108.179705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross R. Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 3.Ridker P.M., Hennekens C.H., Buring J.E., Rifai N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N. Engl. J. Med. 2000;342:836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 4.Friedman G.D., Klatsky A.L., Siegelaub A.B. The leukocyte count as a predictor of myocardial infarction. N. Engl. J. Med. 1974;290:1275–1278. doi: 10.1056/NEJM197406062902302. [DOI] [PubMed] [Google Scholar]
- 5.Liebetrau C., Hoffmann J., Dörr O., Gaede L., Blumenstein J. Release kinetics of inflammatory biomarkers in a clinical model of acute myocardial infarction. Circ. Res. 2015;116:867–875. doi: 10.1161/CIRCRESAHA.116.304653. [DOI] [PubMed] [Google Scholar]
- 6.Castell J.V., Gómez-Lechón M.J., David M., Andus T., Geiger T. Interleukin-6 is the major regulator of acute phase protein synthesis in adult human hepatocytes. FEBS Lett. 1989;242:237–239. doi: 10.1016/0014-5793(89)80476-4. [DOI] [PubMed] [Google Scholar]
- 7.Torzewski J., Torzewski M., Bowyer D.E., Fröhlich M., Koenig W. C-reactive protein frequently colocalizes with the terminal complement complex in the intima of early atherosclerotic lesions of human coronary arteries. Arter. Thromb. Vasc. Biol. 1998;18:1386–1392. doi: 10.1161/01.atv.18.9.1386. [DOI] [PubMed] [Google Scholar]
- 8.Carter A.M., Prasad U.K., Grant P.J. Complement C3 and C-reactive protein in male survivors of myocardial infarction. Atherosclerosis. 2009;203:538–543. doi: 10.1016/j.atherosclerosis.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Muscari A., Bozzoli C., Puddu G.M., Sangiorgi Z., Dormi A. Association of serum C3 levels with the risk of myocardial infarction. Am. J. Med. 1995;98:357–364. doi: 10.1016/S0002-9343(99)80314-3. [DOI] [PubMed] [Google Scholar]
- 10.Fearon D.T. Identification of the membrane glycoprotein that is the C3b receptor of the human erythrocyte, polymorphonuclear leukocyte, B lymphocyte, and monocyte. J. Exp. Med. 1980;152:20–30. doi: 10.1084/jem.152.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schifferli J.A., Ng Y.C., Paccaud J.P., Walport M.J. The role of hypocomplementaemia and low erythrocyte complement receptor type 1 numbers in determining abnormal immune complex clearance in humans. Clin. Exp. Immunol. 1989;75:329–335. [PMC free article] [PubMed] [Google Scholar]
- 12.Klop B., van de Geijn G.-J.M., Bovenberg S.A., van der Meulen N., Elte J.W.F. Erythrocyte-bound apolipoprotein B in relation to atherosclerosis, serum lipids and ABO blood group. PLoS One. 2013;8:e75573. doi: 10.1371/journal.pone.0075573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klop B., van der Pol P., van Bruggen R., Wang Y., de Vries M.A. Differential complement activation pathways promote C3b deposition on native and acetylated LDL thereby inducing lipoprotein binding to the complement receptor 1. J. Biol. Chem. 2014;289:35421–35430. doi: 10.1074/jbc.M114.573840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries M.A., Klop B., van der Meulen N., van de Geijn G.-J.M., Prinzen L. Leucocyte-bound apolipoprotein B in the circulation is inversely associated with the presence of clinical and subclinical atherosclerosis. Eur. J. Clin. Invest. 2016;46:690–697. doi: 10.1111/eci.12650. [DOI] [PubMed] [Google Scholar]
- 15.Klickstein L.B., Bartow T.J., Miletic V., Rabson L.D., Smith J.A. Identification of distinct C3b and C4b recognition sites in the human C3b/C4b receptor (CR1, CD35) by deletion mutagenesis. J. Exp. Med. 1988;168:1699–1717. doi: 10.1084/jem.168.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cockburn I.A., Mackinnon M.J., O'Donnell A., Allen S.J., Moulds J.M. A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proc. Natl. Acad. Sci. U. S. A. 2004;101:272–277. doi: 10.1073/pnas.0305306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson J.G., Murphy E.E., Wong W.W., Klickstein L.B., Weis J.H. Identification of a restriction fragment length polymorphism by a CR1 cDNA that correlates with the number of CR1 on erythrocytes. J. Exp. Med. 1986;164:50–59. doi: 10.1084/jem.164.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ripoche J., Sim R.B. Loss of complement receptor type 1 (CR1) on ageing of erythrocytes. Studies of proteolytic release of the receptor. Biochem. J. 1986;235:815–821. doi: 10.1042/bj2350815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F.S., Chu F.L., Jin L., Li Y.G., Zhang Z. Acquired but reversible loss of erythrocyte complement receptor 1 (CR1, CD35) and its longitudinal alteration in patients with severe acute respiratory syndrome. Clin. Exp. Immunol. 2005;139:112–119. doi: 10.1111/j.1365-2249.2005.02681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senbagavalli P., Geetha S.T., Karunakaran K., Banu Rekha V.V., Venkatesan P. Reduced erythrocyte CR1 levels in patients with pulmonary tuberculosis is an acquired phenomenon. Clin. Immunol. 2008;128:109–115. doi: 10.1016/j.clim.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Ruuska P.E., Ikäheimo I., Silvennoinen-Kassinen S., Käär M.L., Tiilikainen A. Normal C3b receptor (CR1) genomic polymorphism in patients with insulin-dependent diabetes mellitus (IDDM): is the low erythrocyte CR1 expression an acquired phenomenon? Clin. Exp. Immunol. 1992;89:18–21. doi: 10.1111/j.1365-2249.1992.tb06870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen J.H., Lutz H.U., Pennaforte J.L., Bouchard A., Kazatchkine M.D. Peripheral catabolism of CR1 (the C3b receptor, CD35) on erythrocytes from healthy individuals and patients with systemic lupus erythematosus (SLE) Clin. Exp. Immunol. 1992;87:422–428. doi: 10.1111/j.1365-2249.1992.tb03013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arakelyan A., Zakharyan R., Khoyetsyan A., Poghosyan D., Aroutiounian R. Functional characterization of the complement receptor type 1 and its circulating ligands in patients with schizophrenia. BMC Clin. Pathol. 2011;11:10. doi: 10.1186/1472-6890-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham B.-N., Kisserli A., Donvito B., Duret V., Reveil B. Analysis of complement receptor type 1 expression on red blood cells in negative phenotypes of the Knops blood group system, according to CR1 gene allotype polymorphisms. Transfusion. 2010;50:1435–1443. doi: 10.1111/j.1537-2995.2010.02599.x. [DOI] [PubMed] [Google Scholar]
- 25.Zorzetto M., Ferrarotti I., Trisolini R., Lazzari Agli L., Scabini R. Complement receptor 1 gene polymorphisms are associated with idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2003;168:330–334. doi: 10.1164/rccm.200302-221OC. [DOI] [PubMed] [Google Scholar]
- 26.Shepherd J., Blauw G.J., Murphy M.B., Cobbe S.M., Bollen E.L. The design of a prospective study of pravastatin in the elderly at risk (PROSPER). PROSPER study group. PROspective study of pravastatin in the elderly at risk. Am. J. Cardiol. 1999;84:1192–1197. doi: 10.1016/s0002-9149(99)00533-0. [DOI] [PubMed] [Google Scholar]
- 27.Shepherd J., Blauw G.J., Murphy M.B., Bollen E.L.E.M., Buckley B.M. Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 2002;360:1623–1630. doi: 10.1016/s0140-6736(02)11600-x. [DOI] [PubMed] [Google Scholar]
- 28.Shiffman D., Trompet S., Louie J.Z., Rowland C.M., Catanese J.J. Genome-wide study of gene variants associated with differential cardiovascular event reduction by pravastatin therapy. PLoS One. 2012;7:e38240. doi: 10.1371/journal.pone.0038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor J., Tibshirani R. A tail strength measure for assessing the overall univariate significance in a dataset. Biostatistics. 2006;7:167–181. doi: 10.1093/biostatistics/kxj009. [DOI] [PubMed] [Google Scholar]
- 31.Böhringer S. Dynamic parallelization of R functions. R. J. 2013;5:88–96. [Google Scholar]
- 32.Herrera A.H., Xiang L., Martin S.G., Lewis J., Wilson J.G. Analysis of complement receptor type 1 (CR1) expression on erythrocytes and of CR1 allelic markers in Caucasian and African American populations. Clin. Immunol. Immunopathol. 1998;87:176–183. doi: 10.1006/clin.1998.4529. [DOI] [PubMed] [Google Scholar]
- 33.Buraczynska M., Ksiazek P., Wacinski P., Zukowski P., Dragan M. Complement receptor 1 gene polymorphism and cardiovascular disease in dialyzed end-stage renal disease patients. Hum. Immunol. 2010;71:878–882. doi: 10.1016/j.humimm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Boiocchi C., Zorzetto M., Sbarsi I., Pirotta A., Schirinzi S. CR1 genotype and haplotype involvement in coronary artery disease: the pivotal role of hypertension and dyslipidemia. Int. J. Mol. Med. 2009;24:181–187. doi: 10.3892/ijmm_00000221. [DOI] [PubMed] [Google Scholar]
- 35.Kullo I.J., Ding K., Shameer K., McCarty C.A., Jarvik G.P. Complement receptor 1 gene variants are associated with erythrocyte sedimentation rate. Am. J. Hum. Genet. 2011;89:131–138. doi: 10.1016/j.ajhg.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Black S., Kushner I., Samols D. C-reactive protein. J. Biol. Chem. 2004;279:48487–48490. doi: 10.1074/jbc.R400025200. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan M.H., Volanakis J.E. Interaction of C-reactive protein complexes with the complement system. I. Consumption of human complement associated with the reaction of C-reactive protein with pneumococcal C-polysaccharide and with the choline phosphatides, lecithin and sphingomyelin. J. Immunol. 1974;112:2135–2147. [PubMed] [Google Scholar]
- 38.Mold C., Gurulé C., Otero D., Du Clos T.W. Complement-dependent binding of C-reactive protein complexes to human erythrocyte CR1. Clin. Immunol. Immunopathol. 1996;81:153–160. doi: 10.1006/clin.1996.0171. [DOI] [PubMed] [Google Scholar]
- 39.Bovenberg S.A., Klop B., Alipour A., Martinez-Hervas S., Westzaan A. Erythrocyte-associated apolipoprotein B and its relationship with clinical and subclinical atherosclerosis. Eur. J. Clin. Invest. 2012;42:365–370. doi: 10.1111/j.1365-2362.2011.02591.x. [DOI] [PubMed] [Google Scholar]
- 40.Bovenberg S.A., Alipour A., Elte J.W.F., Rietveld A.P., Janssen J.W. Cell-mediated lipoprotein transport: a novel anti-atherogenic concept. Atheroscler. Suppl. 2010;11:25–29. doi: 10.1016/j.atherosclerosissup.2010.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.