Abstract
The coiled-coil motif is found in approximately 10% of all protein sequences and is responsible for the oligomerization of proteins in a highly specific manner. Coiled-coil proteins exhibit a large diversity of function (e.g. gene regulation, cell division, membrane fusion, drug extrusion) thus demonstrating the significance of oligomerization in biological systems. The classical coiled-coil domain comprises a series of consecutive heptad repeats in the protein sequence that are readily identifiable by the location of hydrophobic residues at the ‘a’ and ‘d’ positions. This gives rise to an α-helical structure in which between 2 and 7 helices are wound around each other in the form of a left-handed supercoil. More recently, structures of coiled-coil domains have been solved that have an 11 residue (undecad) or a 15 residue (pentadecad) repeat, which show the formation of a right-handed coiled-coil structure. The high stability of coiled coils, together with the presence of large internal cavities in the pentameric coiled-coil domain of cartilage oligomerization matrix protein (COMPcc) and the tetrameric right-handed coiled coil of Staphylothermus marinus (RHCC) has led us and others to look for therapeutic applications. In this review, we present evidence in support of a vitamin A and vitamin D3 binding activity for the pentameric COMPcc molecule. In addition, we will discuss exciting new developments which show that the RHCC tetramer is capable of binding the major anticancer drug cisplatin and the ability to fuse it to an antigenic epitope for the development of a new generation of vaccines.
Keywords: Coiled coil, Drug delivery, COMPcc, RHCC, Vitamin D3, Vitamin A, Cisplatin
1. Introduction
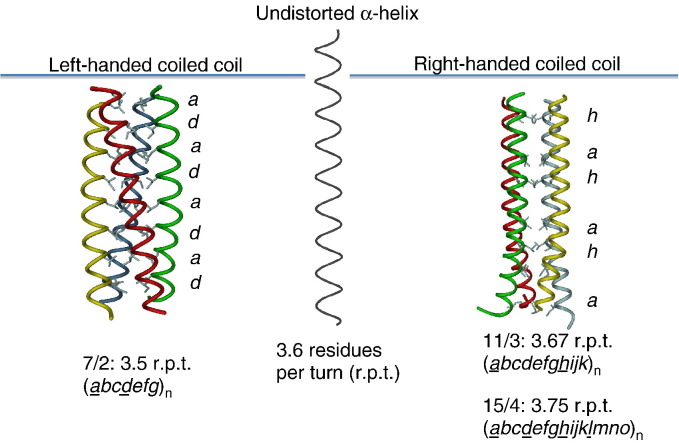
The α-helical coiled coil is the most frequently encountered subunit oligomerization motif found in proteins (Cohen and Parry, 1990, Lupas, 1996, Kohn et al., 1997), with an analysis of the protein database revealing that approximately 10% of all proteins contain the signature heptad repeat (abcdefg)n which is characteristic of coiled-coil proteins (Walshaw and Woolfson, 2001). Its existence was first predicted in a series of seminal papers by Crick (Crick, 1952, Crick, 1953a, Crick, 1953b) and supported by an observation by Perutz and others that the long fibrous protein tropomyosin and other members of the k – m – e–f proteins exhibited diffraction peaks with a repeated spacing of 5.15 Å. Crick proposed that this could be explained by an arrangement in which two α-helices wrapped around each other at an angle of approximately 20° such that their side chains were interlocked together in a motif that repeated itself every 7 residues, or 2 turns of the helix. This arrangement would result in a helix with 3.5 residues per turn compared to the usual 3.6 for an undistorted α-helix, with the driving force for the distortion of the helices coming from the hydrophobic residues at the core ‘a’ and ‘d’ positions (Fig. 1 ). Crick referred to this packing as a “knobs into holes” arrangement in which one residue (knob) fits into a space generated by four residues of the opposing helix (hole). The 4,3 spacing observed in coiled coils is the minimum spacing that allows for the formation of an amphipathic α-helix that has the core hydrophobic residues positioned on the same side of the helix, thereby facilitating the “knobs into holes” packing. Due to the supercoiling of the two helices around each other in a coiled coil, the handedness of the coiled coil is the opposite handedness to that of the helix which formed it. The simplest coiled-coil structure is the dimer, but trimers, tetramers, pentamers (Efimov et al., 1996, Malashkevich et al., 1996), and heptamers (Liu et al., 2006) have been reported. In addition, if the helices come from the same protein chain, the resultant coiled coil is described as a homo-oligomer. Conversely, if the helices originate from different protein chains, the coiled coil is referred to as a hetero-oligomer.
Fig. 1.
From α-helices to left- and right-handed coiled coils. In an undistorted α-helix one amino acid residue is rotated 100° along the screw axis and subsequently 3.6 residues are required for a full turn (centre). Coiled coils, which are pairs of α-helices winding around each other, show deviations from this rule. In left-handed coiled coils 7 residues are required for 2 turns and therefore are more tightly wound around each other in a left-handed supercoil to compensate for this missing 20° (left). In contrast, naturally occurring right-handed coiled coils reveal repeats of 11residues per 3 turns or 15 residues per 4 turns of the helix (right). As a consequence they are supercoiled either 20° or 60° to form a right-handed supercoil. Characteristic in all cases are highly repetitive sequence motifs of 7, 11, or 15 amino acid residues (in italics) with conserved aliphatic core residues (underlined) forming the hydrophobic core of coiled-coil channels.
Given that the α-helix is right-handed (Ramachandran et al., 1963) and that the majority of coiled-coil proteins display a heptad repeat, it follows that the majority of coiled coils will adopt a left-handed supercoiled structure. However, more recent examples of right-handed coiled coils have been reported in the literature (Peters et al., 1996, Stetefeld et al., 2000, Kühnel et al., 2004, Hoiczyk et al., 2000, Nooren et al., 1999). This is not entirely unexpected as in addition to the classical 7/2 repeat suggested by Crick, Pauling proposed that α-helices could undergo deformations that resulted in repeats with an 11/3 (11 residues in 3 turns of the helix), 15/4, or 18/5 periodicity (Pauling et al., 1951). More recently, a protein which displays an additional 25/7 periodicity has been reported (Tarbouriech et al., 2000). Of interest with respect to this review, was the prediction that the 7/2 and 15/4 repeats would have the opposite handedness (Pauling and Corey, 1953). This has since been confirmed with the determination of coiled-coil structures that have a 15/4 (Kühnel et al., 2004, Hoiczyk et al., 2000, Nooren et al., 1999) and an 11/3 periodicity (Stetefeld et al., 2000), which revealed a right-handed coiled-coil structure (Fig. 1). The 18/5 and 25/7 repeats both form left-handed coiled coils (Pauling et al., 1951, Tarbouriech et al., 2000).
The prevalence of the coiled-coil motif across biological systems indicates its significance in many key biological processes. Indeed, coiled-coil motifs are found in proteins involved in transcription (Landschulz et al., 1988), cell/cell communication, membrane fusion, and proteins which act as molecular spacers and motors (see review Lupas and Gruber, 2005). Such a diversity of function requires a simple design that is both robust and highly adaptable. Research is now being directed towards the use of these properties with applications in the field of nanotechnology. Due to the ability of certain coiled-coil structures to bind heavy metals they may also have an important environmental function (Stetefeld et al., 2000). A promising new role relates to the ability of certain coiled-coil domains to bind biologically relevant molecules such as the major anticancer drug cisplatin, which raises the possibility that they could be used in the treatment of cancers. In addition, the use of coiled-coil domains for the production of a new generation of vaccines is another recent and exciting development in the field that will be watched with interest.
2. Structure of coiled coils
The determination of the protein sequence for tropomyosin in 1975 showed that a heptad repeat (abcdefg)n could be identified from analysis of the protein sequence alone (Stone et al., 1975, McLachlan and Stewart, 1975). This spawned a whole field in its own right aimed at correctly predicting coiled-coil motifs from an analysis of any given protein sequence (Parry, 1975, McLachlan and Stewart, 1975, McLachlan and Karn, 1983, Lupas et al., 1991, Berger et al., 1995, Wolf et al., 1997, Delorenzi and Speed, 2002). However, it was not until the publication of the influenza haemagglutinin structure (Wilson et al., 1981) that the coiled coil was first visualized showing the core hydrophobic residues adopting the “knobs into holes” packing as predicted by Crick at positions ‘a’ and ‘d’ in the heptad repeat. Since then much effort has been directed towards a better description of the coiled coil both in terms of the driving forces that determine the oligomeric state and also what additional forces stabilize the coiled-coil oligomer.
The 33 residue leucine zipper region of the yeast transcription factor GCN4 has proven a useful experimental tool to address the issue of oligomerization state. GCN4 has the standard heptad repeat with a 7/2 periodicity and has Ile predominantly at the ‘a’ and Leu at the ‘d’ positions, respectively. In this configuration the crystal structure shows that the protein adopts a dimeric coiled coil (O'Shea et al., 1991). However, if the positions of the Ile and Leu are reversed such that Leu is found at the ‘a’ and Ile at the ‘d’ position then the protein becomes a tetramer (Harbury et al., 1994). Any other combination of Ile, Leu, or Val at the ‘a’ and ‘d’ positions in the same study was shown to result in a trimeric state (Harbury et al., 1993).
The classical left-handed coiled coil displays a heptad repeat with a so-called 7/2 periodicity, which places the core hydrophobic residues at an average spacing of 3.5 residues per turn of the helix. The next nearest number larger than 3.6 which allows residues to assume quasi-equivalent positions after a small number of turns is 3.67. This is achieved with an undecad repeat (abcdefghijk)n that has a periodicity of 11/3. In this motif, hydrophobic residues are present at the ‘a’ and ‘d’ positions as for the heptad repeat but also additionally at the ‘e’ and ‘h’ positions. Such a coiled-coil motif was identified from a naturally occurring archeal protein (RHCC) from Staphylothermus marinus (Peters et al., 1996) and its structure solved in 2000 (Stetefeld et al., 2000). Of interest, is the fact that the undecad motif gives rise to a coiled coil that adopts a right-handed configuration. In this structure, the hydrophobic residues at the ‘a’ and ‘h’ positions are equivalent to the ‘a’ and ‘d’ positions of left-handed coiled coils and form the expected “knobs into holes” packing, whereas the hydrophobic residues at the ‘d’ and ‘e’ positions pack in a “knobs into knobs” arrangement. Given that the number of residues per turn for the 11/3 right-handed coiled coil at 3.67 is only just above the 3.6–3.64 expected for an undistorted α-helix then it is perhaps not surprising to find that the RHCC is only weakly supercoiled to the right.
A final coiled-coil motif discussed here is the pentadecad repeat (abcdefghijklmno)n which is obtained by having fifteen residues over four turns of the helix and has an average spacing between hydrophobic residues of 3.75. The tetramerization domain of VASP adopts such a 15/4 motif and its structure has been solved to atomic resolution (Kühnel et al., 2004). It forms a right-handed coiled coil, with the hydrophobic core residues located at positions ‘a’, ‘d’, ‘e’, ‘h’, and ‘l’ of the repeat. In the VASP structure the residues at positions ‘a’ and ‘h’ form the expected “knobs into holes” packing as in the RHCC structure, whereas the residues at positions ‘d’, ‘e’, and ‘l’ pack in a “knobs into knobs” arrangement. The larger spacing between hydrophobic residues is suggestive of a higher degree of supercoiling of the VASP right-handed coiled coil and this is reflected in the structure. Thus right-handed coiled coils with a 15/4 periodicity form coiled-coil structures that supercoil as much to the right as the more numerous left-handed coiled coils with a 7/2 periodicity coil to the left.
3. Stability, helix packing, and cavities
A key feature for the stability and oligomerization states of coiled coils is the hydrophobic core formed by the “knobs into holes” interactions between residues at the ‘a’ and ‘d’ positions of the heptad repeat (Fig. 1). In left-handed coiled coils, the β-branched side chains Leu, Ile, and Val together with Ala are found almost exclusively at the ‘a’ and ‘d’ positions within the heptad repeat and polar helix favouring residues are generally found elsewhere (Sodek et al., 1972, McLachlan and Stewart, 1975). In a 4,3-hydrophobic pattern they form either parallel or perpendicular “knobs into holes” with amino acid side chains of the adjacent helix (Burkhard et al., 2001). In the RHCC from S . marinus, the “knobs into holes” interactions between helices are formed by the side chains at positions ‘a’ and ‘h’ of the undecad repeat, which is consistent with a 7,4-hydrophobic repeat (Fig. 1). This was an unexpected result, as formation of an artificial right-handed coiled coil (Harbury et al., 1998) was achieved using a 4,4,3-repeat. A further feature that stabilizes the RHCC tetramer is provided by the “knobs into knobs” packing between the hydrophobic residues at the ‘d’ position of one helix and the ‘e’ position of the neighbouring helix to form the de-layer (Fig. 2A). The additional hydrophobic interactions present in the RHCC result in a large buried surface of ~ 9.500 Å2 which accounts for ~ 50% of the total solvent-accessible area of the four isolated helices which compares with an expected value of 20–25% for a non coiled-coil protein. As a consequence, the RHCC is extremely thermostable and resists heating to temperatures > 90 °C (Peters et al., 1995). In contrast, the hydrophobic pattern of the right-handed VASP tetramer revealed a 4,3,4,4-hydrophobic pattern of “knobs into holes” that is equivalent to a heptad repeat and two 4-residue insertions (Kühnel et al., 2004).
Fig. 2.
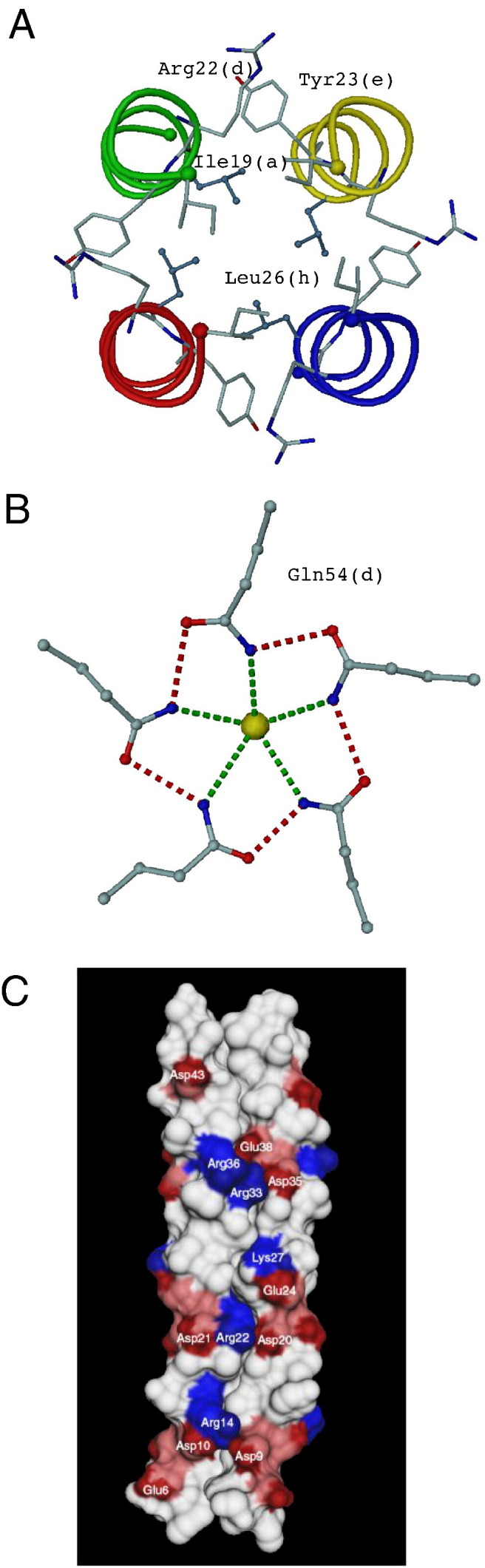
Inside and outside forces for coiled coil stabilization. (A) Inside forces are classical "knobs into holes" formations of aliphatic side chains oriented inside the coiled-coil channel. In addition to a and h positioned "knobs into holes" the right-handed coiled coil from Staphylothermus marinus reveals a so far unknown inter-helical hydrophobic core composed of residues in d and e positions (de-layer). (B) In the case of the pentameric COMPcc (PBD-code 1VDF), a ring of five Gln54 residues in position d is arranged to form an inter-helical network of ionic interactions (red dotted lines) and contributes to the binding (green dotted lines) of a chloride ion (highlighted in yellow). (C) Molecular surface model of the tetrameric RHCC structure. Only the front two helices of the RHCC tetramer are shown. Hydrophobic residues are depicted in grey, whereas positively- and negatively-charged residues, which form the major i-i′ + 2 (inter-helical) and i−i + 3 (intra-helical) salt bridges are depicted in blue and red, respectively.
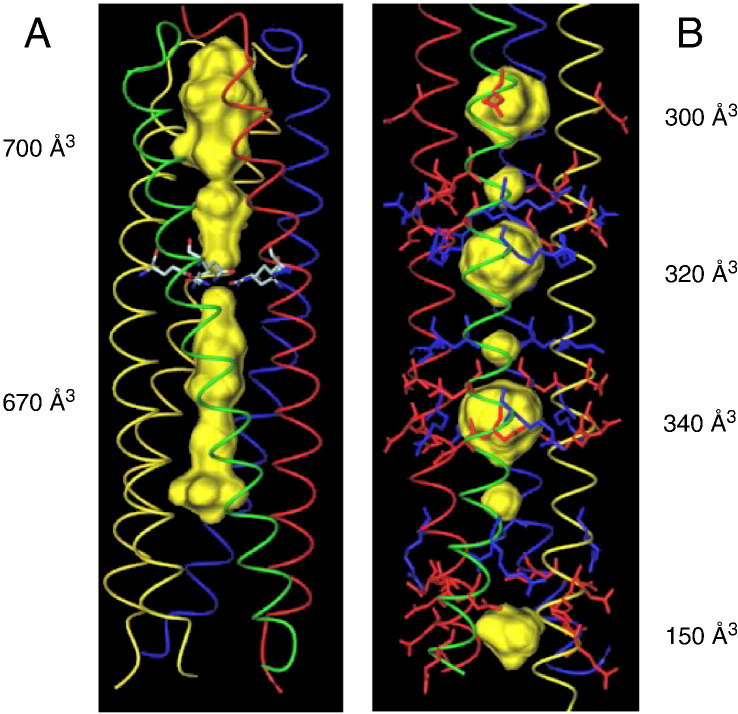
Of particular interest with respect to the topic of this review is that the left-handed pentameric coiled-coil domain of cartilage oligomerization matrix protein (COMPcc) and the right-handed tetrameric RHCC from S . marinus both form large internal cavities. To date, no other examples exist of coiled coils that form such large cavities and so these two domains are unique. COMPcc forms a pentameric coiled-coil structure (Efimov et al., 1996, Malashkevich et al., 1996) that contains a 73 Å-long axial pore that has a diameter of between 2 and 6 Å along its length, with the exception of the constriction formed by a ring of five conserved glutamine residues at position 54 which divides the channel into two (Fig. 2, Fig. 3 ). The positioning of the glutamine residue at a ‘d’ position of the heptad repeat is somewhat surprising, but further analysis shows that this is related to its ligand binding activities which are discussed in detail in Section 5. By contrast, the RHCC molecule contains four large cavities on the inside of the RHCC tetramer (Fig. 3B), a feature which is absent from all known left-handed tetrameric coiled-coil structures deposited in the protein database (PDB). The cavities are connected to a continuous central channel which is exclusively lined with aliphatic side chains. The diameter of the channel as defined by the van der Waals radii varies between 2.0 and 8.4 Å, which exceeds the 6 Å maximum diameter of the pentameric COMPcc pore. The cavities are located in the gap between the ‘a’ and ‘h’ layers. In the native structure, the cavities are occupied with water molecules that form water clusters due to the lack of polar groups. Analysis of the heavy atom derivatives used for phasing the structure showed that the major heavy atom binding sites are located within these cavities. The presence of the heavy atoms in the cavities is almost certainly responsible for the excellent quality of the derivative parameters. The narrow 2 Å-wide constriction formed at the ‘a’ and ‘h’ layers suggests that the derivatives do not enter the cavities via the termini. Instead it would appear likely that the heavy atoms penetrate directly into the cavities through gates formed by a complex network of surface salt bridges and ionic interactions (Stetefeld et al, 2000).
Fig. 3.
Cavities inside tetrameric and pentameric coiled-coil channels. Extraordinary features of COMPcc (A) and RHCC (B) are large cavities along the aliphatic channel core. These cavities (highlighted as yellow van der Waals spheres) can function as storage spaces for a great variety of different cargo systems with large biomedical importance. The volumes of individual cavities are shown in cubic Angstroms (Å3). Salt bridges surrounding individual cavities in RHCC are highlighted in red (acidic resdiues) and blue (basic residues). The Gln54 ring in COMPcc is drawn in colour atom type. Remarkably, both channels allow for different shapes of interior spaces. Whereas RHCC cavities are regularly-arranged separated balls, both cavities in COMPcc are elongated and separated by the Gln54 ring in the d-position.
4. Electrostatics
After the hydrophobic forces, the electrostatic interactions that are formed within (intra-) or between (inter-) the helices of the coiled coil are the next most significant factors involved in stabilization of coiled-coil domains. This is facilitated by an exceptionally high content (up to ~ 27%) of charged residues. Such a high frequency of favourable electrostatic interactions is not observed in non coiled-coil protein structures and may explain in part the high thermostability of many coiled-coil peptides. The inter-helical salt bridges are typically formed between the ‘e’ position of one helix and the ‘g’ position of the adjacent helix (i–i′ + 2) with the residues involved having the opposite charges (McLachlan and Stewart, 1975). With regard to the intra-helical salt bridges, these are typically between residues in an i–i + 3 or an i–i + 4 arrangement (Meier and Burkhard, 2006). In the RHCC structure, besides the expected i–i′ + 2 inter-helical salt bridges, a unique i–i′ + 5 interaction is also observed. Furthermore, in the RHCC tetramer the salt bridges are organized into three networks of complex salt bridges involving residues that are flanked by polar interactions (Fig. 2C).
5. Coiled coils and drug delivery
5.1. COMPcc
The design of efficient drug delivery systems, especially for anticancer therapies, is of great interest as many of these drugs cause toxic side effects due to their lack of specificity for cancerous versus healthy cells. Coiled coils are attractive candidates for drug delivery systems due to their ability to bind a number of biologically relevant molecules. The five-stranded coiled-coil domain of cartilage oligomeric matrix protein (COMPcc) is one such example, where the storage function of the protein has been studied. Through the use of X-ray crystallography and in-solution assays, the ability of COMPcc to reversibly bind various cargo molecules has been thoroughly characterized (Guo et al., 1998, Özbek et al., 2002).
COMPcc is a homopentamer consisting of five α-helices that are joined by interchain disulfide bridges. It forms an internal, hydrophobic pore that is 73 Å-long and has a diameter of 2–6 Å. This pore is divided into two by a ring of conserved glutamines at position 54, which via an intricate network of hydrogen bonds act as a biological magnet to attract electronegative ligands (Efimov et al., 1996, Malashkevich et al., 1996). The ability of the pore to bind a wide spectrum of ligands is directly related to its physicochemical properties. For example, small electronegative ions such as chloride are held in the channel by virtue of an electrostatic interaction with the glutamine 54 ring, whereas vitamins A and D3 are bound by a combination of hydrophobic forces along the length of the pore and an electrostatic interaction with the glutamine 54 ring (Malashkevich et al., 1996, Guo et al., 1998, Özbek et al., 2002). In addition, small hydrophobic compounds such as benzene can also bind the channel due to its hydrophobicity (Guo et al., 1998). Thus the COMPcc channel has the potential to act as a “Trojan horse” to deliver therapeutic compounds in the treatment of disease.
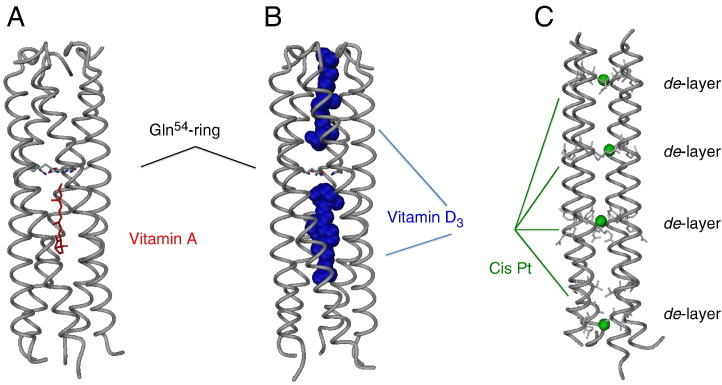
A possible storage and delivery function of COMPcc was first suggested by Guo et al. with the observation that the coiled-coil protein was able to bind a number of important cell signalling molecules, including vitamin A (all-trans retinol) and vitamin D3. They were able to determine a dissociation constant of K D = 0.6 μM for all-trans retinol using fluorescence titration. Furthermore, they solved the crystal structure of the COMPcc-all-trans retinol complex to validate the ligand binding within the hydrophobic channel. A single molecule of retinol is bound in the “lower” N-terminal cavity of COMPcc, with its β-ionone ring oriented towards the N-terminus and the isoprene tail oriented towards the C-terminus (Fig. 4A). The hydroxyl group of retinol forms hydrogen bonds with two of the five glutamine residues (Guo et al., 1998). A subsequent study by Özbek et al. confirmed the binding of vitamin D3 to COMPcc by X-ray crystallography of the COMPcc–vitamin D3 complex. An earlier study by Guo et al. had been suggestive of vitamin D3 binding as the midpoint transition temperature T m for unfolding increased by 8 °C in the presence of the molecule (Guo et al., 1998). Interestingly, the crystal structure by Özbek et al. shows that two molecules of vitamin D3 are bound to COMPcc. One molecule of vitamin D3 is bound in each cavity, with the molecules oriented in a head-to-head fashion in the hydrophobic channel (Fig. 4B). Of significance is that upon binding to vitamin D3, the hydrophobic cavities increase in volume by ~ 30%. This is achieved by a reorientation of the β-branched side chains at positions ‘a’ and ‘d’ of the coiled coil, which are responsible for forming the “knobs into holes” (Özbek et al., 2002) and shows the dynamic nature of the channel when binding ligands. Both the studies by Guo et al. and Özbek et al. are supportive of a storage and delivery function for COMPcc.
Fig. 4.
Storage properties of coiled-coil domains. COMPcc has been shown to store vitamin A (A) and vitamin D3 (B). In both cases the elongated cargo is oriented such that its hyroxyl group can form an electrostatic interaction with the Gln54 ring system. In contrast to vitamin A where only molecule is bound, COMPcc can store two vitamin D3 molecules, one in each cavity. (C) The larger cavities in RHCC can store one molecule of cisplatin each. Whereas aliphatic, elongated cargos in COMPcc are diffusing via both N- and C-terminus into the hydrophobic channel, the ionic network in RHCC allows for a diffusion of cisplatin via the inter-helical interface.
5.2. RHCC
Another coiled-coil domain of interest is the right-handed coiled-coil domain (RHCC) from S . marinus. This domain forms a tetrameric right-handed coiled coil, which contains four large internal cavities that are capable of binding large metals such as gold and platinum (Stetefeld et al., 2000). Of relevance to this review, a recent study by Eriksson et al. demonstrated that the anticancer drug cisplatin could bind to RHCC. Furthermore, the complex was delivered to tumor cell lines in vitro (Eriksson et al., 2009). Cisplatin is a widely-used chemotherapeutic drug that is used to treat a number of cancers including those of the reproductive system, as well as stomach and bladder (Boulikas and Vougiouka, 2004). Unfortunately, treatment with cisplatin leads to numerous toxic side effects, thus the development of an effective drug delivery system aimed at selectively targeting cancerous cells is highly desirable.
Eriksson et al. incubated RHCC with cisplatin and the excess was removed by gel filtration. The amount of bound cisplatin was determined by inductively-coupled plasma optical emission spectroscopy (ICP-OES), and it was found that on average one molecule of cisplatin is bound per cavity in each RHCC tetramer (Fig. 4C). They also demonstrated that cisplatin-bound RHCC was stable for up to 12 h in solution. Fluorescently-labelled RHCC was incubated with FADu carcinoma cells and the uptake of the protein into the cells was monitored by fluorescence microscopy. It was determined that the cell lines were able to take up the protein into intracellular vesicles. The cytotoxicity of RHCC and cisplatin-bound RHCC was analyzed by incubating the proteins with various carcinoma cell lines. RHCC exerted no cytotoxic effect on cells, while cisplatin-bound RHCC was found to either show an equivalent effect or a significantly higher effect than cisplatin alone. Perhaps the most encouraging findings of the study by Eriksson et al. were in the trials of cisplatin-bound RHCC in vivo. Mice treated with the drug-loaded protein exhibited no significant immune response (Eriksson et al., 2009). These findings indicate the great potential of RHCC as a carrier system for cisplatin.
5.3. Future prospects
One of the main challenges of drug delivery is selectively targeting toxic compounds to their desired locations. In particular with chemotherapeutic drugs, the cytotoxicity of these compounds causes many harsh side effects, thus the development of a targeted system to bypass healthy cells and deliver therapies directly to cancer cells is of great interest. The unique properties of coiled coils make them highly suitable for use in these systems. Since the major function of coiled coils is to act as oligomerization domains, the N- and C-terminals of the proteins can be easily conjugated to numerous epitopes that can bind specific cell surface markers, thereby creating a powerful targeted drug delivery system. The ability to fuse coiled-coil domains to other protein domains was first shown by Tomschy et al. who conjugated five extracellular CAD domains of E-cadherin to the N-terminus of COMPcc (Tomschy et al., 1996). In addition, Ahrens et al. fused five CAD domains of P-cadherin via a linker region to the N-terminus of COMPcc (Ahrens et al., 2002). The classical cadherin domains (E-, N-, P-, and C-cadherin) function in Ca2+-dependent selective cell recognition and adhesion (Takeichi, 1988, Rimm and Morrow, 1994). Therefore, by attaching COMPcc to molecules that are involved in cell recognition and adhesion, this raises the possibility that the complex can be directed to a target cell where it would release its cargo. These studies illustrate the potential of coiled coils in the development of new targeted drug delivery systems.
A final therapeutic application discussed here is the fusion of coiled-coil domains to epitopes for the production of therapeutic vaccines. As coiled coils form oligomers, such fusions should result in the repetitive display of antigens thereby generating a strong immune response. This property was examined by Schroeder et al. and Pimentel et al., who generated a peptide nanoparticle for the presentation of actin antigens and severe acute respiratory syndrome (SARS) virus epitopes, respectively (Schroeder et al., 2009, Pimentel et al., 2009). The core polypeptide of the nanoparticle contains a modified sequence of the pentameric COMPcc connected to a de novo designed trimeric coiled coil via two glycine residues. The desired epitope can then be linked to the C-terminus of the core polypeptide for antigenic display. Schroeder et al. linked a short sequence corresponding to the hydrophobic loop of actin and Pimentel et al. linked the HRC1 epitope from the SARS coronavirus spike protein to the trimeric coiled coil, respectively (Schroeder et al., 2009, Pimentel et al., 2009). The resultant polypeptide self-assembles into an icosahedron and resembles a virus-like particle (VLP) (Raman et al., 2006). Both studies demonstrated that this repetitive arrangement of epitopes on the surface of the peptide nanoparticle was able to elicit a specific immunogenic response to the projected epitopes, indicating the potential of this system in the design of vaccines (Schroeder et al., 2009, Pimentel et al., 2009).
6. Conclusion
Coiled coils are primarily oligomerization domains that are capable of forming well defined higher order multimers, which are extremely stable. They were initially identified in proteins of the k – m–e–f series, which includes the keratins, tropomyosin, and fibrinogen; therefore it was believed that they would simply provide rigidity to proteins. Despite the fact that these proteins led to the proposal of a coiled-coil structure, they are only poorly characterized at the structural level due to their biochemical characteristics. For example, the full length tropomyosin has only been solved to 7 Å resolution (Whitby and Phillips, 2000) although the N-terminal (Brown et al., 2001) and C-terminal fragments (Li et al., 2002) have been solved separately to 2 Å and 2.7 Å, respectively. Of all the k – m–e–f proteins, the only one whose structure has been completely solved is fibrinogen (Brown et al., 2000, Yang et al., 2000, Yang et al., 2001). However, since their initial discovery they have been shown to be involved in key biological processes that include gene transcription (Landschulz et al., 1988), regulation (Nooren et al., 1999), cytokinesis (Lee et al., 2008), molecular motors (Yun et al., 2003), muscle fibres (Strelkov et al., 2002), multidrug resistance (Higgins et al., 2004, Koronakis et al., 2000), membrane fusion (Deng et al., 2006), protein stability in harsh environments (Peters et al., 1995, Peters et al., 1996), and more recently as potential delivery systems either for the treatment of cancer (Eriksson et al., 2009) or in the development of novel vaccines (Schroeder et al., 2009, Pimentel et al., 2009). Despite this diversity of function, the one common characteristic is the requirement for a stable oligomerization domain and this is provided by the coiled-coil domain.
It is perhaps not surprising to find that the α-helical coiled coil is derived from one of the two stable secondary structure elements found in proteins, namely the α-helix. Thus, nature has taken an already stable structural element and improved its stability by virtue of a simple repeating motif that takes advantage of the inherent properties of the α-helix. This has the major advantage that such domains can be readily incorporated into existing structural motifs. Furthermore, all that is required to adjust the oligomeric state of the coiled coil is a substitution of the core residues at the ‘a’ and ‘d’ positions in the heptad repeat (Harbury et al., 1993, Harbury et al., 1994). Therefore, the coiled coil can be easily adapted to suit the end function, which may explain why this domain is so prevalent in biological systems. Interestingly, all the right-handed coiled coils identified to date (Peters et al., 1996, Nooren et al., 1999, Stetefeld et al., 2000) are tetrameric, so it remains to be seen if other oligomeric states of such domains are identified in the future or if the tetrameric state is preferred above all other combinations. As a simple rule of thumb, it would appear that when the average spacing between hydrophobic residues is < 3.6 Å the coiled coil is left-handed, whereas when the spacing is > 3.6 Å it adopts a right-handed configuration. Thus it seems that biological systems have devised an easy way in which to differentiate between these two coiled-coil forms.
With regard to this review, the two coiled-coil domains of the most interest are the left-handed COMPcc from Homo sapiens and the right-handed RHCC from S . marinus. The shared feature of these two proteins is the presence of large internal cavities that are absent from all other coiled-coil structures solved to date. COMPcc contains two internal cavities that have been shown to bind the signalling molecules vitamins A and D3. These molecules are largely hydrophobic with polar groups at one end and bind readily inside the hydrophobic cavity of COMPcc. Many drugs used in the treatment of cancers are hydrophobic, which makes drug delivery by conventional means problematic due to solubility issues. Therefore, this problem could be overcome by loading the therapeutic drugs inside the cavity. This is exactly the approach that Eriksson et al. (2009) used when they demonstrated that the cavities of RHCC could be loaded with the anticancer drug cisplatin. There are two main advantages to using coiled-coil domains as “Trojan horses” for the delivery of therapeutic compounds. Firstly, by shielding the therapeutic drug inside the cavity it reduces the possibility of the drug being metabolized before reaching the cancerous site. Secondly, it avoids the recognition of the drug by one of the many membrane-bound drug exporters, which actively pump the drug back out of the cell before it reaches a high enough concentration to kill the cancer cell.
An additional property of coiled coils that is beneficial for drug delivery relates to its oligomeric state. Studies in the late 1990s and early 2000s showed that it was possible to fuse coiled coils to other protein domains involved in cell recognition and adhesion (Tomschy et al., 1996, Ahrens et al., 2002). These proofs of principle experiments are important as they raise the possibility that a targeting molecule can be attached to a coiled coil in order to direct it to a specific location in the cell for delivery of a target cargo. Following on from this, more recent experiments have shown that coiled-coil domains can be fused to various epitopes for the production of therapeutic antibodies. This is possible as a result of the experiments by Eriksson et al. (2009) which showed that the coiled coil itself did not elicit an immune response, and also because any immune response generated by fusion to an antigenic epitope will be amplified due to the multiple copies of protein chains coming from the coiled coil. Preliminary results (Schroeder et al., 2009, Pimentel et al., 2009) are encouraging and further developments in this area are eagerly awaited.
Acknowledgements
This work was supported by a grant from the University of Manitoba Research and IPM Program. Dr. Jörg Stetefeld is supported by the Canada Research Chair Program. We thank Dr. Simone Karrasch and Mr. Michael McFarlane for their editorial assistance.
References
- Ahrens T., Pertz O., Häussinger D., Fauser C., Schulthess T., Engel J. Analysis of heterophilic and homophilic interactions of cadherins using the c-Jun/c-Fos dimerization domains. J. Biol. Chem. 2002;277:19455–19460. doi: 10.1074/jbc.M200606200. [DOI] [PubMed] [Google Scholar]
- Berger B., Wilson D.B., Wolf E., Tonchev T., Milla M., Kim P.S. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T., Vougiouka M. Recent clinical trials using cisplatin, carboplatin and their combination chemotherapy drugs. Oncol. Rep. 2004;11:559–595. [PubMed] [Google Scholar]
- Brown J.H., Volkmann N., Jun G., Henschen-Edman A.H., Cohen C. The crystal structure of modified bovine fibrinogen. Proc. Natl. Acad. Sci. USA. 2000;97:85–90. doi: 10.1073/pnas.97.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.H., Kim K.H., Jun G., Greenfield N.J., Dominiquez R., Volkmann N., Hitchcock-DeGregori S.E., Cohen C. Deciphering the design of the tropomyosin molecule. Proc. Natl. Acad. Sci. USA. 2001;98:8496–8501. doi: 10.1073/pnas.131219198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P., Stetefeld J., Strelkov S.V. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- Cohen C., Parry D.A.D. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- Crick F.H.C. Is alpha-keratin a coiled coil? Nature. 1952;170:882–883. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]
- Crick F.H.C. The Fourier transform of a coiled-coil. Acta Crystallogr. 1953;6:685–689. [Google Scholar]
- Crick F.H.C. The packing of alpha-helices: simple coiled-coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- Delorenzi M., Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- Deng Y., Liu J., Zheng Q., Yong W., Lu M. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure. 2006;14:889–899. doi: 10.1016/j.str.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov V.P., Engel J., Malashkevich V.N. Crystallization and preliminary crystallographic study of the pentamerizing domain from cartilage oligomeric matrix protein: a five-stranded alpha-helical bundle. Proteins. 1996;24:259–262. doi: 10.1002/(SICI)1097-0134(199602)24:2<259::AID-PROT13>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Hassan S., Larsson R., Linder S., Ramqvist T., Lövborg H., Vikinge T., Figgemeier E., Müller J., Stetefeld J., Dalianis T., Özbek S. Utilization of a right-handed coiled-coil protein from archaebacteria Staphylothermus marinus as a carrier for cisplatin. Anticancer Res. 2009;29:11–18. [PubMed] [Google Scholar]
- Guo Y., Bozic D., Malashkevich V.N., Kammerer R.A., Schulthess T., Engel J. All-trans retinol, vitamin D, and other hydrophobic compounds bind in the axial pore of the five-stranded coiled coil domain of cartilage oligomeric matrix protein. EMBO J. 1998;17:5265–5272. doi: 10.1093/emboj/17.18.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury P.B., Zhang T., Kim P.S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Harbury P.B., Kim P.S., Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- Harbury P.B., Plecs J.J., Tidor B., Alber T., Kim P.S. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- Higgins M.K., Bokma E., Koronakis E., Hughes C., Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA. 2004;101:9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiczyk E., Roggenkamp A., Reichenbecher M., Lupas A., Heesemann J. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 2000;19:5989–5999. doi: 10.1093/emboj/19.22.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn W.D., Mant C.T., Hodges R.S. Alpha-helical protein assembly motifs. J. Biol. Chem. 1997;272:2583–2586. doi: 10.1074/jbc.272.5.2583. [DOI] [PubMed] [Google Scholar]
- Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- Kühnel K., Jarchau T., Wolf E., Schlichting I., Walter U., Wittinghofer A., Strelkov S.V. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17027–17032. doi: 10.1073/pnas.0403069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W.H., Johnson P.F., McKnight S.L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lee H.H., Elia N., Ghirlando R., Lippincott-Schwartz J., Hurley J.H. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Mui S., Brown J.H., Strand J., Reshetnikova L., Tobacman L.S., Cohen C. The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc. Natl. Acad. Sci. USA. 2002;99:7378–7383. doi: 10.1073/pnas.102179999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Deng Y., Zheng Q., Cheng C.S., Kallenbach N.R., Lu M. A seven-helix coiled coil. Proc. Natl. Acad. Sci USA. 2006;103:15457–15462. doi: 10.1073/pnas.0604871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Vandyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends Biochem. Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- Lupas A., Gruber M. The structure of alpha-helical coiled coils. Adv. Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- Malashkevich V.N., Kammerer R.A., Efimov V.P., Schulthess T., Engel J. The crystal structure of a five-stranded coiled coil in COMP: a prototype ion channel? Science. 1996;274:761–765. doi: 10.1126/science.274.5288.761. [DOI] [PubMed] [Google Scholar]
- McLachlan A.D., Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J. Mol. Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- McLachlan A.D., Karn J. Periodic features in the amino acid sequence of nematode myosin rod. J. Mol. Biol. 1983;164:605–626. doi: 10.1016/0022-2836(83)90053-0. [DOI] [PubMed] [Google Scholar]
- Meier M., Burkhard P. Statistical analysis of intrahelical ionic interactions in alpha-helices and coiled coils. J. Struct. Biol. 2006;155:116–129. doi: 10.1016/j.jsb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Nooren I.M.A., Kaptein R., Sauer R.T., Boelens R. The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils. Nat. Struct. Biol. 1999;6:755–759. doi: 10.1038/11531. [DOI] [PubMed] [Google Scholar]
- O'Shea E.K., Klemm J.D., Kim P.S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Özbek S., Engel J., Stetefeld J. Storage function of cartilage oligomeric matrix protein: the crystal structure of the coiled-coil domain in complex with vitamin D3. EMBO J. 2002;21:5960–5968. doi: 10.1093/emboj/cdf628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D.A.D. Analysis of the primary sequence of alpha-tropomyosin from rabbit skeletal muscle. J. Mol. Biol. 1975;98:519–535. doi: 10.1016/s0022-2836(75)80084-2. [DOI] [PubMed] [Google Scholar]
- Pauling L., Corey R.B., Branson H.R. The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L., Corey R.B. Compound helical configurations of polypeptide chains: structure of proteins of the alpha-keratin type. Nature. 1953;171:59–61. doi: 10.1038/171059a0. [DOI] [PubMed] [Google Scholar]
- Peters J., Nitsch M., Kühlmorgen B., Golbik R., Lupas A., Kellermann J., Engelhardt H., Pfander J.-P., Müller S., Goldie K., Engel A., Stetter K.-O., Baumeister W. Tetrabrachion: a filamentous archaebacterial surface protein assembly of unusual structure and extreme stability. J. Mol. Biol. 1995;245:385–401. doi: 10.1006/jmbi.1994.0032. [DOI] [PubMed] [Google Scholar]
- Peters J., Baumeister W., Lupas A. Hyperthermostable surface layer protein tetrabrachion from the archaebacterium Staphylothermus marinus: evidence for the presence of a right-handed coiled coil derived from the primary structure. J. Mol. Biol. 1996;257:1031–1041. doi: 10.1006/jmbi.1996.0221. [DOI] [PubMed] [Google Scholar]
- Pimentel T.A., Yan Z., Jeffers S.A., Holmes K.V., Hodges R.S., Burkhard P. Peptide nanoparticles as novel immunogens: design and analysis of a prototypic severe acute respiratory syndrome vaccine. Chem. Biol. Drug Des. 2009;73:53–61. doi: 10.1111/j.1747-0285.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G.N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Raman S., Machaidze G., Lustig A., Aebi U., Burkhard P. Structure-based design of peptides that self-assemble into regular polyhedral nanoparticles. Nanomedicine. 2006;2:95–102. doi: 10.1016/j.nano.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Rimm D.L., Morrow J.S. Molecular cloning of human E-cadherin suggests a novel subdivision of the cadherin superfamily. Biochem. Biophys. Res. Commun. 1994;200:1754–1761. doi: 10.1006/bbrc.1994.1656. [DOI] [PubMed] [Google Scholar]
- Schroeder U., Graff A., Buchmeier S., Rigler P., Silvan U., Tropel D., Jockusch B.M., Aebi U., Burkhard P., Schoenenberger C.A. Peptide nanoparticles serve as a powerful platform for the immunogenic display of poorly antigenic actin determinants. J. Mol. Biol. 2009;386:1368–1381. doi: 10.1016/j.jmb.2008.11.023. [DOI] [PubMed] [Google Scholar]
- Sodek J., Hodges R.S., Smillie L.B., Jurasek L. Amino-acid sequence of rabbit skeletal tropomyosin and its coiled-coil structure. Proc. Natl. Acad. Sci. U.S.A. 1972;69:3800–3804. doi: 10.1073/pnas.69.12.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetefeld J., Jenny M., Schulthess T., Landwehr R., Engel J., Kammerer R.A. Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat. Struct. Biol. 2000;7:772–776. doi: 10.1038/79006. [DOI] [PubMed] [Google Scholar]
- Stone D., Sodek J., Johnson P., Smillie L.B. Tropomyosin: correlation of amino acid sequence and structure. Proc. IX FEBS Meeting. 1975;31:125–136. [Google Scholar]
- Strelkov S., Herrmann H., Geisler N., Wedig T., Zimbelmann R., Aebi U., Burkhard P. Conserved segments 1A and 2B of the intermediate filament dimer: their atomic structures and role in filament assembly. EMBO J. 2002;21:1255–1266. doi: 10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell–cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Tarbouriech N., Curran J., Ruigrok R.W.H., Burmeister W.P. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 2000;7:777–781. doi: 10.1038/79013. [DOI] [PubMed] [Google Scholar]
- Tomschy A., Fauser C., Landwehr R., Engel J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- Walshaw J., Woolfson D.N. SOCKET: a program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 2001;307:1412–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- Whitby F.G., Phillips G.N. Crystal structure of tropomyosin at 7 Å resolution. Proteins. 2000;38:49–59. [PubMed] [Google Scholar]
- Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Wolf E., Kim P.S., Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Science. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Mochalkin I., Veerapandian L., Riley M., Doolittle R.F. Crystal structure of native chicken fibrinogen at 5.5 Å resolution. Proc. Natl. Acad. Sci. USA. 2000;97:3907–3912. doi: 10.1073/pnas.080065697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Kollman J.M., Pandi L., Doolittle R.F. Crystal structure of native chicken fibrinogen at 2.7 Å resolution. Biochemistry. 2001;40:12515–12523. doi: 10.1021/bi011394p. [DOI] [PubMed] [Google Scholar]
- Yun M., Bronner C.E., Park C.-G., Cha S.-S., Park H.-W., Endow S.A. Rotation of the stalk/neck and one head in a new crystal structure of the kinesin motor protein, Ncd. EMBO J. 2003;22:1–8. doi: 10.1093/emboj/cdg531. [DOI] [PMC free article] [PubMed] [Google Scholar]