Abstract
SARS‐CoV is a newly identified coronavirus that causes severe acute respiratory syndrome (SARS). Currently, there is no effective method available for prophylaxis and treatment of SARS‐CoV infections. In the present study, the influence of small interfering RNA (siRNA) on SARS‐CoV nucleocapsid (N) protein expression was detected in cultured cells and mouse muscles. Four siRNA expression cassettes driven by mouse U6 promoter targeting SARS‐CoV N gene were prepared, and their inhibitory effects on expression of N and enhanced green fluorescence protein (EGFP) fusion protein were observed. A candidate siRNA was proved to down‐regulate N and EGFP expression actively in a sequence‐specific manner. The expression vector of this siRNA was constructed and confirmed to reduce N and EGFP expression efficiently in both cultured cells and adult mouse muscles. Our findings suggest that the siRNA should provide the basis for prophylaxis and therapy of SARS‐CoV infection in human.
Keywords: Severe acute respiratory syndrome-coronavirus, Nucleocapsid protein, RNA interference, Small interfering RNA
1. Introduction
Severe acute respiratory syndrome (SARS), an emerging infectious disease of humans, appeared in China in November 2002 and spread to 30 countries in early 2003. The etiologic agent of SARS was identified as a coronavirus (CoV) and sequence analysis reveals that the SARS‐CoV is distinct from all known human coronaviruses [1, 2, 3]. The two previously identified human CoVs are associated only with mild upper respiratory tract diseases, whereas the novel SARS‐CoV appears to be the first human coronavirus responsible for severe disease in humans [4, 5]. All CoVs encode a common set of structural components consisting of a nucleocapsid protein (N) and three integral membrane proteins, namely the spike protein (S), the membrane protein (M), and the envelope protein (E) [6, 7]. The S protein plays essential roles in mediating receptor binding and internalization of the virus and is one of the major antigens of the virus [8]. The M and E proteins are essential for virion assembly, and the N protein binds to a defined packaging signal on viral genome RNA, leading to the formation of the helical nucleocapsid [6, 7].
RNA interference (RNAi) is a process in which double‐stranded RNA (dsRNA) induces the posttranscriptional degradation of homologous transcripts, and has been observed in a variety of organisms including plants, fungi, insects, Drosophila, and mammals [9, 10, 11]. It is believed to have evolved as a host defense mechanism directed at transposable elements and infectious viruses. Many studies have proved that siRNA can significantly suppress gene expression when exposed to mammalian cells in vitro [12, 13, 14]. Moreover, a number of groups demonstrated effective silencing of both endogenous gene and transgene expression in vivo [15, 16, 17, 18]. These findings raised the possibility that RNAi might advance the field of therapeutic approaches for virus infection. In this study, we screened an expression cassette‐based siRNA that targets at the N sequence of SARS‐CoV, and it can significantly suppress the expression of SARS‐CoV N protein in cultured cells in a sequence‐specific manner. Furthermore, the siRNA expression vector was constructed and proved to be effective in silencing the expression of SARS‐CoV N protein in adult mouse muscles.
2. Materials and methods
2.1. Construction of SARS‐CoV N and EGFP fusion protein expression plasmid
The 1266 base pair, none‐stop codon fragment of SARS‐CoV N gene was synthesized according to the published sequence (GenBank accession number: AY278554, the nucleoprotein coding region is from nt 28105 to nt 29373 in the full length SARS‐CoV genome) and used as the template to amplify the DNA fragment for construction of N and enhanced green fluorescence protein (EGFP) fusion protein expression plasmid. The forward primer was 5′‐GAATTCGCCACCATGTCTGATAATGGACCCCAATC‐3′; and the reverse primer was 5′‐GGATCCATTGCCTGAGTTGAATCAGCAG‐3′ (the underlined sequences were EcoRI and BamHI sites, respectively). Then the EcoRI–BamHI fragment was transferred into pEGFP‐N1 vector (Clontech), yielding the recombinant pN‐EGFP construct, in which N gene was located upstream of EGFP. The inserted fragment was verified by restriction endonucleases digestion and DNA sequencing.
2.2. Selection of the siRNA target sites and preparation of siRNA expression cassettes
The siRNAs targeting SARS‐CoV N sequence were selected based on the following guidelines: (1) the 19–21 nucleotides that following AA dinucleotides, (2) the sequence begins with G that preferring RNA polymerase III transcript, (3) the sequence contains about 50% GC, (4) the sequence does not contain a run of 4 or 5 A's or T's for avoiding early termination of RNA polymerase III transcript and (5) the sequence has no homology with any known human genes. The selected siRNA target sites were shown in Table 1 . The S1184 target site is specific for SARS‐CoV S gene, and muN388 target site is a mutant sequence for N388, in which bi‐nucleotides of the 6th site C and 7th site G are reversed from N388. Both of the S1184 and muN388 were served as controls for analysis of the specificity of the siRNA. PCR based siRNA expression cassettes, containing U6 promoter, a 19–21‐nt sense strand of siRNA, a 9‐nt loop, a 19–21‐nt antisense strand of siRNA, and a stretch of six deoxythymidines (poly(T)6), were amplified by a two‐step PCR described by Castanotto [19] and Gou [20] (Fig. 1 ). The 5′ primer mU6F 5′‐GAATTGGGTACCCGCTCTAG‐3′, is complementary to 20 nt at the 5′ end of the murine U6 promoter and was used for all PCR procedures. In the first round of PCR, the plasmid pSilence1.0‐U6 containing mouse U6 promoter (Ambion) was used as template, and the reverse primers contain the 9‐nt loop (5′‐TTCAAGAGA‐3′), a 19–21‐nt antisense sequence of siRNA target site and the complementary sequence to the last 14 nts of the mouse U6 promoter (5′‐GCCTTGTTTGGGCC‐3′). The PCR amplification using Pfu DNA Polymerase was carried out as follows: 94 °C for 4 min; 94 °C for 30 s, 58 °C for 30 s, 72 °C for 40 s, 30 cycles; 72 °C for 10 min. One microliter of the first PCR product was re‐amplified using the mixture of Taq and Pfu DNA Polymerase. The amplification conditions and forward primer were the same as those in the first round PCR, while the reverse primers contain poly(A)6, a 19–21‐nt sense strand of siRNA target site, and a complementary sequence to the 9‐nt loop. In addition, the mouse U6 promoter was amplified as a control for siRNA screening. The second round PCR products were purified using QIAquick PCR purification kit (Qiagen). All the reverse primers were listed in Table 1.
Table Table 1.
The siRNA target sequences and the reverse primers for preparation of siRNA expression cassettes
| siRNA | Sequence | Reverse primer sequence a |
|---|---|---|
| N186 | 5′‐GGAGGAACTTAGATTCCCT‐3′ | 3′ primer 1:5′‐TCTCTTGAAAGGGAATCTAAGTTCCTCCGGCCCAAACAAGGC‐3′ |
| 3′ primer 2:5′‐AAAAAAGGAGGAACTTAGATTCCCTTCTCTTGAAAGG‐3′ | ||
| N388 | 5′‐GGCATCGTATGGGTTGCAACT‐3′ | 3′ primer 1:5′‐TCTCTTGAAAGTTGCAACCATACGATGCGGCCCAAACA AGGC‐3′ |
| 3′ primer 2:5′‐AAAAAAGGCATCGTATGGGTTGCAACTTCTCTTGAAAGTTG‐3′ | ||
| N587 | 5′‐GAAATTCAACTCCTGGCAG‐3′ | 3′ primer 1:5′‐TCTCTTGAACTGCCAGGAGTTGAATTTCGGCCCAAACAAGGC‐3′ |
| 3′ primer 2:5′‐AAAAAAGAAATTCAACTCCTGGCAGTCTCTTGAACTG‐3′ | ||
| N811 | 5′‐GTCACTCAAGCATTTGGGAGA‐3′ | 3′ primer 1:5′‐TCTCTTGAATCTCCCAAATGCTTGAGTGACGGCCCAAACAAGGC‐3′ |
| 3′ primer 2:5′‐AAAAAAGTCACTCAAGCATTTGGGAGATCTCTTGAATCT‐3′ | ||
| S1184 | 5′‐GACAAATAGCGCCAGGACA‐3′ | 3′ primer 1:5′‐TCTCTTGAATGTCCTGGCGCTATTTGTCGGCCCAAACAAGGC‐3′ |
| 3′ primer 2:5′‐AAAAAAGACAAATAGCGCCAGGACATCTCTTGAATGT‐3′ | ||
| muN388 | 5′‐GGCATGCTATGGGTTGCAACT‐3′ | 3′ primer 1:5′‐TCTCTTGAAAGTTGCAACCATAGCATGCGGCCCAAACA AGGC‐3′ |
| 3′ primer 2:5′‐AAAAAAGGCATGCTATGGGTTGCAACTTCTCTTGAAAGTTG‐3′ |
3′ primer 1 denotes the reverse primer for the first round PCR, 3′ primer 2 denotes the reverse primer for the second round PCR.
Figure 1.
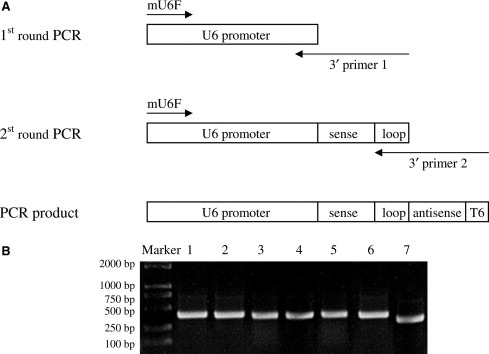
Preparation of PCR‐based siRNA expression cassettes. (A) Two‐step PCR procedure for the siRNA expression cassettes was described in the text, the PCR products consist of mouse U6 promoter, short hairpin DNA and terminator sequence, six thymidines (T6). (B) The siRNA expression cassettes were prepared by two rounds of PCR. Lanes 1–4: PCR products of siRNA expression cassettes of N186, N388, N587 and N811, respectively. Lane 5: PCR product of muN388 expression cassettes. Lane 6: PCR product of S1184 expression cassettes. Lane 7: PCR product of mouse U6 promoter.
2.3. Cell culture and transfection
293T, CHO and Vero E6 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 1 mM l‐glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Twenty‐four hours before transfection, 60–80% confluent cells were trypsinized, and 1 × 105 cells were plated into a 24‐well format in 500 μl of fresh culture medium without antibiotics. The plasmid pN‐EGFP (0.4 μg) was co‐transfected with the siRNA expression cassettes (0.4 μg) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At various time points, the cells were examined for the expression of EGFP with a fluorescence microscope (Olympus) and the expression of SARS‐CoV N protein was detected by Western blot and real‐time PCR analyses.
2.4. Construction of siRNA expression vector
After observation of the inhibitory effects of the siRNA expression cassettes on the expression of SARS‐CoV N and EGFP protein, the most active siRNA expression cassette that targets 388–408‐nt of SARS‐CoV N sequence was inserted into the pMD‐18T vector (TaKaRa) using TA‐cloning technique, and the correct sequence was verified by DNA sequencing. The resulting siRNA expression plasmid pU6‐shN388 was used for the further study.
2.5. Effect of siRNA expression vector on expression of SARS‐CoV N protein in cultured cells
For the sequence specificity analysis of the siRNA, a plasmid pCI‐HBc was used as an unrelated viral gene expression control, which was constructed by inserting of hepatitis B virus core antigen (HBcAg) DNA sequence into a eukaryotic expression vector pCI‐neo (Promega). 293T cells plated in 35 mm dishes were co‐transfected respectively with plasmids (1) 0.3 μg pN‐EGFP, 0.3 μg pCI‐HBc and 0.6 μg pUC18, (2) 0.3 μg pN‐EGFP, 0.3 μg pCI‐HBc, 0.3 μg pU6‐shN388 and 0.3 μg pUC18, or (3) 0.3 μg pN‐EGFP, 0.3 μg pCI‐HBc and 0.6 μg pU6‐shN388. At 48 h post‐transfection, the cells were harvested for Western blot analysis.
2.6. Effect of siRNA on expression of SARS‐CoV N protein in murine skeletal muscles
Plasmids pN‐EGFP, pU6‐shN388 and pUC18 were purified using QIAfilter™ Plasmid Mega kit (Qiagen) and mixed respectively with 3.4% polyvinyl pyrrolidone (PVP, MW 40 000, Sigma) in 0.01 M PBS to a final concentration of 2 g/l. These plasmids were later used for muscle injection of mice. Six‐week old female BALB/c mice (Xiper‐Bikai Experimental Animal Co., LTD, Shanghai) were randomly divided into two groups (four mice in each group). The experimental group were injected with 15 μl pN‐EGFP and 45 μl pU6‐shN388 at both sides of tibialis anterior muscles, and the control group received 15 μl pN‐EGFP and 45 μl pUC18. At day 4, 8, 12 and 16 post‐injection, one mouse per group was sacrificed, and the tibialis anterior muscles were isolated for N and EGFP expression analyses.
2.7. Reverse transcription and real‐time PCR
Total RNA from the transfected cells was extracted using Trizol reagent (GIBCO) according to the manufacturer's instruction and then treated with RNase‐free DNase 1 (Promega). To make sure that no plasmid pN‐EGFP derived DNA presented in the total RNA, 0.1 μg of each total RNA was checked by PCR using the 5′ primer (5′‐CCTCGCGCTATTGC‐3′) and 3′ primer (5′‐GATGCCTCAG CAGCAG‐3′) prior to reverse transcription, and no a 111‐bp fragment corresponding to the nt 657–766 of SARS‐CoV N gene was obtained. One microgramme total RNA was subjected to reverse transcription reaction with 5 U AMV reverse transcriptase (TaKaRa) and 2.5 pmol oligo(dT)18 primer in 25 μl reaction mixture at 42 °C for 40 min. The RT product was then inactivated by heating at 99 °C for 5 min and subjected to real‐time PCR analysis using SYBR Premix Ex Taq™ system (TaKaRa). For real‐time PCR, 3 μl RT product was amplified in a 20 μl reaction volume containing 5 pmol of 5′ primer and 3′ primer, and 10 μl SYBR Premix Ex Taq™ 2× buffer (including SYBR green I reagent, Taq DNA polymerase, and dNTP). The primers for SARS‐CoV N are the same as those used in PCR for total RNA. Following a denaturation step at 95 °C for 10 s, the amplification reaction was performed 40 cycles at 95 °C for 5 s, 60 °C for 20 s in a real‐time quantitative Lightcycler™ system (Roche). Each sample was run in triplicate. Six serially 10‐fold diluted samples of EcoRI–BamHI fragment of N sequence (varied from 107 to 102 copies in each reaction) were used to create standard curve for the quantitation of SARS N cDNA concentration in RT product. For analysis of SARS‐CoV N mRNA in mouse skeletal muscles, the tibialis anterior muscles were freeze–thawed and homogenized, and the cellular RNA was extracted as described above.
2.8. Western blot
Cells were lyzed using lysis buffer (1% Triton X‐100, 2 mM EDTA, 50 Mm Tris–HCl, pH 7.4, 200 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride), and the cell lysate was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrophoretically transferred to nitrocellulose membrane. The primary antibodies used for detection of specific proteins were as follows: anti‐SARS‐CoV N positive sera from convalescent SARS patients (1:200 dilution), anti‐HBcAg positive sera from acute hepatitis B patients (1:500 dilution) and anti‐β‐actin monoclonal antibody (SantaCruz, 1:1000 dilution). The secondary antibody detection was performed using alkaline phosphatase‐conjugated goat anti‐human IgG, or horseradish peroxidase‐conjugated goat anti‐mouse IgG, and immunoreactive bands were visualized with nitroblue tetrazolium and 5‐bromo‐4‐chloro‐3‐indolylphosphate (NBT/BCIP; Pierce), or DAB and H2O2. For detection of SARS‐CoV N protein in mouse skeletal muscles, the isolated tibialis anterior muscles were freeze–thawed, homogenized, and the extracted protein was analyzed by the procedure similar to that described above.
3. Results
3.1. Preparation of mouse U6 promoter‐derived siRNA expression cassettes
For constructing the siRNA expression cassettes, a procedure consisting of two rounds of PCR was used as described by Castanotto [19] and Gou [20], and the resulting PCR products include the mouse U6 promoter, sense sequence, loop, antisense sequence, and followed by the Pol III terminator sequence poly(T)6 (Fig. 1A). Using this method, six siRNA expression cassettes were prepared with the correct predicted molecular weights (Fig. 1B).
3.2. Efficient inhibition of SARS‐CoV N‐EGFP expression by siRNA expression cassettes
The plasmid pN‐EGFP was transfected into 293T, CHO and Vero E6 cells either alone or along with siRNA expression cassettes, respectively. At various time points post‐transfection, the effect of siRNA on EGFP expression was monitored by fluorescence microscope. The results showed that all four siRNA expression cassettes targeting N gene could inhibit the expression of EGFP to some extent compared with that in cells transfected with plasmid pN‐EGFP alone, especially for N388 expression cassette. The N388 expression cassette targeting nt 388–407 of N sequence was most effective, and only very weak fluorescence was observed at 48–72 h post‐transfection. No significant inhibition of EGFP expression was observed at any time in cells that co‐transfected with the control constructs (the mouse U6 promoter or S1184 siRNA expression cassette). Given that the inhibition of N388 expression cassette on the EGFP expression, its mutant version (muN388) with two nucleotides reversed was prepared. The muN388 gave little influence on EGFP expression.
Western blot and semi‐quantitative real‐time PCR were carried out for determination of the inhibitory effect of the siRNA on SARS‐CoV N gene expression. Western blot demonstrated that the inhibition of N gene shRNA on the N protein was similar as that on EGFP expression. The suppressive effect of N388 siRNA was most significant, while the control constructs and mutant muN388 had little effect (Fig. 2 A). The real‐time PCR assay also showed that the four siRNA expression cassettes could down‐regulate the mRNA transcription of N gene. The three control constructs co‐transfected with the pN‐EGFP only had marginal effect on the mRNA of N gene (Fig. 2B). Therefore, the N388 siRNA was selected for further studies.
Figure 2.
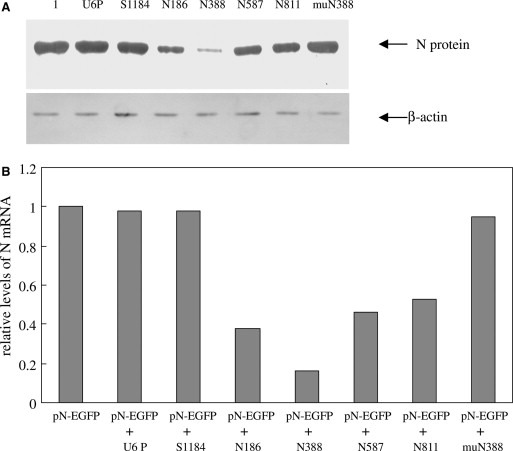
Inhibition of SARS‐CoV N protein expression by siRNA expression cassettes. The 293T cells were co‐transfected with N‐EGFP expression plasmid and various PCR products. The SARS‐CoV N protein expression was measured. (A) N protein expression was detected by Western blot at 48 h post‐transfection. The upper panel shows N protein expression (lane 1 shows N expression in cells transfected with pN‐EGFP alone, other lanes show that in cells co‐transfected with pN‐EGFP and various PCR products, U6P denotes PCR product of mouse U6 promoter only). The lower panel shows β‐actin expression (as an endogenous control). (B) At 48 h post‐transfection, the total RNA was isolated and subjected to reverse transcription and real‐time PCR analysis. The mRNA levels of cells transfected with plasmid pN‐EGFP along with various PCR products were quantitated relative to that in the cells transfected with plasmid pN‐EGFP alone.
3.3. Silencing of N‐EGFP by siRNA expression vector in cultured cells
The N388 siRNA expression vector was constructed by inserting the expression cassette into the vector pMD18‐T (Fig. 3 A). The recombinant vector was co‐transfected into cultured cells with the N‐EGFP expression plasmid. The hepatitis B virus core antigen expression plasmid was adopted as an exogenous gene control in this assay. As shown in Fig. 3B and C, the siRNA expression vector inhibits N and EGFP expression in 293 cells in a dose‐dependent manner. With 0.6 μg pU6‐shN388 administered, N and EGFP expression were significantly suppressed, while HBcAg expression was not influenced.
Figure 3.
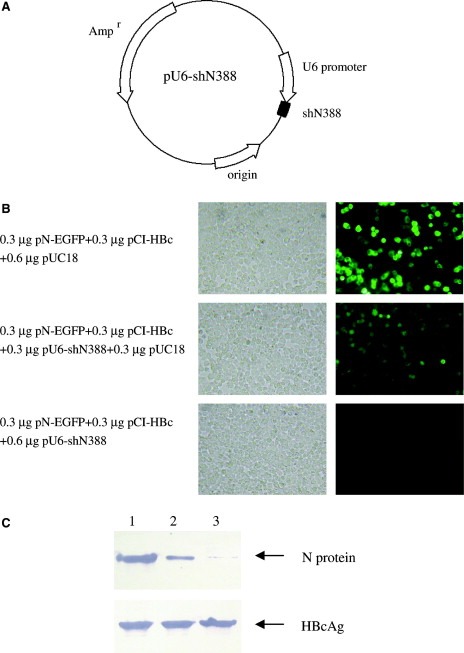
Inhibition of SARS‐CoV N protein expression by siRNA expression vector. The N388 siRNA expression vector was constructed and its influence on the N‐EGFP expression was detected in cultured 293T cells. (A) The PCR product of N388 siRNA expression cassette was inserted into the vector pMD‐18T, and the resulting plasmid was used as siRNA expression vector. (B) 293T cells were co‐transfected with plasmid pN‐EGFP and N388 siRNA expression vector, along with HBcAg expression plasmid as an unrelated control. The images show the EGFP expression at 48 h post‐transfection. The left panel shows the cells under a bright field. (C) The expression of N protein and HBcAg were detected by Western blot at 48 h post‐transfection. Lane 1: 293T cells transfected with 0.3 μg pN‐EGFP, 0.3 μg pCI‐HBc and 0.6 μg pUC18. Lane 2: 293T cells transfected with 0.3 μg pN‐EGFP, 0.3 μg pCI‐HBc and 0.3 μg pU6‐shN388. Lane 3: 293T cells transfected with 0.3 μg pN‐EGFP, 0.3 μg pCI‐HBc and 0.6 μg pU6‐shN388. In fluorescence and Western blot analysis, plasmid pUC18 was used to standardize the plasmid dosage for transfection. The experiment was repeated three times, and similar results were obtained.
3.4. Silencing of N‐EGFP by siRNA expression vector in mouse muscles
A mouse model expressing N‐EGFP fusion protein in skeletal muscles was used to test the potential anit‐SARS‐CoV activity of the siRNA in vivo. When intramuscularly co‐injected the N‐EGFP expression plasmid and N388 siRNA expression plasmid pU6‐shN388 to mice, we found that the plasmid pU6‐shN38 could reduce the expression of EGFP and N protein significantly, and this inhibitory effect lasted 16 days of the entire observation period (Fig. 4 ). Real‐time PCR analysis showed that the mRNA level of SARS‐CoV N in mouse muscles was reduced to 19%, 17%, 21% and 23%, relative to that co‐injected with pN‐EGFP and control plasmid pUC18 at day 4, 8, 12 and 16 after siRNA delivery, respectively.
Figure 4.
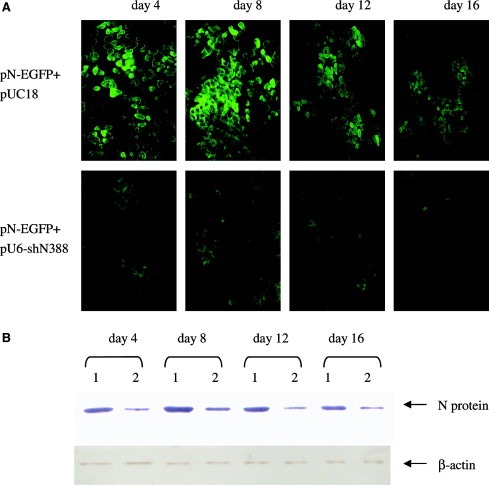
Silencing of N‐EGFP expression by siRNA in mouse muscles. Mice were injected with N‐EGFP expression plasmid and the siRNA expression vector in both tibialis anterior muscle. The N and EGFP expression was detected at day 4, 8, 12 and 16 post‐injection. (A) The muscles were isolated and frost slices were prepared, and EGFP expression was visualized under fluorescence microscope. The upper panel shows EGFP expression in mice injected with plasmid pN‐EGFP along with siRNA expression vector at various time post‐injection. The lower panel shows EGFP expression in mice injected with plasmid pN‐EGFP along with pUC18. (B) The muscles were isolated and N protein expression was detected by Western blot at different times. The upper panel indicates N protein expression in mouse muscles injected with plasmid pN‐EGFP along with: (1) pUC18 or (2) siRNA expression vector. The lower panel shows β‐actin expression as a loading control. The plasmid pUC18 used was to standardize the plasmid dosage for gene delivery in vivo. This figure shows the results of one representative experiment of two.
4. Discussion
Small interfering RNAs (siRNAs), mediators of RNAi, are short (21–26 nt), dsRNA duplexes that inhibit gene expression by inducing sequence‐specific degradation of homologous RNA, which may stand for a new promising method for protection against viral infection [12, 13, 14, 16, 18]. Several methods have been introduced for siRNA preparation, such as chemical synthesis, T7 RNA polymerase directed in vitro transcription, and short hairpin RNAs (shRNAs) expressed from DNA vectors. A PCR‐based shRNA expression cassette may represent one of the most time‐ and labor‐saving methods, especially for the identification of specific and efficient target sites [19, 20]. In addition, PCR products can be easily inserted into vectors by T‐A cloning, and if suitable restriction enzyme site tags are present in the primers, directional cloning into plasmid vectors is also feasible.
The N protein of SARS‐CoV is a key protein for the formation of the helical nucleocapsid in the cytoplasm, and is more conserved than other structural protein, which makes it an ideal target for RNA interference [2, 3]. In this study, four siRNA candidate sites targeting SARS‐CoV N sequence were selected, and mouse U6 promoter regulated siRNA expression cassettes were constructed. All four candidate siRNAs gave some reduction of EGFP expression in cultured cells, and N388 showed the most significant effect. Western blot demonstrated the similar inhibitory effect on N protein and EGFP, and the semi‐quantitative PCR confirmed that N388 down‐regulated mRNA of N gene more efficiently than other three siRNAs did. For gene intervention, sequence specificity is very important for its practical application. It is know that, in mammalian cells, dsRNA 30 base pairs or longer can trigger interferon responses that are intrinsically sequence‐nonspecific [21, 22, 23]. In our observation, the control PCR products, including the U6 promoter, SARS‐CoV S‐targeted siRNA expression construct, have little influence on the N‐EGFP expression. More importantly, the bi‐nucleotide reversed mutant of N388 siRNA had no inhibitory activity for N‐EGFP expression. These results indicate that the inhibition of N‐EGFP expression by siRNAs is sequence‐specific, requiring homology between the siRNAs and gene targets, and is not the consequence of interferon responses induced by dsRNA.
The siRNA expression vector of N388 was constructed by inserting the PCR product into the T vector for further observation. In cultured cells, this plasmid could significantly suppress N‐EGFP expression in a dose‐dependent manner, while the expression of another viral protein, HBcAg, did not change. As the expression of both N‐EGFP and HBcAg are driven by the same transcriptional control element (CMV immediate/early promoter/enhancer) and that down‐regulation of viral RNA by cytokines is not sequence‐specific, our results suggest that the inhibitory effect of siRNA or the SARS‐CoV N expression is not due to IFN response, but sequence‐dependent siRNA mechanism. To determine the potential biological activities of siRNA in vivo, we established a mouse model expressing N‐EGFP in skeletal muscles. Muscle cells are excellent for exogenous gene expression, for example, direct intramuscular DNA vaccine injection could lead to antigen expression in muscle cells and induction of systemic immune responses in hosts [24]. Analysis on protein and mRNA levels showed that just as in cultured cells, the siRNAs could down‐regulate N and EGFP expression in mouse muscles, and their effects lasted the entire observation period. Though Lower dose of siRNA develop less suppression activities on N and EGFP expression, they still behave significant inhibition. When 0.4 μg, instead of 0.6 μg pU6‐shN388 was administered, the mRNA of N gene was reduced to 28%, 24%, 31%, and 36% at different time points, respectively (data not shown). Although we have not evaluated the activities of the siRNA in respiratory tract, of the relevant target for SARS‐CoV infection, the siRNA seems to be functional for silencing N protein expression in vivo based on the data from mouse muscles and the cultured cells, especially from Vero E6 cells, which is supportive for SARS‐CoV replication. The siRNAs are promising for the development of antiviral agents for human use, especially when combining several siRNAs specific for different gene regions, and delivering with an appropriate carrier, such as cationic polymers [25] or virus vectors, or with proper modification, such as linkage with cholesterol [26]. For the characterization of SARS‐CoV infection, the recombinant vaccinia virus vector may be an attractive alternative for siRNA delivery. Recently, the siRNAs targeting the polymerase and S sequence were reported to silence gene expression and inhibit SARS‐CoV replication in mammalian cells efficiently [27, 28, 29, 30, 31]. Together with our data, it is considerable that siRNA may be possible for developing anti‐SARS agents.
In summary, we screened some siRNA candidates targeting SARS‐CoV N gene based on the siRNA expression cassettes, and identified a siRNA N388, that could efficiently and specifically inhibit the expression of SARS‐CoV N protein in human 293T cells and monkey Vero E6 cells. The inhibitory activity and durable effects of siRNA N388 on N expression were further confirmed in mouse muscles. Our present data suggest that siRNA should be of potential values in prophylaxis and treatment of SARS‐CoV infection in humans.
Acknowledgements
This work was supported by the Shanghai Science and Technology Research Project, China (No. 04DZ19221).
Zhao Ping,Qin Zhao-Ling,Ke Jin-Shan,Lu Yang,Liu Min,Pan Wei,Zhao Lan-Juan,Cao Jie and Qi Zhong-Tian(2005), Small interfering RNA inhibits SARS-CoV nucleocapsid gene expression in cultured cells and mouse muscles, FEBS Letters, 579, doi: 10.1016/j.febslet.2005.02.080
References
- 1. Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W., Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med., 348, (2003), 1967– 1976. [DOI] [PubMed] [Google Scholar]
- 2. Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples GA., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L., The Genome sequence of the SARS-associated coronavirus. Science, 300, (2003), 1399– 1404. [DOI] [PubMed] [Google Scholar]
- 3. Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J., Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science, 300, (2003), 1394– 1399. [DOI] [PubMed] [Google Scholar]
- 4. Wang G., Deering C., Macke M., Shao J., Burns R., Blau D.M., Holmes K.V., Davidson B.L., Perlman S., McCray P.B. Jr., Human coronavirus 229E infects polarized airway epithelia from the apical surface. J. Virol., 74, (2000), 9234– 9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F., An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis., 36, (2003), 985– 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lai M.M., Molecular biology of coronavirus. Adv. Exp. Med. Biol., 218, (1987), 7– 13. [DOI] [PubMed] [Google Scholar]
- 7. Masters P.S., Sturman L.S., Background paper. Functions of the coronavirus nucleocapsid protein. Adv. Exp. Med. Biol., 276, (1990), 235– 238. [DOI] [PubMed] [Google Scholar]
- 8. Sanchez C.M., Izeta A., Sanchez-Morgado J.M., hAlonso S., Sola I., Balasch M., Plana-Duran J., Enjuanes L., Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol., 73, (1999), 7607– 7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bass B.L., Double-stranded RNA as a template for gene silencing. Cell, 101, (2000), 235– 238. [DOI] [PubMed] [Google Scholar]
- 10. Kennerdell J.R., Carthew R.W., Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol., 18, (2000), 896– 898. [DOI] [PubMed] [Google Scholar]
- 11. Meister G., Tuschl T., Mechanisms of gene silencing by double-stranded RNA. Nature, 431, (2004), 343– 349. [DOI] [PubMed] [Google Scholar]
- 12. Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J., Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol., 20, (2002), 500– 505. [DOI] [PubMed] [Google Scholar]
- 13. Kapadia S.B., Brideau-Andersen A., Chisari F.V., Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA, 100, (2003), 2014– 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee M.T., Coburn G.A., McClure M.O., Cullen B.R., Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J. Virol., 77, (2003), 11964– 11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCaffrey A.P., Meuse L., Pham T.T., Conklin D.S., Hannon G.J., Kay M.A., RNA interference in adult mice. Nature, 418, (2002), 38– 39. [DOI] [PubMed] [Google Scholar]
- 16. Giladi H., Ketzinel-Gilad M., Rivkin L., Felig Y., Nussbaum O., Galun E., Small interfering RNA inhibits hepatitis B virus replication in mice. Mol. Ther., 8, (2003), 769– 776. [DOI] [PubMed] [Google Scholar]
- 17. Sorensen D.R., Leirdal M., Sioud M., Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J. Mol. Biol., 327, (2003), 761– 766. [DOI] [PubMed] [Google Scholar]
- 18. Tompkins S.M., Lo C.Y., Tumpey T.M., Epstein S.L., Protection against lethal influenza virus challenge by RNA interference in vivo. Proc. Natl. Acad. Sci. USA, 101, (2004), 8682– 8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Castanotto D., Li H., Rossi J.J., Functional siRNA expression from transfected PCR products. RNA, 8, (2002), 1454– 1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gou D., Jin N., Liu L., Gene silencing in mammalian cells by PCR-based short hairpin RNA. FEBS Lett., 548, (2003), 113– 118. [DOI] [PubMed] [Google Scholar]
- 21. Svoboda P., Stein P., Hayashi H., Schultz R.M., Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development, 127, (2000), 4147– 4156. [DOI] [PubMed] [Google Scholar]
- 22. Stark G.R., Kerr I.M., Williams B.R., Silverman R.H., Schreiber R.D., How cells respond to interferons. Annu. Rev. Biochem., 67, (1998), 227– 264. [DOI] [PubMed] [Google Scholar]
- 23. Clemens M.J., Elia A., The double-stranded RNA-dependent protein kinase PKR: structure and function. J. Interferon Cytokine Res., 17, (1997), 503– 524. [DOI] [PubMed] [Google Scholar]
- 24. Barry M.A., Johnston S.A., Biological features of genetic immunization. Vaccine, 15, (1997), 788– 791. [DOI] [PubMed] [Google Scholar]
- 25. Ge Q., Filip L., Bai A., Nguyen T., Eisen H.N., Chen J., Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl. Acad. Sci. USA, 101, (2004), 8676– 8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., John M., Kesavan V., Lavine G., Pandey R.K., Racie T., Rajeev K.G., Rohl I., Toudjarska I., Wang G., Wuschko S., Bumcrot D., Koteliansky V., Limmer S., Manoharan M., Vornlocher H.P., Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature, 432, (2004), 173– 178. [DOI] [PubMed] [Google Scholar]
- 27. He M.L., Zheng B., Peng Y., Peiris J.S., Poon L.L., Yuen K.Y., Lin M.C., Kung H.F., Guan Y., Inhibition of SARS-associated coronavirus infection and replication by RNA interference. JAMA, 290, (2003), 2665– 2666. [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y., Li T., Fu L., Yu C., Li Y., Xu X., Wang Y., Ning H., Zhang S., Chen W., Babiuk L.A., Chang Z., Silencing SARS-CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett., 560, (2004), 141– 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z., Ren L., Zhao X., Hung T., Meng A., Wang J., Chen Y.G., Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J. Virol., 78, (2004), 7523– 7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu A., Zhang H., Zhang X., Wang H., Hu Q., Shen L., Schaffhausen B.S., Hou W., Li L., Attenuation of SARS coronavirus by a short hairpin RNA expression plasmid targeting RNA-dependent RNA polymerase. Virology, 324, (2004), 84– 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng B.J., Guan Y., Tang Q., Du C., Xie F.Y., He M.L, Chan K.W., Wong K.L., Lader E., Woodle M.C., Lu P.Y., Li B., Zhong N., Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus. Antivir. Ther., 9, (2004), 365– 374. [PubMed] [Google Scholar]


