Abstract
Serum biomarkers associated with Fasciola hepatica infection of Corriedale sheep were analysed during the first 12 weeks of infection using surface-enhanced laser desorption ionisation time of flight mass spectrometry (SELDI-TOF MS). In the discovery phase of analysis, pooled sera collected at week 0 and at each week p.i. to week 12 were fractionated by anion-exchange chromatography and the protein mass fingerprints obtained in individual fractions were in the M/z range 1.5–150 kDa. A total of 2302 protein clusters (peaks) were identified that varied between time-points following infection with peaks increasing or decreasing in intensity, or showing transient variation in intensity, during the 12 weeks of parasite challenge. In the validation phase, candidate biomarkers in sera of individual sheep at weeks 3 and 9 p.i. were analysed, identifying 100 protein peaks, many of which are small peptides <10 kDa in size: 54% of these peaks were up-regulated in intensity at week 3 or 9 p.i. Twenty-six biomarkers were chosen for further study, ranging in size from 1832 to 89,823 Da: six biomarkers were up-regulated at weeks 3 and 9 p.i., 16 biomarkers were up-regulated only at week 9 p.i. and four biomarkers were down-regulated at week 9 p.i. Two biomarkers up-regulated at week 9 were identified as transferrin (77.2 kDa) and Apolipoprotein A-IV (44.3 kDa), respectively. The results show that the interaction between the host and F. hepatica is complex, with changes in biomarker patterns beginning within 3 weeks of infection and either persisting to weeks 9–12 or showing transient changes during infection. Identification of biomarkers expressed during ovine fasciolosis may provide insights into mechanisms of pathogenesis and immunity to Fasciola and may assist in the rational development and delivery of vaccines.
Keywords: Fasciola hepatica, Liver fluke, Fasciolosis, Sheep, SELDI-TOF MS, Biomarkers, Serum, Transferrin, Apolipoprotein A-IV
1. Introduction
Fasciolosis is a disease affecting sheep, cattle and humans caused by Fasciola hepatica in temperate climates and Fasciola gigantica in the tropics. It is estimated that 250 million sheep, 350 million cattle and 180 million humans worldwide are at risk of infection, with production losses of over USD 3 billion per year (Hillyer and Apt, 1997, Mas-Coma et al., 1999, Mas-Coma et al., 2005, Spithill et al., 1999). Fasciola spp. parasites have complex interactions with their host. The initial phase of the disease includes parasite excystment in the small intestine and migration to the liver where subsequent movement and feeding in the liver parenchyma causes extensive tissue inflammation and necrosis. This acute phase of disease occurs during the first 5 weeks of infection in sheep and, at high worm burdens, can cause death. Sexually mature parasites migrate to the bile ducts of sheep at about 8–10 weeks after infection, where they feed on the blood and duct mucosa of the host. This chronic phase is characterised by liver fibrosis, anaemia and subsequent production losses.
Proteomic studies of Fasciola spp. have recently expanded from the analysis of subclasses of proteins such as cathepsins (Dalton et al., 2003) and glutathione S-transferases (Hillyer, 2005, Chemale et al., 2006), among others, to the identification of subsets of proteins of certain parasitic products such as ES products released by adult F. hepatica in vitro (Jefferies et al., 2001) and a comparison between ES products detected in vitro and in bile of infected sheep (Morphew et al., 2007) by two-dimensional gel electrophoresis.
Recent advances in mass spectrometry, namely the development of electrospray ionisation (ESI) and matrix-assisted laser desorption ionisation (MALDI), have extended our ability to unravel proteomes. Surface-enhanced laser desorption ionisation time of flight mass spectrometry (SELDI-TOF MS) allows sample binding to chemically active ProteinChip® surfaces, such as ion-exchange or immobilized metal affinity capture (IMAC) surfaces. As with MALDI, different matrices can be used to facilitate the uniform ionisation and desorption of molecules from the ProteinChip array surface (Merchant and Weinberger, 2000). SELDI technology allows the rapid and quantitative profiling of proteins in complex samples under different biological conditions and identification of markers of a biological state, termed ‘biomarkers’ (Xiao et al., 2005).
SELDI analyses were initially applied to the discovery of early diagnostic or prognostic biomarkers of cancer, such as prostate cancer (Petricoin et al., 2002b, Lehrer et al., 2003, Semmes et al., 2005), ovarian cancer (Petricoin et al., 2002a, Kozak et al., 2003, Zhang et al., 2004), pancreatic cancer (Rosty et al., 2002, Koopmann et al., 2004) and renal cancer (Won et al., 2003, Tolson et al., 2004; reviewed in Xiao et al., 2005). Recently, this technique has been applied to the study of serum biomarkers of infectious diseases such as Severe Acute Respiratory Syndrome (Poon et al., 2004, Yip et al., 2005), African Trypanosomiasis (Papadopoulos et al., 2004) and several blood borne protozoa (Ndao et al., unpublished data). Such studies have focused on identifying a distinctive configuration of circulating serum proteins that are indicative of a specific pathophysiological state, a so-called “proteomic fingerprint”.
Here, we believe we report the first proteomic study of biomarkers in serum of sheep infected with F. hepatica. The aim of the study was to establish the proteomic fingerprints in sheep serum at intervals during the first 12 weeks of infection with the goals of identifying diagnostic biomarkers for early parasite invasion and gaining insights into host–parasite interactions during establishment of infection and the transition from acute to chronic infection. As acquired resistance to Fasciola spp. is expressed during the first few weeks of infection, we were particularly interested to define biomarkers expressed within 5 weeks of infection since these could theoretically be involved in establishment or suppression of host immunity (Spithill et al., 1997, Piedrafita et al., 2004, Piedrafita et al., 2007). Our results highlight the complexity of the changes that occur in the sheep serum protein fingerprint during fasciolosis and reveal multiple biomarkers that are expressed during the acute and chronic phases of disease.
2. Materials and methods
2.1. Sheep serum samples
Serum samples were obtained from eight male Corriedale sheep (2 years old) housed in a paddock with an artificial water supply. The animals were purchased from a fluke-free area and shown to be free of infection by fecal analysis and ELISA using F. hepatica cathepsin L1 as a specific antigen (Piacenza et al., 1999). The animals were from the control group of a vaccination trial carried out in Uruguay where eight sheep were immunized with PBS in FCA, followed 4 weeks later by Freund’s incomplete adjuvant. After 2 weeks, each sheep was orally challenged with a gelatin capsule containing 300 metacercariae of F. hepatica and humanely slaughtered at 12 weeks p.i. The experimental protocol was performed in compliance with national regulations and approved and supervised by the Animal Experimentation Honorary Committee, University of the Republic, Uruguay. Flukes in the main bile ducts and gall bladder were removed. The worm burdens (sheep number) were: 18 (#12), 19 (#24), 20 (#16), 24 (#13), 49 (#22), 50 (#25), 61 (#20) and 70 (#30). Blood was collected from all sheep prior to infection and then weekly until the time of slaughter. The serum was obtained and then stored at −80 °C.
2.2. Serum fractionation
Sera were fractionated using a Ciphergen Q HyperD F strong anion-exchange resin filtration plate. The filtration plate was re-equilibrated by adding 200 μl of rehydration buffer (50 mM Tris–HCl, pH 9.0) and placed on a MicroMix 5 orbital vortex (Beckman Coulter) (form 20 and amplitude 7) for 60 min at room temperature (RT). The rehydration buffer was removed by vacuum and the resin was washed four times with 200 μl rehydration buffer and four times with 200 μl U1 solution (1 M urea, 0.2% 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM Tris–HCl, pH 9.0). Serum samples were thawed on ice and centrifuged at high speed (17,300g) for 5 min at RT to remove particulates. Twenty microlitres of sample were added to a v-bottom 96-well microplate (Costar Corning) with 30 μl of U9 buffer (9 M urea, 2% CHAPS, 50 mM Tris–HCl, pH 9). The microplate was sealed and placed on a MicroMix 5 orbital vortex (form 20, amplitude 5) for 20 min at RT. Fifty microlitres of sample were added to the equilibrated resin with 50 μl of U1 buffer. The filtration plate was sealed and placed on the MicroMix 5 orbital vortex (form 20, amplitude 7) for 30 min at RT. The fraction was collected by vacuum. One hundred microlitres of pH 9 buffer [50 mM Tris–HCl, 0.1% octyl β-d-glucopyranoside (OGP), pH 9] were added to the wells of the filtration plate using the Biomek robot automation system (Beckman Coulter), the microplate was placed on the MicroMix 5 orbital vortex at RT for 10 min and the fraction collected by vacuum. One hundred microlitres of the following buffers were added in two consecutive applications and collected by vacuum: pH 9 buffer, pH 7 buffer (50 mM 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), 0.1% OGP, pH 7), pH 5 buffer (100 mM sodium acetate, 0.1% OGP, pH 5), pH 4 buffer (100 mM sodium acetate, 0.1% OGP, pH 4), pH 3 buffer (50 mM sodium citrate, 0.1% OGP, pH 3), and organic wash buffer (33.3% isopropanol, 16.7% acetonitrile (ACN), 0.1% trifluoroacetic acid (TFA)). The two 100 μl eluants from each pH fraction were pooled, aliquoted and stored at −80 °C.
2.3. Binding of fractions to ProteinChip® arrays
The six pH fractions of serum were profiled on a weak cation-exchange (CM10) ProteinChip® Array according to the manufacturer’s instructions. All steps were performed at RT. Briefly, the ProteinChip® arrays were placed in a Ciphergen bioprocessor (C503-0006) and washed twice with 200 μl low-stringency binding buffer (0.1 M sodium acetate, 0.1% Triton X-100, pH 4) and placed on a multi-tube vortexer (VWR VX-2500) at speed 1 for 5 min. Each of the fractions was bound to the chip by adding 10 μl of sample in 90 μl of binding buffer; the bioprocessor was placed on the multi-tube vortexer for 60 min. The samples were discarded, the ProteinChip® arrays washed three times with 200 μl of binding buffer, placed on the multi-tube vortexer for 5 min and washed twice with 200 μl 1 mM HEPES pH 7.4 for 1 min. The ProteinChip® arrays were air-dried prior to matrix application. In the first set of analyses, samples were not spotted randomly on chips: the eight samples from one time-point were spotted on the same array. In the second analyses, the samples from different time-points were spotted randomly on ProteinChip® arrays.
2.4. Preparation and application of matrix
For peptide analysis (1.5–10 kDa), a 50% saturated solution of α-cyano-4-hydroxy-cinnamic acid (CHCA) was prepared by adding 200 μl of 50% ACN, 0.25% TFA to 5 mg of CHCA and placed on a tube vortex (Fisher 12–812) at a high setting for 5 min, left to stand for 5 min and centrifuged for 10 min at 17,300g. One hundred microlitres of CHCA solution was diluted to 50% saturation with 100 μl 50% ACN/0.25% TFA solution. For protein analysis (7–150 kDa), a saturated sinapinic acid (SPA) solution was similarly prepared by adding 50% ACN/0.5% TFA solution. Matrix (0.5 μl) was added to each spot on the ProteinChip array and air-dried prior to adding an additional 0.5 μl of matrix.
2.5. SELDI-TOF MS analysis
ProteinChip® arrays were read using a Ciphergen PBSIIc SELDI-TOF MS reader. Profiles were collected in the ranges of 1.5–10, 7–30 and 29–150 kDa. The intensity and sensitivity of the instrument were adjusted for each of these ranges on each day of analysis. The instrument was calibrated for dataset collection using an all-in-one peptide standard (Ciphergen Biosystems) when collecting data in the 1.5–10 kDa range and all-in-one protein standard (Ciphergen Biosystems) when collecting data in the 7–30 or 30–150 kDa ranges using the intensity and sensitivity of collection. Spectra from profiling experiments are an average of data from 110 laser shots.
2.6. Ciphergen Express software analysis
Spectra were normalised by total ion current intensity starting and ending at the M/z of the collection ranges (1.5–10, 7–30, 3–150 kDa). The spectra were aligned to a spectrum with the normalisation factor nearest 1.0. The spectra were only aligned if the percentage coefficient of variation was reduced after the alignment. Peaks from the different spectra were aligned using the cluster wizard function of Ciphergen Express 3.0.6 software. The peak detection was completely automated within the M/z range of analysis: peaks were automatically detected on the first pass when the signal-to-noise (S/N) ratio was 5, and the peak was five times the valley depth. User-detected peaks below threshold were deleted and all first-pass peaks were preserved. Clusters were created within 0.3% of M/z for each peak detected in the first pass. The clusters were completed by adding peaks with a S/N ratio of 2 and two times the valley depth. When no peaks were detected, the peak intensity was estimated at the centre of the cluster. In the discovery study, peaks were screened for potential biomarkers using the following criteria: difference in intensity must be observed in two or more consecutive weeks compared with the control sample and peaks must show at least a 2-fold increase in intensity (preferably a difference of presence/absence of intensity). The peaks were inspected to determine if they were multi-charged entities. In the validation study, P-values and receive operation characteristic (ROC) values were calculated by using the P-value wizard by comparing 2 weeks at a time. P-values below 0.05 were considered statistically significant.
2.7. Purification of 77.2 and 44.3 kDa biomarkers
The proteins from the pH 9 anion-exchange fraction in weeks 0 and 9 p.i. pooled samples were separated by stepwise hydrophobic fractionation at RT. RPC PolyBio beads (BioSepra) were washed three times with 10 times bead volumes of 80% ACN/0.1% TFA for 5 min on a rotational vortex (Barnstead/Thermolyne Lab Quake). Fifty microlitres of beads were equilibrated with 10% ACN/0.1% TFA for 5 min on the rotational mixer. The pH 9 anion-exchange fraction sample was adjusted to 10% ACN/0.1% TFA with a final volume of 500 μl, mixed using a rotational vortex for 5 min, added to the equilibrated RPC PolyBio beads and mixed for 30 min at RT. The tube was centrifuged for 1 min at 2300g and the supernatant removed by aspiration. Bound proteins were eluted successively using this method, adding 400 μl of 10%, 20%, 30%, 40%, 50% and 60% ACN in 0.1% TFA. Proteins in each fraction were profiled by adding 5 μl of hydrophobic fraction to a normal phase (NP20) array (Ciphergen) with SPA matrix and analysed with the PBSIIc reader. The 40% ACN/0.1% TFA sample and 60% ACN/0.1% TFA sample were concentrated using SpeedVac dehydration, reconstituted in NuPAGE® LDS sample buffer (4×) under denaturing reducing conditions and analysed on a 4–12% Bis–Tris NuPAGE gel using MOPS running buffer (Invitrogen) according to the manufacturer’s instructions. The gels were stained using Coomassie R250 and the 77.2 and 44.3 kDa bands excised from the 40% ACN/0.1% TFA and 60% ACN/0.1% TFA fractions, respectively, from week 9 p.i. lanes. Trypsin digestion and liquid chromatography (LC)-MS/MS analysis were carried out on an Agilent micro LC connected to an ABI Q-STAR mass spectrometer at the Sheldon Biotechnology Centre, McGill University. The resulting tryptic peptides were searched against both mammalian and other eukaryotic NCBI databases using MASCOT search engine (http://www.matrixscience.com/) for product ion confirmation.
2.8. Western blot analysis of biomarkers in sera
An aliquot (1 μl) of pooled sheep sera (diluted 1/25 in water) or 25 ng bovine apo-transferrin (Sigma) was prepared under denaturing reducing conditions and separated using a 4–12% Bis–Tris NuPAGE gel using MOPS running buffer (Invitrogen) as per the manufacturer’s instructions. The gel and 0.45 μm nitrocellulose membrane were equilibrated for 15 min with transfer buffer (48 mM Tris, 39 mM glycine, 20% v/v methanol, pH 9.2). The transfer conditions were 100 V for 1 h at 4 °C using a Mini Trans-blot cell (Bio-Rad). The membrane was blocked with 5% skim milk in 0.05% PBST (0.05% Tween 20 in PBS) for 1 h at RT. The membrane was incubated with 1/2000 horseradish-peroxidase conjugated sheep anti-bovine transferrin polyclonal antibody (Bethyl Laboratories) in 0.05% skim milk overnight at 4 °C. The membrane was washed five times with 0.05% PBST for 30 s, and four times with 0.05% PBST for 5 min. Two ml of WestPico luminescent substrate (Pierce) was added to the membrane for 5 min, blotted, and the membrane exposed to photographic film (Kodak). The same method was used for individual sera samples but a 1/10 dilution of the serum was used. Immunodetection of Apo A-IV was performed essentially as described above using 2 μl of pooled or individual serum. The membrane was washed with PBS three times for 5 min prior to incubation with 1/250 dilution of rabbit anti-human Apo A-IV antibody (Atlas antibodies) in PBS overnight at 4 °C. The membrane was washed three times for 5 min and incubated with horseradish peroxidase-conjugated sheep anti-rabbit IgG antibodies (Amersham) at 1/50,000 dilution for 1 h at RT for pooled samples and 1/25,000 in individual samples. The membrane was washed and developed as described above.
2.9. Densitometry analysis
The photographic film from the Western blot analysis was scanned using a Molecular imager FX (Bio-Rad) and the bands of interest were quantitated with Quantity One 4.4.1 software [Bio-Rad] using the densitometry function grey type. The density of each of the bands from the individual serum analysis (CNT/mm2) was compared using GraphPad Prism 4 software (GraphPad Software) using a one-tailed parametric t-test after testing the data for normality. A P-value of less than 0.05 was considered significant.
3. Results
3.1. Reproducibility study
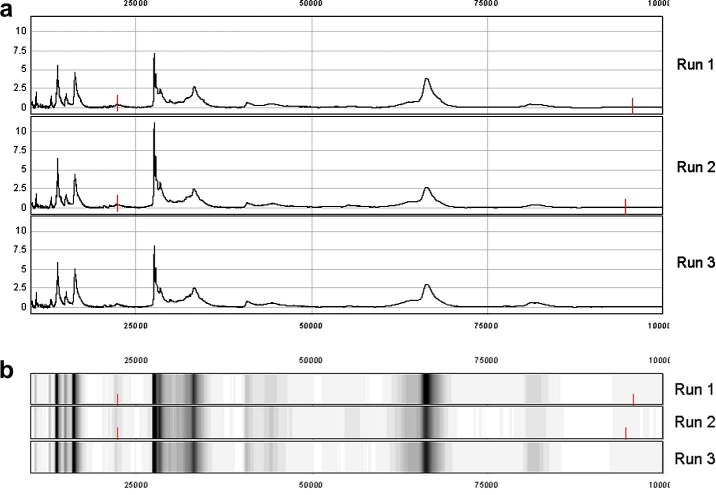
Aliquots of pooled sera from eight sheep collected at week 1 p.i. were independently fractionated three times. The organic wash fractions were each bound to CM10 ProteinChip® arrays at the same time and analysed by SELDI-TOF MS on the same day under identical conditions. Seventeen peaks were detected in the range of 10–100 kDa in experiments 1 and 2, and 15 peaks were detected in experiment 3. Fig. 1 shows the output both as a mass spectrum (Fig. 1a) and gel view (Fig. 1b). The variation between samples in the number of peaks detected in the 10–100 kDa range with a S/N ratio of at least 5 was 12% (2/17) and the mean variation in peak intensities was 13.54%. In a previous study by Semmes et al. (2005), the Coefficient of Variation for the S/N between institutions were 34–40% and peak intensity variation was found to vary from 15% to 36% for three individual peaks.
Fig. 1.
Reproducibility of SELDI-TOF MS analysis of sheep serum. A pool of sheep serum collected at week 1 p.i. was fractionated three times on consecutive days and then spotted and analysed on the same day by SELDI-TOF MS in the range 10–100 kDa. (a) Spectral view of the three protein profiles; (b) gel view of the same profiles. The tick marks indicate the peaks that were automatically detected in runs 1 and 2 but not run 3.
3.2. SELDI-TOF MS discovery study
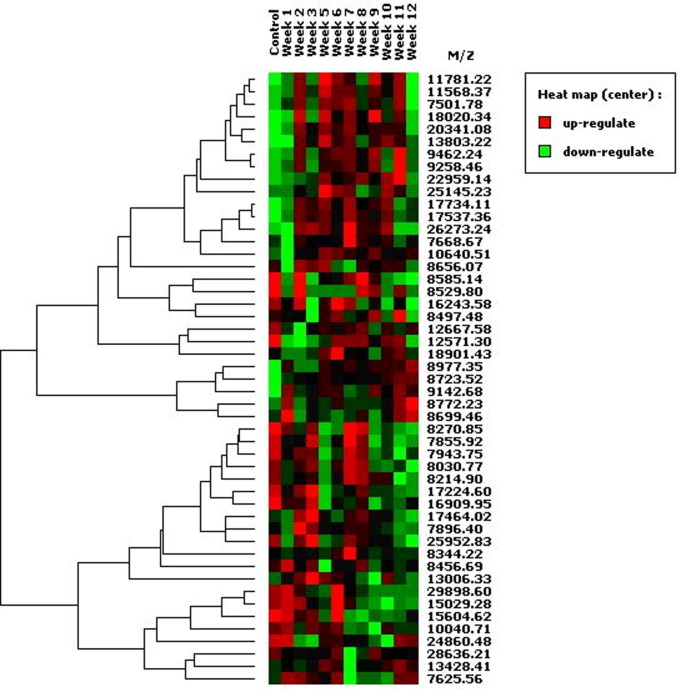
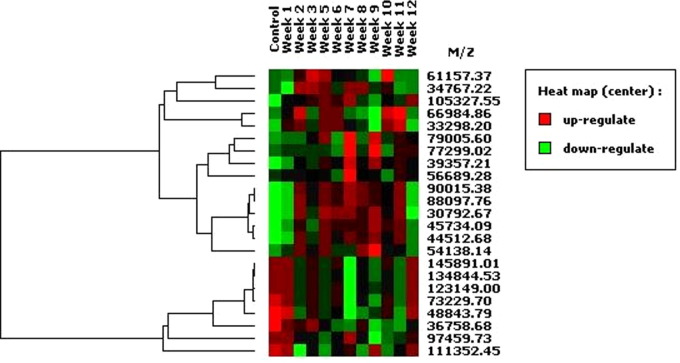
For the discovery phase, two independent fractionations of pooled sera from each time-point were performed to identify biomarkers whose patterns of expression were reproducible. Each of the anion-exchange fractions (pH 9, pH 7, pH 5, pH 4, pH 3 and organic wash) were analysed in the following calibration ranges: 1.5–10 kDa (CHCA), 7–30 kDa (SPA) and 30–150 kDa (SPA). From an analysis of all the fractions using pooled sera (data not shown) a total of 2302 clusters were identified by Ciphergen Express software (Table 1 ). Of these clusters, 1694 (74%) were in the range of 1.5–10 kDa, 301 (13%) in the range of 7–30 kDa and 307 (13%) in the range of 30–150 kDa. Broad patterns of variation in peak intensity are evident in the pooled sera with peaks either increasing (on) or decreasing (off) in intensity over the 12 weeks of the experiment. In addition, some peaks show transient variation in intensity (ie. on-off-on: cluster at 145,891–48,843 Da; or off-on-off: cluster at 11,781–8656 Da; Supplementary Fig. 1a, b) during the 12 weeks of infection. Peaks were scanned for differences in intensity that occurred in two or more consecutive weeks and which demonstrated more than a 2-fold difference in intensity or presence/absence compared with the control (week 0) sample: 46 markers were targeted for further validation at weeks 3 and 9 p.i., corresponding to the early stage of migration in the liver parenchyma (week 3 p.i.) and establishment in the bile ducts (weeks 7–9 p.i.).
Table 1.
Number of clusters detected by Ciphergen Express software in all six fractions within the three M/z ranges analysed
| 1.5–10 kDa | 7–30 kDa | 30–150 kDa | Total | |
|---|---|---|---|---|
| pH 9 | 203 | 80 | 81 | 364 |
| pH 7 | 178 | 65 | 44 | 287 |
| pH 5 | 563 | 29 | 38 | 630 |
| pH 4 | 440 | 23 | 33 | 496 |
| pH 3 | 0 | 14 | 21 | 35 |
| Organic wash | 310 | 90 | 90 | 490 |
| Total | 1694 | 301 | 307 | 2302 |
3.3. SELDI-TOF MS validation study
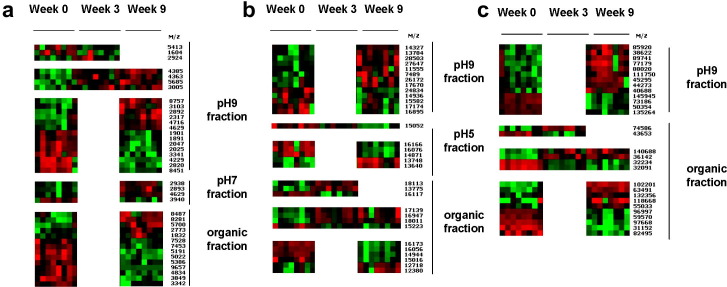
Sera from eight individual sheep collected at weeks 0, 3 and 9 p.i. were independently fractionated twice and analysed by SELDI- TOF MS. Fig. 2 a–c shows the heat maps of all 100 peaks that were found to show statistically significant differences in intensity (P < 0.05) in both experiments in the 1.5–10, 7–30 and 30–150 kDa ranges, respectively. The data were clustered to assist visualisation of patterns of variation between the different time-points. Four broad patterns of variation in peak intensity are evident: peaks that are up- or down-regulated early or late p.i., with 54% of the markers being up-regulated (Supplementary Table 1). Relative to week 0 sera, eight peaks were significantly different only at week 3 p.i., 79 peaks different only at week 9 p.i. and 13 peaks were different at weeks 3 and 9 p.i. in the two independent analyses. Note that the actual number of unique markers may be less than 100 since a mass spectrum usually contains more peaks than the number of different molecular species in the sample, due to the fact that molecules form complexes or carry multiple charges and appear as several peaks in the spectrum (Dijkstra et al., 2007). These statistically significant peaks are summarised in Supplementary Table 1. Many of the 100 markers are <10 kDa in size (Supplementary Table 1).
Fig. 2.
Heat maps of SELDI-TOF MS profiles of single sheep sera at week 0 and weeks 3 and 9 p.i. with Fasciola hepatica. Heat maps of all 100 statistically significant clusters from the pH 9, pH 7, pH 5 and organic wash fractions are shown. Biomarkers found to be statistically significant in two independent analyses were determined by comparison of individual spectra from weeks 0, 3 and 9 p.i. using Ciphergen Express software. At each time-point the data are presented in rank order of increasing worm burden (sheep 12, 24, 16, 13, 22, 25, 20 and 30). Data are shown in the range 1.5–10 kDa (a), 7–30 kDa (b) and 30–150 kDa (c). Heat maps are derived from analysis of randomly spotted samples. Peaks are clustered using dendograms (not shown) to reveal patterns of up-regulation (red) and down-regulation (green).
Twenty-six of the markers targeted in the discovery phase were statistically validated at weeks 3 and 9 p.i. and considered validated biomarkers: 13 biomarkers in the 1.5–10 kDa range (Fig. 3 a), five biomarkers in the 7–30 kDa range (Fig. 3b) and eight biomarkers in the 30–150 kDa range (Fig. 3c). Note that biomarkers at 3341/3342 Da and 4629 Da are found in two different fractions (pH 9/organic wash and pH 9/pH 7, respectively) and may represent duplications of the same biomarker: in this case, we have validated a total of 24 biomarkers. A summary of the P-value statistics and corresponding ROC values of these 26 biomarkers are included in Table 2 . ROC values represent the joint values of the true positive ratio (sensitivity) and the false positive ratio (1-specificity) for the relative intensity of the biomarker. ROC area values close to 1 or 0 indicate that the test has high sensitivity and specificity, whereas a value close to 0.5 indicates the test cannot reliably distinguish between positive and negative cases. Biomarkers with ROC area values nearing 1 are positively associated with infection (up-regulated), whereas values nearing 0 are negatively associated with infection (down-regulated). As shown in Table 2, six biomarkers were found to be significantly increased at weeks 3 and 9 p.i., 16 biomarkers were significantly increased only at week 9 p.i. and four biomarkers were significantly decreased only at week 9 p.i.
Fig. 3.
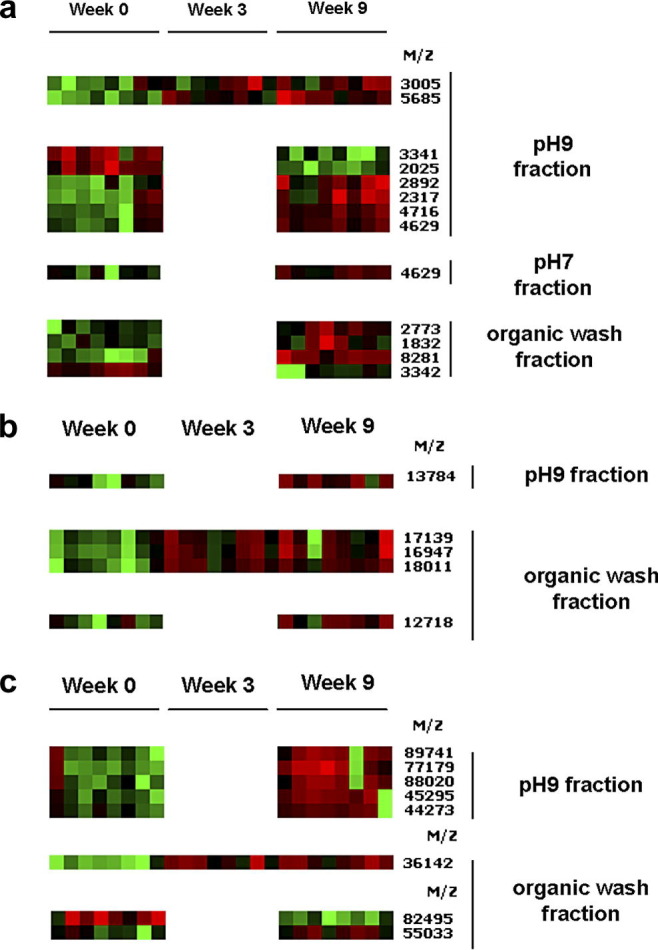
Heat maps of SELDI-TOF MS profiles of single sheep sera at week 0 and weeks 3 and 9 p.i. with Fasciola hepatica. At each time-point the data are presented in rank order of increasing worm burden (sheep 12, 24, 16, 13, 22, 20 and 30). Heat maps show the validated biomarkers from the pH 9, pH 7 and organic wash fractions in the range 1.5–10 kDa (a), 7–30 kDa (b) and 30–150 kDa (c). Peaks are clustered using dendograms to reveal patterns of up-regulation (red) and down-regulation (green).
Table 2.
Summary of the 26 validated biomarkers identified in two separate fractionations of sheep sera ordered by M/z from both the first analysis (non-randomized spotting) and second analysis (randomized spotting) of individual serum fractions
| Fraction | M/z | 1st run |
2nd run |
||
|---|---|---|---|---|---|
| P-value | ROC | P-value | ROC | ||
| Week 3 | |||||
| pH 9 | 3005.7∗ | 0.039 | 0.797 | 0.019 | 0.750 |
| pH 9 | 5684.9∗ | 0.013 | 0.984 | 0.013 | 0.938 |
| Organic | 16,946.5∗ | 0.013 | 0.984 | 0.019 | 0.938 |
| Organic | 17,138.4∗ | 0.013 | 0.984 | 0.019 | 0.891 |
| Organic | 18,011.0∗ | 0.013 | 0.984 | 0.019 | 0.891 |
| Organic | 36,144.5∗ | 0.019 | 0.797 | 0.013 | 0.984 |
| Week 9 | |||||
| Organic | 1832.4 | 0.013 | 0.984 | 0.013 | 0.703 |
| pH 9 | 2025.2 | 0.019 | 0.063 | 0.013 | 0.016 |
| pH 9 | 2317.7 | 0.013 | 0.938 | 0.013 | 0.844 |
| Organic | 2773.0 | 0.013 | 0.938 | 0.019 | 0.938 |
| pH 9 | 2892.9 | 0.013 | 0.891 | 0.013 | 0.891 |
| pH 9 | 3005.9∗ | 0.013 | 0.891 | 0.013 | 0.891 |
| Organic | 3342.9 | 0.013 | 0.063 | 0.013 | 0.016 |
| pH 9 | 3341.2 | 0.013 | 0.063 | 0.013 | 0.063 |
| pH 9 | 4629.4 | 0.013 | 0.938 | 0.013 | 0.938 |
| pH 7 | 4629.3 | 0.027 | 0.891 | 0.027 | 0.844 |
| pH 9 | 4716.2 | 0.013 | 0.891 | 0.013 | 0.891 |
| pH 9 | 5685.6∗ | 0.013 | 0.984 | 0.013 | 0.984 |
| Organic | 8281.5 | 0.013 | 0.984 | 0.013 | 0.984 |
| Organic | 12,718.6 | 0.013 | 0.938 | 0.013 | 0.891 |
| pH 9 | 13,784.7 | 0.013 | 0.844 | 0.019 | 0.891 |
| Organic | 16,948.7∗ | 0.013 | 0.984 | 0.019 | 0.844 |
| Organic | 17,144.2∗ | 0.013 | 0.984 | 0.027 | 0.844 |
| Organic | 18,012.8∗ | 0.013 | 0.984 | 0.013 | 0.984 |
| Organic | 36,150.3∗ | 0.013 | 0.984 | 0.013 | 0.984 |
| pH 9 | 44,273.9 | 0.039 | 0.844 | 0.019 | 0.891 |
| pH 9 | 45,295.6 | 0.039 | 0.844 | 0.013 | 0.938 |
| Organic | 55,033.6 | 0.019 | 0.938 | 0.027 | 0.844 |
| pH 9 | 77,179.1 | 0.019 | 0.891 | 0.027 | 0.844 |
| Organic | 82,495.9 | 0.019 | 0.063 | 0.013 | 0.016 |
| pH 9 | 88,020.3 | 0.027 | 0.891 | 0.039 | 0.844 |
| pH 9 | 89,741.3 | 0.027 | 0.859 | 0.039 | 0.844 |
P-values < 0.05 are considered statistically significant. Biomarkers with ROC values above 0.5 are termed up-regulated biomarkers and biomarkers with ROC values below 0.5 are termed down-regulated biomarkers. Week 3 and week 9 biomarkers are shown. Peaks marked with ∗ indicate that the peak is present at both weeks 3 and 9 as their M/z are within the error of the instrument (0.1%).
3.4. Biomarker screening and validation
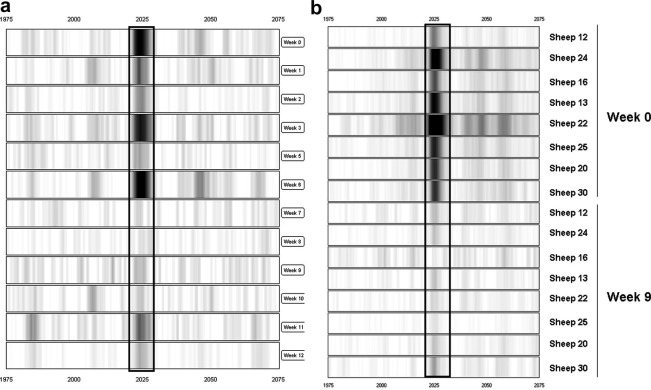
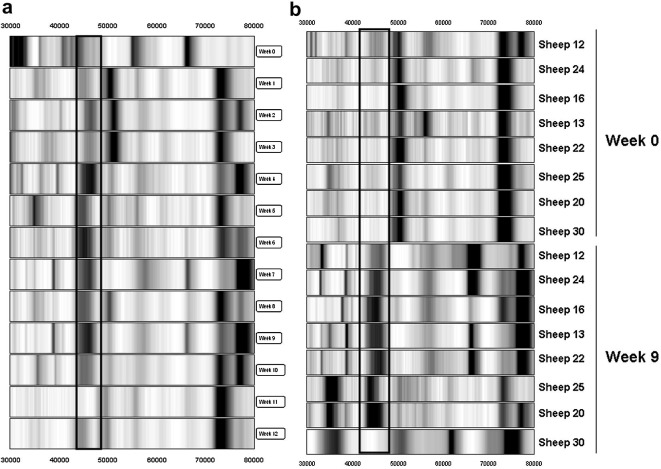
The 2025 Da biomarker represents a down-regulated biomarker which demonstrates a decrease in intensity in pooled sera from approximately week 7 p.i. relative to week 0 p.i. sera (Fig. 4 a). The intensity was significantly lowered (6.6-fold) at week 9 p.i., based on relative intensity data from individual sheep sera (Fig. 4b). The mean relative intensity decreased from 0.96 at week 0 p.i. to 0.14 at week 9 p.i.
Fig. 4.
Gel view of SELDI-TOF MS profiles of the 2025 Da biomarker in sheep sera at different times p.i. with Fasciola hepatica. (a) 2025 Da biomarker in pooled fractionated samples at weeks 0 through 12 p.i. (relative intensity scale of 1/−0.1). (b) 2025 Da biomarker in individual sheep at weeks 0 and 9 p.i. (relative intensity scale of 1/−0.1). At each time-point the results are presented in rank order of increasing worm burden (18–70 worms). The biomarker is boxed.
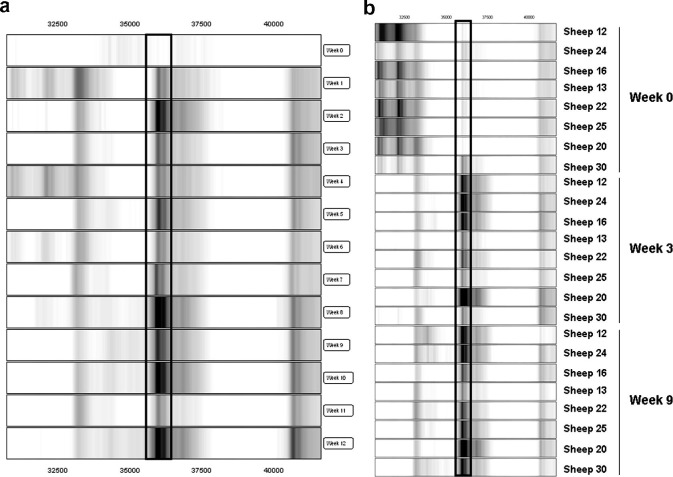
The 36.2 kDa biomarker exemplifies a biomarker with an early and persistent up-regulation of intensity in pooled sera that begins at week 1 p.i. (Fig. 5 a). The relative intensity of this biomarker in individual sheep sera was significantly up-regulated 3.4-fold at week 3 p.i. and 3.6-fold at week 9 p.i. (Table 2), based on relative intensity data from individual sheep sera (Fig. 5b). The mean relative intensity increases from 0.49 at week 0 p.i. to 1.29 at week 3 p.i. and 1.51 at week 9 p.i.
Fig. 5.
Gel view of SELDI-TOF MS profiles of the 36.2 kDa biomarker in sheep sera at different times p.i. with Fasciola hepatica. (a) 36.2 kDa biomarker in pooled fractionated samples at weeks 0 through 12 (relative intensity scale of 3/−0.1). (b) 36.2 kDa biomarker in individual sheep at week 0 and weeks 3 and 9 p.i. (relative intensity scale of 5/−0.1). At each time-point the results are presented in rank order of increasing worm burden (18–70 worms). The biomarker is boxed.
The 44.3 kDa biomarker is an up-regulated biomarker that showed a transient increase in relative intensity in pooled sera from about weeks 4 to 9 p.i. (Fig. 6 a) and was validated at week 9 p.i. (Table 2). From an analysis of individual sheep sera, the relative intensity of this biomarker was significantly elevated (3.8-fold) at week 9 p.i. (Fig. 6b). The mean relative intensity increases from 0.13 at week 0 p.i. to 0.49 at week 9 p.i.
Fig. 6.
Gel view of SELDI-TOF MS profiles of the 44.3 kDa biomarker in sheep sera at different times p.i. with Fasciola hepatica. (a) 44.3 kDa biomarker in pooled fractionated samples at weeks 0 through 12 p.i. (relative intensity scale of 1/−0.1). (b) 44.3 kDa biomarker in individual sheep at weeks 0 and 9 p.i. (relative intensity scale of 1/−0.1). At each time-point the results are presented in rank order of increasing worm burden (18–70 worms). The biomarker is boxed.
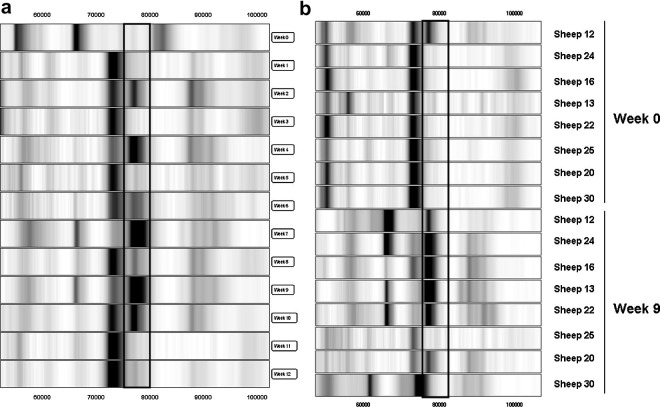
The 77.2 kDa biomarker is an up-regulated biomarker that was observed to increase in relative intensity in pooled sera during infection (Fig. 7 a). From an analysis of individual sheep sera, the relative intensity of the biomarker was significantly increased (5.3-fold) at week 9 p.i. compared with week 0 sera although there was variation in marker intensity among the individual sheep (Fig. 7b). The mean relative intensity increased from 0.23 at week 0 p.i. to 1.21 at week 9 p.i.
Fig. 7.
Gel view of SELDI-TOF MS profiles of the 77.2 kDa biomarker in sheep sera at different times p.i. with Fasciola hepatica. (a) 77.2 kDa biomarker in pooled fractionated samples at weeks 0 through 12 p.i. (relative intensity scale of 1/−0.1). (b) 77.2 kDa biomarker in individual sheep at weeks 0 and 9 p.i. At each time-point the results are presented in rank order of increasing worm burden (18–70 worms). The biomarker is boxed.
3.5. Identification of biomarkers
The 77.2 kDa biomarker was further purified from the pH 9 fraction and its tryptic peptides analysed by LC–MS/MS. One hundred and ten peptides matched to bovine transferrin (GenBank Accession No. Q29443, M r 79,870 Da) with percentage coverage of 32% and a MOWSE (molecular weight search; http://www.matrixscience.com/help/scoring_help.html#MOW) score of 1164. Sixteen peptides matched the incomplete sheep transferrin sequence with a MOWSE score of 109. The 44.3 kDa biomarker was similarly purified and its tryptic peptides analysed by LC–MS/MS. Twenty-nine peptides matched to a protein annotated as a hypothetical cattle protein (GenBank Accession No. NP_001032557, M r 42,991 Da). By BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/), this protein shows 77% identity and 86% similarity to human Apolipoprotein A-IV (data not shown). The percentage coverage of the hypothetical bovine protein was 46% with a MOWSE score of 1172.
3.6. Validation of transferrin and Apo A-IV biomarkers by Western blot
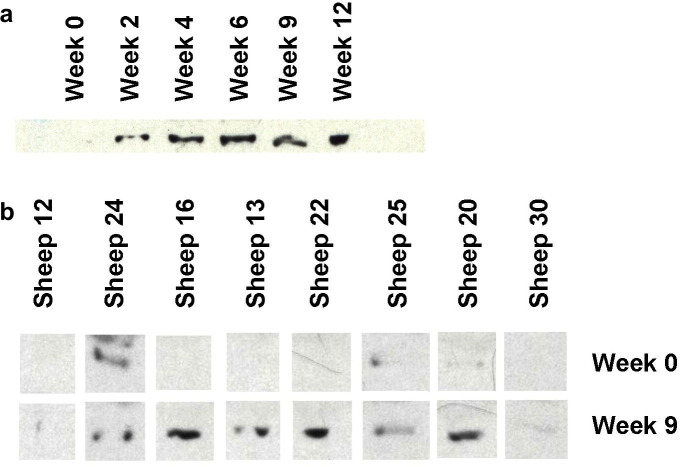
In Western blot analysis with antisera to bovine transferrin, native bovine apo-transferrin appears as a doublet at about 78 and 72 kDa (Fig. 8 a) (Tsuji et al., 1984). The band observed in the pooled sheep samples corresponds to approximately 77 kDa and demonstrates an increase in intensity at week 9 p.i. (Fig. 8a). Analysis of individual sheep serum samples at weeks 0 and 9 p.i. showed that the level of transferrin varied between animals at both time-points. Six sheep had an elevated level of serum transferrin by Western blotting (sheep #24, 16, 13, 22, 20 and 30) whereas no apparent change was observed in the sera of sheep 12 and 25 (Fig. 8b). The same six sheep also showed an apparent increase in serum transferrin peak intensity at week 9 by SELDI-TOF MS analysis (Fig. 3, Fig. 7b). By densitometry, the intensity of the Western blot bands at 77 kDa for week 9 p.i. samples from the paired individual sera were found to be significantly increased (P < 0.05) compared with week 0 p.i.,using a one-tail paired parametric t-test (data not shown).
Fig. 8.
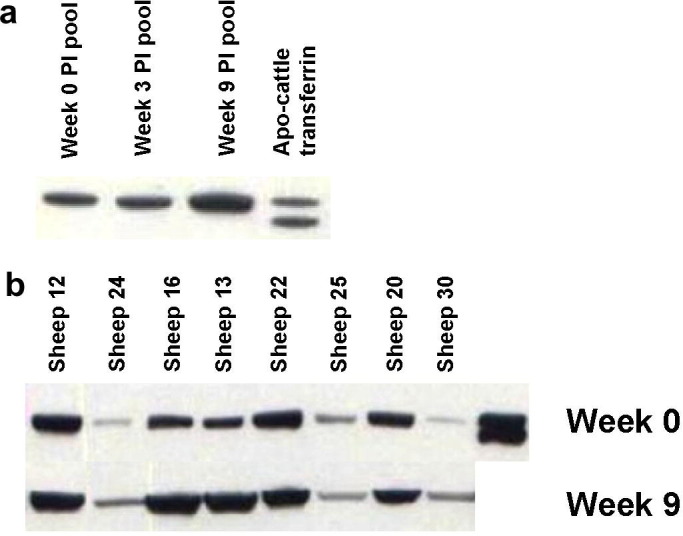
Western blot analysis of transferrin levels in sheep. (a) Western blot using anti-bovine transferrin of pooled sera from weeks 0, 3 and 9 p.i. with cattle apo-transferrin as a positive control. (b) Western blot of 1/10 dilution of individual sera from weeks 0 and 9 p.i. Data is presented in rank order of increasing worm numbers. The worm burdens (sheep number) were: 18 (#12), 19 (#24), 20 (#16), 24 (#13), 49 (#22), 50 (#25), 61 (#20) and 70 (#30).
Western blot analysis using antisera to human Apo A-IV detected a band at approximately 44 kDa; the intensity of this 44 kDa band increased at weeks 2, 4, 6, 9 and 12 p.i. (Fig. 9 a). Western blotting of sera from individual sheep showed that the intensity of the 44 kDa Apo A-IV band was elevated in sheep #13, 16, 20, 22 and 25 (Fig. 9b). By densitometry, the intensity of the bands at 44 kDa of week 9 p.i. samples from the paired individual sera were found to be significantly increased (P = 0.019) compared with week 0 p.i., using a one-tail parametric t-test (data not shown).
Fig. 9.
Western blot analysis of Apo A-IV levels in sheep. (a) Western blot using anti-human Apo A-IV of 2 μl of pooled sera from weeks 0, 2, 4, 6, 9 and 12 p.i. with 1/50,000 secondary antibody. (b) Western blot of 2 μl of individual sera from weeks 0 and 9 p.i. with 1/25,000 secondary antibody. Data is presented in rank order of increasing worm numbers.
4. Discussion
In this study, we believe we report for the first time an analysis of biomarkers in serial collections of sheep serum during the first 12 weeks of F. hepatica infection. The analysis of pooled sera provided a broad snapshot of the variation in peak intensity during infection, with peaks increasing or decreasing in intensity, or showing transient variation in intensity, during the 12 weeks of parasite challenge. It is clear that dramatic changes occur in the sheep serum proteome during fasciolosis with at least 2302 clusters shown to vary in peak intensity during infection. The two particularly informative time-points identified for further study (weeks 3 and 9 p.i.) corresponded to the migration of the parasite in the liver parenchyma and establishment in the bile ducts, respectively. From the 2302 clusters identified, 100 peaks (4.3%) in the size range 1604–145,945 Da showed significant differences in intensity in individual sera at weeks 3 and/or 9 p.i. relative to week 0 sera, with a similar number of peaks being up-regulated or down-regulated. Seventy-nine peaks showed differences in intensity only at week 9 p.i. (Fig. 2; Supplementary Table 1). This bias of peaks towards later time-points may reflect the impact on the serum proteome of the larger adult parasite biomass at week 9 p.i. relative to the much smaller biomass of immature parasites at week 3 p.i.
The main bottle-neck of SELDI-TOF MS profiling technology is the purification and identification of individual proteins. In order to concentrate our resources on biomarkers that are most likely to be biologically significant, a two-step approach was used. This study had the benefit of having serial bleeds from eight sheep during the course of infection so that the inherent sheep-to-sheep variation in serum protein levels was theoretically minimised. The initial discovery phase of the study allowed us to target peaks that would be of biological relevance because they appeared in pooled sera over several consecutive weeks of infection. The second step was to statistically validate these differences. Using the conditions that had been determined in the discovery phase, the individual sheep sera at weeks 0, 3 and 9 p.i. were fractionated and analysed. This validated the biomarkers that differed significantly between time-points and verified that the differences observed in the discovery phase were not attributable to large variations in sera from a few sheep in the group. A second independent fractionation and randomized binding experiment confirmed these results and also controlled for day-to-day differences in manipulations using the SELDI-TOF MS platform. The biomarkers were only considered to be validated if the cluster was calculated as statistically significant in both data sets.
From the 26 biomarkers chosen for study, the validation results showed that 22 biomarkers increased in intensity and four biomarkers decreased in intensity over the first 9 weeks of infection. These biomarkers cover a wide size range (1832–89,741 Da) with many biomarkers being less than 10 kDa, suggesting that small peptides are a rich source of relevant biomarkers using the SELDI-TOF MS approach as noted earlier in human sera (Hortin, 2006). These results show that the interaction between the host and F. hepatica is complex, with significant changes in biomarker patterns beginning within 3 weeks of infection, as observed with the 36.2 kDa biomarker. The analysis of pooled sera suggests that biomarker patterns can change within 2 weeks of infection (see data for the pH 9 fraction, Supplementary Fig. 1). This complexity reflects the nature of the migration path of the parasite from the gut, through the intestinal wall, peritoneum, liver parenchyma and bile duct wall, with final residence within the bile ducts. We would anticipate different biomarkers to be generated in different host tissues or produced by different developmental stages of the parasite.
Purification of several of the 26 biomarkers was attempted. Given the limited sequence database information for sheep and F. hepatica, biomarker identities derived from MS/MS analysis were confirmed using antibody reagents against specific proteins. One biomarker at 77.2 kDa was identified as transferrin which increased in intensity during the biliary phase of infection at week 9 p.i. The increase in the 77.2 kDa transferrin marker at week 9 p.i. detected by SELDI-TOF MS analysis, from being almost absent at week 0 p.i., is not reflected in the Western blot results where a more modest change was observed. This weak signal at week 0 p.i. in the SELDI-TOF MS spectra appears to be an artefact attributed to peak suppression by the neighbouring 73.2 kDa peak, the double-charge peak of IgG (146 kDa) (Fig. 7). As observed in Supplementary Table 1, the 146 and 73.2 kDa peaks decrease at week 9 p.i. (ROC = 0.016, ROC = 0.063/0.016, respectively). Suppression of the 77.2 kDa signal at week 0 p.i. in the SELDI analysis may account for the less marked difference in marker intensity observed in the transferrin Western blot analysis. Peak suppression in SELDI-TOF MS analysis by blood components has been previously observed (van Breemen et al., 2006, Roche et al., 2006) and is a factor that must be carefully considered in interpretation of SELDI-TOF MS data.
Transferrin is a negative acute phase protein (McNair et al., 1998), hypothesised to decrease the bioavailability of iron to micro-organisms, whereas we observed an increase in transferrin levels in our study. Fasciolosis, however, is characterised by both an acute (peritoneal and parenchymal phase) and a chronic (biliary) phase of disease. In a study of F. hepatica in sheep, with similar worm burdens to those present in our study (10–62 flukes/sheep), a loss of 9.3–23.6 ml of plasma per day was estimated and the onset of anaemia was associated with the entry of the flukes into the biliary system, which occurs at approximately week 6–7 p.i. (Sinclair, 1972). Accelerated erythropoiesis occurs in response to the developing anaemia during fasciolosis (Sinclair, 1972) and a significant negative correlation has previously been observed between the concentration of haemoglobin and serum transferrin in calves (Martinsson and Mollerberg, 1973). Transferrin levels have also been shown to increase during iron deficiency in calves (Moser et al., 1994). Thus, an increase in serum transferrin levels in response to induced anaemia in sheep during fasciolosis is consistent with these observations. The variation in relative transferrin levels between individual sheep at weeks 0 and 9 p.i. is consistent with data in cattle where transferrin concentrations have been shown to vary over a 3- to 6-fold range under different physiological conditions but the basis for this variation is not clear (Moser et al., 1994, McNair et al., 1998).
The biomarker at 44.3 kDa has been identified as Apolipoprotein A-IV, based on MS/MS analysis. The increase in intensity of the Apo A-IV marker by SELDI-TOF MS analysis was confirmed by Western blot analysis of pooled sera where an increase in Apo A-IV levels was observed at selected time-points from week 2–12 p.i. Apo A-IV is a 46 kDa glycoprotein and one of the minor apolipoproteins in triglyceride-rich apo B-containing lipoproteins (chylomicron, very low density proteins [VLDL]) and in Apo A-I containing lipoproteins (HDL) in blood: it is synthesised in the small intestine in humans and the liver and small intestine in rodents (Stan et al., 2003, Takahashi et al., 2004). In cattle, Apo A-IV levels vary during lactation and decrease during fasting (Takahashi et al., 2004); variations in the intralumenal availability of bile acids may affect the regulation of Apo A-IV synthesis (Stan et al., 2003). Apo A-IV may play a role in the control of food consumption and gastric acid secretion, as well as protection against lipoprotein oxidation (Stan et al., 2003). The regulation of Apo A-IV levels is complex and the basis for the apparent up-regulation of serum Apo A-IV during fasciolosis remains to be determined but may be related to pathological changes in the liver and bile ducts accompanying fluke infection.
While this manuscript was in preparation, a study was published identifying four biomarkers of F. hepatica infection in sheep bile which varied in intensity between control and infected bile: chain A of the trypsin inhibitor complex (up-regulated), regucalcin (down-regulated), enolase 1 (down-regulated) and transferrin (down-regulated) (Morphew et al., 2007) (Supplementary Table 2). This apparent discrepancy between the serum transferrin levels observed in our study and bile transferrin levels may be related to iron homeostasis during fasciolosis. The host may alter transferrin distribution in bile to enhance iron uptake from plasma. Of the other three host bile biomarkers, we observed serum biomarkers which may correspond to chain A of the trypsin inhibitor complex (26,169 Da) and regucalcin (31,152, 32,091, 32,234 Da) since their pattern of expression, apparent M r and elution profile correspond with the expression, M r and pI of the bile proteins (Supplementary Table 2). Work is in progress to confirm these observations. No serum biomarker corresponded to Enolase 1. We did not detect a serum biomarker corresponding to the main up-regulated parasite biomarker in bile, isotypes of F. hepatica cathepsin L (observed M r 23,588–24,497 Da, pI 4.87–6.50) (Morphew et al., 2007). This may be related to the relatively low parasite burden in the sheep used in this study.
Increased aspartate aminotransferase (AST) (E.C. 2.6.1.1) and glutamate dehydrogenase (GLDH) (EC 1.4.1.2) activity in plasma is associated with the liver damage caused by fasciolosis and appear in serum at weeks 4–6 p.i. (Sexton et al., 1990). Sheep AST exists as two isoforms of 87.1 kDa (pI of 9.14) (Orlacchio et al., 1979) and 86–88.9 kDa (Campos-Cavieres and Munn, 1973). The up-regulated biomarkers at 88.0 and 89.7 kDa in the pH 9 fraction observed in week 9 sera may represent AST (Fig. 2, Table 2). Bovine GLDH is 55.4 kDa (pI 6.77) (Moon and Smith, 1973) and a 55.0 kDa up-regulated biomarker was observed in the organic wash fraction in week 9 p.i. sera (Fig. 2, Table 2). Gamma-glutamyl transferase (GGT) (EC 2.3.2.2) is a 92 kDa serum biomarker of bile duct damage in sheep (Zelazo and Orlowski, 1976, Ferre et al., 1994) but we did not observe a biomarker of this size in serum. Work is in progress to confirm these identities.
We were interested in identifying a biomarker that could be diagnostic of early infection and whether the intensity of any biomarker was correlated with worm burden, such that a biomarker may act as a marker of intensity of infection. We identified 13 biomarkers significantly up-regulated and another seven biomarkers whose intensity was significantly down-regulated, at week 3 p.i. (Supplementary Table 1). Several markers were up-regulated in all sheep at week 3, e.g. markers at 13,775 Da, 18,113 Da (Fig. 2b, organic fraction) and 74,586 Da (Fig. 2c, organic fraction). Down-regulated markers were observed at 32,091 Da and 32,234 Da (Fig. 2c, organic fraction). Although the intensity of these biomarkers changes early p.i., further work is required to determine if these biomarkers are specific for fasciolosis. This will require comparative analysis of sera from other parasite infections of sheep and we are attempting such studies. Other changes were observed in all sheep at week 9 such as up-regulated markers at 63,491 Da, 102,201 Da (Fig. 2c, organic fraction) and down-regulated markers at 5022 Da (Fig. 2a, organic fraction), 31,152 Da, 32,091 Da and 97,688 Da (Fig. 2c, organic fraction). A regression analysis comparing biomarker intensity and worm burdens was performed on all 100 biomarkers but no significant correlations were observed in either data set (data not shown). This may reflect the small sample size in our study as well as the relatively low worm burden observed. Biomarkers of intensity of fluke infection may be more easily identified in sera from sheep carrying a larger worm burden since the impact of such an infection on the serum proteome would be more dramatic.
Only one published study has applied SELDI-TOF MS to serum biomarker discovery in parasitic infections. This study described a serum biomarker pattern associated with human African trypanosome infections but none of the putative biomarkers were identified (Papadopoulos et al., 2004). Recent work has also applied this technology to profile human sera from Chagas disease patients to identify five biomarkers that are diagnostic for this disease (Ndao et al., unpublished data). In this case several proteolytic fragments of proteins were found to be useful in various diagnostic algorithms for Trypanosoma cruzi infection, suggesting that analysis of peptides derived from intact proteins will prove to be a rich source of diagnostic information. It is possible that some biomarkers we observed, particularly those smaller than 10 kDa, may represent fragments of larger proteins. These results demonstrate that protein profiling of parasite infection sera may reveal new insights into how parasites and hosts interact.
Future work will involve the further identification of the biomarkers validated herein to gain a fuller understanding of the effect of fasciolosis on the serum proteome. Identification of Fasciola and sheep biomarkers has proven to be difficult, mainly because of the low levels of many markers (which complicates purification), the sparseness of completely annotated Fasciola and sheep genome sequences and the lack of homologous antibody reagents. A sheep genome project is in progress (http://www.sheephapmap.org/) and cDNAs from F. hepatica are being generated (http://www.sanger.ac.uk/Projects/Helminths/). A focus on the less abundant proteins present in the sheep serum proteome is possible by depletion of the higher abundance proteins using the human ProteomeLab IgY-12 depletion system (Beckman Coulter; Roche et al., 2006) which is also effective at binding abundant sheep proteins (Rioux et al., unpublished data). This may allow us to further identify proteins involved in pathology and host immune responsiveness, such as biomarkers of acquired resistance to Fasciola or biomarkers associated with immunosuppression, which is a feature of fasciolosis (reviewed in Piedrafita et al., 2004). It is notable that certain sheep breeds can acquire resistance to F. gigantica but not F. hepatica. However, the precise nature of the responses determining acquired resistance to fluke infection is not known (Piedrafita et al., 2004). Indonesian sheep acquire resistance to F. gigantica within 2–4 weeks of infection and recent results suggest that a superoxide-mediated IgG-dependent effector response mediated by monocytes/macrophages and eosinophils from these sheep is effective at killing juvenile F. gigantica in vitro (Piedrafita et al., 2007). A biomarker analysis of Indonesian sheep sera within 4 weeks of infection may reveal peptides/proteins whose expression correlates with acquired resistance, such as proteins involved in macrophage/eosinophil activation. The identification of such functional biomarkers will provide insights into host resistance pathways which may assist the design of rational delivery systems to improve the efficacy of vaccines for fasciolosis (Hillyer, 2005).
Acknowledgements
C. Carmona and D. Acosta were supported by CSIC, UdelaR, Uruguay. M.-C. Rioux was the recipient of a travel award from the Centre for Host–Parasite Interactions to attend a training course at Ciphergen, UK. This work was supported by operating funds from the Natural Sciences and Engineering Research Council (Canada), the Canada Research Chair program, the Canadian Institutes of Health Research (#200402UOP-UI-130124-B and #200401NTA-126473-DAI-CFAC-41490), as well as infrastructure funds from the Canadian Foundation for Innovation, McGill University and the FQRNT Center for Host–Parasite Interactions. T. Spithill is a recipient of a Canada Research Chair in Immunoparasitology.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ijpara.2007.07.017.
Appendix A. Supplementary data
Supplementary Figure 1.
Heat maps of SELDI-TOF profiles of pooled sheep sera from weeks 0 to 12 p.i. with Fasciola hepatica. Heat maps show all clusters detected from the pH 9 fraction in the range 7–30 kDa (A) and 30–150 kDa (B). Peaks are clustered using dendograms to reveal patterns of up-regulation (red) and down-regulation (green).
Supplementary Figure 2.
References
- Campos-Cavieres M., Munn E.A. Purification and some properties of cytoplasmic aspartate aminotransferase from sheep liver. Biochem. J. 1973;135:683–693. doi: 10.1042/bj1350683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemale G., Morphew R., Moxon J.V., Morassuti A.L., Lacourse E.J., Barrett J., Johnston D.A., Brophy P.M. Proteomic analysis of glutathione transferases from the liver fluke parasite, Fasciola hepatica. Proteomics. 2006;6:6263–6273. doi: 10.1002/pmic.200600499. [DOI] [PubMed] [Google Scholar]
- Dalton J.P., Neill S.O., Stack C., Collins P., Walshe A., Sekiya M., Doyle S., Mulcahy G., Hoyle D., Khaznadji E., Moire N., Brennan G., Mousley A., Kreshchenko N., Maule A.G., Donnelly S.M. Fasciola hepatica cathepsin L-like proteases: biology, function, and potential in the development of first generation liver fluke vaccines. Int. J. Parasitol. 2003;33:1173–1181. doi: 10.1016/s0020-7519(03)00171-1. [DOI] [PubMed] [Google Scholar]
- Dijkstra M., Vonk R.J., Jansen R.C. SELDI-TOF mass spectra: a view on sources of variation. J. Chromatogr. B. 2007;847:12–23. doi: 10.1016/j.jchromb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Ferre I., Barrio J.P., Gonzalez-Gallego J., Rojo-Vazquez F.A. Appetite depression in sheep experimentally infected with Fasciola hepatica L. Vet. Parasitol. 1994;55:71–79. doi: 10.1016/0304-4017(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Hillyer G.V., Apt G. Food-borne trematode infections in the Americas. Parasitol. Today. 1997;13:87–88. [Google Scholar]
- Hillyer G.V. Fasciola antigens as vaccines against fascioliasis and schistosomiasis. J. Helminthol. 2005;79:241–247. doi: 10.1079/joh2005304. [DOI] [PubMed] [Google Scholar]
- Hortin G.L. The MALDI-TOF mass spectrometric view of the plasma proteome and peptidome. Clin. Chem. 2006;52:1223–1237. doi: 10.1373/clinchem.2006.069252. [DOI] [PubMed] [Google Scholar]
- Jefferies J.R., Campbell A.M., van Rossum A.J., Barrett J., Brophy P.M. Proteomic analysis of Fasciola hepatica excretory–secretory products. Proteomics. 2001;1:1128–1132. doi: 10.1002/1615-9861(200109)1:9<1128::AID-PROT1128>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Koopmann J., Zhang Z., White N., Rosenzweig J., Fedarko N., Jagannath S., Canto M.I., Yeo C.J., Chan D.W., Goggins M. Serum diagnosis of pancreatic adenocarcinoma using surface-enhanced laser desorption and ionization mass spectrometry. Clin. Cancer Res. 2004;10:860–868. doi: 10.1158/1078-0432.ccr-1167-3. [DOI] [PubMed] [Google Scholar]
- Kozak K.R., Amneus M.W., Pusey S.M., Su F., Luong M.N., Luong S.A., Reddy S.T., Farias-Eisner R. Identification of biomarkers for ovarian cancer using strong anion-exchange ProteinChips: potential use in diagnosis and prognosis. Proc. Nat. Acad. Sci. USA. 2003;100:12343–12348. doi: 10.1073/pnas.2033602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S., Roboz J., Ding H., Zhao S., Diamond E.J., Holland J.F., Stone N.N., Droller M.J., Stock R.G. Putative protein markers in the sera of men with prostatic neoplasms. BJU Int. 2003;92:223–225. doi: 10.1046/j.1464-410x.2003.04341.x. [DOI] [PubMed] [Google Scholar]
- Martinsson K., Mollerberg L. On the transferrin concentration in blood serum of growing calves and in bovine colostrum. Zentralblatt fur Veterinarmedizin. 1973;20:277–284. doi: 10.1111/j.1439-0442.1973.tb00885.x. [DOI] [PubMed] [Google Scholar]
- Mas-Coma M.S., Esteban J.G., Bargues M.D. Epidemiology of human fascioliasis: a review and proposed new classification. Bull. World Health Organ. 1999;77:340–346. [PMC free article] [PubMed] [Google Scholar]
- Mas-Coma S., Bargues M.D., Valero M.A. Fascioliasis and other plant-borne trematode zoonoses. Int. J. Parasitol. 2005;35:1255–1278. doi: 10.1016/j.ijpara.2005.07.010. [DOI] [PubMed] [Google Scholar]
- McNair J., Elliott C., Bryson D.G., Mackie D.P. Bovine serum transferrin concentration during acute infection with Haemophilus somnus. Vet J. 1998;155:251–255. doi: 10.1016/s1090-0233(05)80020-6. [DOI] [PubMed] [Google Scholar]
- Merchant M., Weinberger S.R. Recent advancements in surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Electrophoresis. 2000;21:1164–1177. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1164::AID-ELPS1164>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Moon K., Smith E.L. Sequence of bovine liver glutamate dehydrogenase. 8. Peptides produced by specific chemical cleavages; the complete sequence of the protein. J. Biol. Chem. 1973;248:3082–3088. [PubMed] [Google Scholar]
- Morphew R.M., Wright H.A., Lacourse E.J., Woods D.J., Brophy P.M. Comparative proteomics of excretory–secretory proteins released by the liver fluke Fasciola hepatica in sheep host bile and during in vitro culture ex host. Mol. Cell. Proteomics. 2007;6:963–972. doi: 10.1074/mcp.M600375-MCP200. [DOI] [PubMed] [Google Scholar]
- Moser M., Pfister H., Bruckmaier R.M., Rehage J., Blum J.W. Blood serum transferrin concentration in cattle in various physiological states, in veal calves fed different amounts of iron, and in cattle affected by infectious and non-infectious diseases. Zentralblatt fur Veterinarmedizin. 1994;41:413–420. doi: 10.1111/j.1439-0442.1994.tb00108.x. [DOI] [PubMed] [Google Scholar]
- Orlacchio A., Campos-Cavieres M., Pashev I., Munn E.A. Some kinetic and other properties of the isoenzymes of aspartate aminotransferase isolated from sheep liver. Biochem. J. 1979;177:583–593. doi: 10.1042/bj1770583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos M.C., Abel P.M., Agranoff D., Stich A., Tarelli E., Bell B.A., Planche T., Loosemore A., Saadoun S., Wilkins P., Krishna S. A novel and accurate diagnostic test for human African trypanosomiasis. Lancet. 2004;363:1358–1363. doi: 10.1016/S0140-6736(04)16046-7. [DOI] [PubMed] [Google Scholar]
- Petricoin E.F., Ardekani A.M., Hitt B.A., Levine P.J., Fusaro V.A., Steinberg S.M., Mills G.B., Simone C., Fishman D.A., Kohn E.C., Liotta L.A. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- Petricoin E.F., 3rd, Ornstein D.K., Paweletz C.P., Ardekani A., Hackett P.S., Hitt B.A., Velassco A., Trucco C., Wiegand L., Wood K., Simone C.B., Levine P.J., Linehan W.M., Emmert-Buck M.R., Steinberg S.M., Kohn E.C., Liotta L.A. Serum proteomic patterns for detection of prostate cancer. J. Nat. Cancer Inst. 2002;94:1576–1578. doi: 10.1093/jnci/94.20.1576. [DOI] [PubMed] [Google Scholar]
- Piacenza L., Acosta D., Basmadjian I., Dalton J.P., Carmona C. Vaccination with cathepsin L proteinases and with leucine aminopeptidase induces high levels of protection against fascioliasis in sheep. Infect. Immun. 1999;67:1954–1961. doi: 10.1128/iai.67.4.1954-1961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedrafita D., Raadsma H.W., Prowse R., Spithill T.W. Immunology of host–parasite relationship in fasciolosis (Fasciola hepatica and Fasciola gigantica) Can. J. Zool. 2004;82:233–250. [Google Scholar]
- Piedrafita D., Estuningsih E., Pleasance J., Prowse R., Raadsma H.W., Meeusen E.N., Spithill T.W. Peritoneal lavage cells of Indonesian thin tail sheep mediate antibody-dependent superoxide radical cytotoxicity in vitro against newly excysted juvenile Fasciola gigantica but not juvenile Fasciola hepatica. Infect. Immun. 2007;75:1954–1963. doi: 10.1128/IAI.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon T.C., Chan K.C., Ng P.C., Chiu R.W., Ang I.L., Tong Y.K., Ng E.K., Cheng F.W., Li A.M., Hon E.K., Fok T.F., Lo Y.M. Serial analysis of plasma proteomic signatures in pediatric patients with severe acute respiratory syndrome and correlation with viral load. Clin. Chem. 2004;50:1452–1455. doi: 10.1373/clinchem.2004.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S., Tiers L., Provansal M., Piva M.T., Lehmann S. Interest of major serum protein removal for surface-enhanced laser desorption/ionization – time of flight (SELDI-TOF) proteomic blood profiling. Proteome Sci. 2006;4:20–26. doi: 10.1186/1477-5956-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosty C., Christa L., Kuzdzal S., Baldwin W.M., Zahurak M.L., Carnot F., Chan D.W., Canto M., Lillemoe K.D., Cameron J.L., Yeo C.J., Hruban R.H., Goggins M. Identification of hepatocarcinoma–intestine–pancreas/pancreatitis-associated protein I as a biomarker for pancreatic ductal adenocarcinoma by protein biochip technology. Cancer Res. 2002;62:1868–1875. [PubMed] [Google Scholar]
- Semmes O.J., Feng Z., Adam B.L., Banez L.L., Bigbee W.L., Campos D., Cazares L.H., Chan D.W., Grizzle W.E., Izbicka E., Kagan J., Malik G., McLerran D., Moul J.W., Partin A., Prasanna P., Rosenzweig J., Sokoll L.J., Srivastava S., Srivastava S., Thompson I., Welsh M.J., White N., Winget M., Yasui Y., Zhang Z., Zhu L. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin. Chem. 2005;51:102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- Sexton J.L., Milner A.R., Panaccio M., Waddington J., Wijffels G., Chandler D., Thompson C., Wilson L., Spithill T.W., Mitchell G.F. Glutathione S-transferase. Novel vaccine against Fasciola hepatica infection in sheep. J. Immunol. 1990;145:3905–3910. [PubMed] [Google Scholar]
- Sinclair K.B. Studies on the anaemia of chronic ovine fascioliasis. Res. Vet. Sci. 1972;13:182–184. [PubMed] [Google Scholar]
- Spithill T.W., Piedrafita D., Smooker P.M. Immunological approaches for the control of fasciolosis. Int. J. Parasitol. 1997;27:1221–1235. doi: 10.1016/s0020-7519(97)00120-3. [DOI] [PubMed] [Google Scholar]
- Spithill T.W., Smooker P.M., Copeman D.B. Fasciola gigantica: epidemiology, control, immunology and molecular biology. In: Dalton J., editor. Fasciolosis. CAB International; Oxford: 1999. pp. 465–525. [Google Scholar]
- Stan S., Delvin E., Lambert M., Seidman E., Levy E. Apo A-IV: an update on regulation and physiologic functions. Biochimica et biophysica acta. 2003;1631:177–187. doi: 10.1016/s1388-1981(03)00004-0. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Konishi H., Sato K., Oohashi T., Miyamoto T. Enzyme-linked immunosorbent assay for bovine apolipoprotein A-IV. J. Vet. Med. Sci. 2004;66:1199–1204. doi: 10.1292/jvms.66.1199. [DOI] [PubMed] [Google Scholar]
- Tolson J., Bogumil R., Brunst E., Beck H., Elsner R., Humeny A., Kratzin H., Deeg M., Kuczyk M., Mueller G.A., Mueller C.A., Flad T. Serum protein profiling by SELDI mass spectrometry: detection of multiple variants of serum amyloid alpha in renal cancer patients. Lab. Invest. 2004;84:845–856. doi: 10.1038/labinvest.3700097. [DOI] [PubMed] [Google Scholar]
- Tsuji S., Kato H., Matsuoka Y., Fukushima T. Molecular weight heterogeneity of bovine serum transferrin. Biochem. Genet. 1984;22:1145–1159. doi: 10.1007/BF00499638. [DOI] [PubMed] [Google Scholar]
- van Breemen M.J., Bleijlevens B., de Koster C.G., Aerts J.M. Limitations in quantitation of the biomarker CCL18 in Gaucher disease blood samples by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Biochimica et biophysica acta. 2006;1764:1626–1632. doi: 10.1016/j.bbapap.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Won Y., Song H.J., Kang T.W., Kim J.J., Han B.D., Lee S.W. Pattern analysis of serum proteome distinguishes renal cell carcinoma from other urologic diseases and healthy persons. Proteomics. 2003;3:2310–2316. doi: 10.1002/pmic.200300590. [DOI] [PubMed] [Google Scholar]
- Xiao Z., Prieto D., Conrads T.P., Veenstra T.D., Issaq H.J. Proteomic patterns: their potential for disease diagnosis. Mol. Cell. Endocrinol. 2005;230:95–106. doi: 10.1016/j.mce.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Yip T.T., Chan J.W., Cho W.C., Yip T.T., Wang Z., Kwan T.L., Law S.C., Tsang D.N., Chan J.K., Lee K.C., Cheng W.W., Ma V.W., Yip C., Lim C.K., Ngan R.K., Au J.S., Chan A., Lim W.W. Protein chip array profiling analysis in patients with severe acute respiratory syndrome identified serum amyloid a protein as a biomarker potentially useful in monitoring the extent of pneumonia. Clin. Chem. 2005;51:47–55. doi: 10.1373/clinchem.2004.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazo P., Orlowski M. Gamma-glutamyl transpeptidase of sheep-kidney cortex. Isolation, catalytic properties and dissociation into two polypeptide chains. Eur. J. Biochem./FEBS. 1976;61:147–155. doi: 10.1111/j.1432-1033.1976.tb10005.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Bast R.C., Jr., Yu Y., Li J., Sokoll L.J., Rai A.J., Rosenzweig J.M., Cameron B., Wang Y.Y., Meng X.Y., Berchuck A., Van Haaften-Day C., Hacker N.F., de Bruijn H.W., van der Zee A.G., Jacobs I.J., Fung E.T., Chan D.W. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.