Abstract
The past decades have witnessed enormous technological improvements towards the development of simple, cost-effective and accurate rapid diagnostic tests for detection and identification of infectious pathogens. Among them is dengue virus, the etiologic agent of the mosquito-borne dengue disease, one of the most important emerging infectious pathologies of nowadays. Dengue fever may cause potentially deadly hemorrhagic symptoms and is endemic in the tropical and sub-tropical world, being also a serious threat to temperate countries in the developed world. Effective diagnostics for dengue should be able to discriminate among the four antigenically related dengue serotypes and fulfill the requirements for successful decentralized (point-of-care) testing in the harsh environmental conditions found in most tropical regions. The accurate identification of circulating serotypes is crucial for the successful implementation of vector control programs based on reliable epidemiological predictions. This paper briefly summarizes the limitations of the main conventional techniques for biomolecular diagnosis of dengue disease and critically reviews some of the most relevant biosensors and rapid diagnostic tests developed, implemented and reported so far for point-of-care testing of dengue infections. The invaluable contributions of microfluidics and nanotechnology encompass the whole paper, while evaluation concerns of rapid diagnostic tests and foreseen technological improvements in this field are also overviewed for the diagnosis of dengue and other infectious and tropical diseases as well.
Abbreviations: CDC, Centers for Disease Control; DENV1–4, dengue virus serotypes (1–4); ssRNA, single-stranded ribonucleic acid; ORF, open-reading frame; NS1, non-structural 1; DHF, dengue hemorrhagic fever; DSS, dengue shock syndrome; WHO, World Health Organization; HI, hemagglutination-inhibition; MAC-EIA, monoclonal antibody capture-enzyme linked immunosorbent assay; RT-PCR, reverse transcription-polymerase chain reaction; 3′-NR, 3′noncoding region; RNA, ribonucleic acid; IgG, immunoglobulin G; IgM, immunoglobulin M; DNA, deoxyribonucleic acid; QCM, quartz-crystal microbalance; MIP, molecularly imprinted polymer; GNP, gold nanoparticle; SAM, self-assembled monolayer; BSA, bovine serum albumin; SPR, surface plasmon resonance; NASBA, nucleic acid sequence-based amplification; S/N, signal-to-noise ratio; CMOS, complementary metal oxide semiconductor; FIA, flow-injection analysis; FCCS, fluorescence cross-correlation spectroscopy; FCS, fluorescence correlation spectroscopy; EIS, electrochemical impedance spectroscopy; BST, barium strontium titanate; FET, field-effect transistor; PNA, peptide nucleic-acid; LOD, limit of detection; cDNA, complementary DNA; TDR, Special Programme for Research and Training in Tropical Diseases; UNDP, United Nations Development Programme; PDVI, Pediatric Dengue Vaccine Initiative; STARD, Standards for Reporting of Diagnostic Accuracy; FIOCRUZ, Fundação Oswaldo Cruz; DPP®, Dual-Path Platform; BLM, bilayer lipid membrane; QD, quantum dot; CNT, carbon nanotube; MS, mass spectrometry; SARS, severe acute respiratory syndrome
Keywords: Biosensor, Dengue, Diagnosis, Evaluation, Rapid test, Tropical disease
1. Introduction
Dengue disease is caused by females of Aedes spp. mosquitoes infected with RNA-containing dengue virus (classified has a category A agent by the Centers for Disease Control and Prevention (CDC, USA) and is the most important arthropod-born disease in the present. It affects more than 100 countries worldwide [1] and threatens around half of the world population [2], mainly urban populations in the tropical world, while being a serious threat to more temperate countries. For this, the deforestation processes, the human population burst and the collapse of vector control programs in tropical and developing countries, as well as the enormous worldwide expansion in air travels, have largely contributed. Symptoms are highly non-specific, usually arising after an incubation period of up to two weeks (coinciding with the sudden of viral particles in the blood) [3]. As a potential cause of fatal hemorrhagic fever, dengue disease may occur in the form of four antigenically related viral serotypes (DENV1–4). Dengue viruses have a capsid with three structural proteins, including a glycoprotein-E responsible for attachment to host cells and reconnaissance by specific antibodies. They also possess a single-stranded RNA (ssRNA) genome, of 11 kbp and 4 kDa, with a single open-reading frame (ORF) flanked by two non-translated regions at each end, plus seven sequences codifying non-structural proteins, the most well-known being non-structural 1 protein (NS1) [4]. Apart the more common and less dangerous dengue fever, dengue hemorrhagic fever (DHF) or even dengue shock syndrome (DSS), both conditions requiring critical healthcare, may also occur. Most infections are asymptomatic, but an enormous increase in the DHF and DSS cases has been reported in the past decades. During 2008, the Americas reported, to the World Health Organization (WHO), around 900,000 suspected cases of dengue (with almost 60,000 confirmed), 25,000 of DHF and more than 306 fatal cases. The Brazilian State of Rio de Janeiro, in which a serious and widely reported outbreak occurred during that period, accounted for more than 32,000 cases of dengue fever (almost 40% of the total number of cases in Brazil) and around 250 cases of DHF [5]. Despite all serotypes cause disease, studies in Southeast Asia evidenced higher probability for occurrence of severe symptoms in secondary infections with DENV2 than with the other serotypes [6], probably the reason why, in most of the biosensors described ahead, DENV2-related nucleic-acid and immunological (antibodies or antigens) biomarkers have been predominantly reported. Although identical seroprevalence rates that have been registered between Asian and Caribbean new mothers and neonates, much higher incidences of DHF and DSS have occurred in Southeast Asia than in Caribbean, probably resulting from different circulating dengue genotypes, genetic resistance in human populations or differences in vector control. Detailed explanation of this fact may be crucial to avoid, in the Americas, the alarming scenario of Southeast Asia [7]. In Africa, notifications of dengue and other arboviral infections have been, most probably, highly underestimated [8]. In the absence of approved vaccines or therapeutics, which should be effective against all the four serotypes, accurate and rapid diagnoses are urgently needed against the most deadly forms of the disease. Longer acute febrile periods caused by dengue infections, when compared to other causes of acute fever syndromes, have been reported [9]. However, clinical diagnosis is usually insufficient for specific identification of the etiological agent, especially for disease surveillance purposes, hence requiring definite serological or virological laboratory confirmation. Highly sensitive virus isolation in culture followed by immunofluorescence microscopy employing dengue-specific antibodies has been considered the “gold-standard” for conventional dengue diagnosis, but requires high-level equipment and manpower and, due to the low replication rate of dengue virus, is not adequate for early diagnosis. Apart the general drawbacks of serological tests, some of them, namely the hemagglutination-inhibition (HI) and the neutralization test, are cumbersome, expensive and significantly non-specific [3]. Monoclonal antibody capture-enzyme linked immunosorbent assay (MAC-EIA) and reverse-transcription polymerase chain-reaction (RT-PCR) have been mainly used for routine laboratorial diagnosis of dengue infections although, like the former, they still fail to accomplish all requirements for successful decentralized diagnosis in tropical and developing countries: simplicity, economy, rapidity and accuracy [10]. These features can be materialized all together in the emerging paradigm of point-of-care (decentralized) testing for bioanalytical clinics, corresponding to near-patient analysis (in the healthcare unit or at home), even in remote regions and by non-skilled individuals (including end-users). Reliable and high-throughput sample processing in a timely manner are ultimately envisaged in point-of-care testing. These tailored tests, many of which consist in the well-known and widely available test strip format (also called immunochromatographic or lateral-flow tests), may thus be considered valuable complements of conventional, in-the-bench analytical techniques, not only for allowing remote detection, but also because rapid information permits improved decision making processes (e.g., in emergence situations), as well as reducing the amount of samples reaching the laboratory, especially when they undertake simultaneous determination of several analytes (multiplexing) [11]. Technological improvements in bioelectronics have driven the appearance of many methodologies and analytical devices, as valuable alternatives to conventional laboratory assays, for rapid and accurate detection of infectious pathogens. Many commercial immunoassay kits for serological diagnosis of dengue infections based on MAC-EIA are already available, but usually require long assay times (in the order of some hours) and time-consuming sample processing (including extensive washing and incubation steps). This is why it is difficult to considerer these immunoassays as true rapid diagnostic tests. This article overviews published literature, especially dated from the last decade, about biosensors and rapid diagnostic tests for biomolecular diagnosis of dengue disease, both with clinical or simply model synthetic samples. Of note, experimental conditions in biosensors and rapid tests assaying synthetic target biomarkers may not differ significantly from those of real samples [12].
2. Routine dengue diagnosis by MAC-EIA and RT-PCR: features and limitations
RT-PCR and MAC-EIA are the main routine laboratory tests more employed worldwide to the diagnosis of dengue infections, as described in Sections 2.1, 2.2.
2.1. RT-PCR
Early diagnosis is important to hinder the development of long-term serious symptoms and the spread of dengue disease. PCR and PCR-related techniques have been employed for a long time to routine diagnosis of viral infections, including dengue. The performance of PCR for the amplification of pre-defined dengue genomic sequences greatly depends on the specificity of pre-designed primer sequences, in order to conveniently target the desired genomic regions. One of the biggest challenges in molecular diagnosis of dengue viruses is the high variability among the genomic sequences of the different serotypes, but the 3′-noncoding region (3′-NR) of dengue virus RNA, of near 400 bp, has been used as the most favorable target for this purpose. The 3′-NR contains the most conserved sequences among all the serotypes, as well as serotype-specific sequences. It can thus be used to simultaneously amplify, by RT-PCR, all dengue serotypes in a given clinical sample, and to build specific primer sets to identify and quantify each one of those serotypes, as shown in a study employing highly sensitive and specific fluorogenic detection [13]. A major challenge for early dengue diagnosis by RT-PCR is the small (up to one week) period available for successful detection of viremia. Infection with a given dengue serotype usually confers life-long immunity against that serotype but enhances susceptibility to the remaining. Thus, accurate identification of prevalent circulating serotypes and genotypes may be crucial for predicting epidemiological implications and patterns during and after epidemic outbreaks. RT-PCR is even able to rapidly detect new infecting serotypes in endemic areas [14]. Therefore, in the context of epidemiological and evolutionary studies, PCR-based techniques (such as nested PCR, single-tube multiplexed PCR or even real-time automated PCR [15]) find extreme usefulness where serology fails [3]. However, PCR-based methods need stringent quality control and meticulous manipulation of biological specimens to avoid false-positive results due to easy unwanted amplification of contaminants. Additionally, serum samples for RT-PCR assays must be stored at extremely low temperatures due to the intrinsic liability of dengue RNA genome, which is not feasible in many endemic areas.
2.2. MAC-EIA
Measurements of non-structural dengue proteins, mainly the NS1, and of specific immunoglobulins G (IgG) or M (IgM) dengue antibodies, have been frequently chosen as the preferred serological tests. Typically, dengue infections elicit stronger and more specific IgM than IgG responses in primary flaviviral infections and in individuals never immunized before with a flaviviral vaccine, while the opposite happens in secondary infections. On this basis, EIA is suitable to distinguish between primary and secondary dengue infections based on the IgM/IgG ratio. On the other hand, serum is a suitable biological fluid for testing since it is highly stable at tropical temperatures even after prolonged exposure [3]. Accordingly, colorimetric IgM MAC-EIA is the most widely used technique for dengue serological diagnosis. Classical MAC-EIA assays are usually cheaper, simpler, faster and more informative than other serological tests [16]. However, the MAC-EIA test is not always available in resource-limited healthcare settings. Moreover, the cost of these lab-based assays, in general, is still too high for purchase by developing countries where dengue exists, especially if point-of-care is envisaged, including by end-users. In particular, IgM-based assays are widely available for utilization in dengue-endemic countries, although they should not be used as confirmatory tests for current disease (especially in the first days of illness [17]), since IgM antibodies only appear in the blood at least five days upon infection and persist thereafter for 2–3 months. Being IgM a biomarker for acute infections and IgG for previous (primary) infections, concomitant detection of both may enhance diagnostic accuracy [18]. In conclusion, classical serological techniques for dengue diagnosis have, in general, low sensitivity, high complexity and long assay times, reducing their usefulness during the first days of illness. Many commercial diagnostic kits based on MAC-EIA for dengue diagnosis are available [19], [20], being the predominant type of immunossays in most clinical settings [21]. Dengue NS1 antigen-based kits, in particular, have proven to be highly valuable for rapid on-site screening of dengue-infected travelers in international airports, including in countries where dengue is not endemic [22]. This is of utmost importance to quickly forward these patients for medical treatment and follow-up in healthcare units, a possibility to drastically diminish the burden of mosquito control measures. Most of these test kits have limited sensitivity in patients with acute febrile illness in endemic areas [23], [24]. Related rapid test kits claiming enhanced rapidity, sensitivity and specificity have also been cited elsewhere [25], although further improvements in detecting low levels of NS1 antigens in acute-phase serum samples from patients with secondary infections and with several strains (of all four serotypes) circulating in different geographical regions are still required to increase the reliability of these tests. Many of these kits may still lack validation and standardization, for which multi-institutional studies may be necessary [26]. Interpretation of serology results frequently is not conclusive because of misjudgment with other febrile illnesses, especially other flaviviroses (e.g., yellow fever and viral encephalitis), in secondary infections (due to cross-reactivity with serotype-specific dengue antibodies from previous infections) and in individuals previously vaccinated against yellow fever or Japanese encephalitis (also due to cross-reactivity) [3]. Serological analysis may be useful for diagnosis of dengue infections and for surveillance purposes when used together with clinical and epidemiological data [27]. To be confirmatory by themselves, although of low utility in early diagnosis, serological tests usually require testing paired serum samples (collected in the acute and convalescence phases), which is more tedious and time-consuming [28]. In general, MAC-EIA and RT-PCR assays require a range of laboratory facilities and skilled manpower that are prohibitive for point-of-care testing in resource-limited settings.
3. Biosensors and rapid tests for biodiagnostics
3.1. Biosensors and rapid tests: clarifying concepts
Fundamental scientists and engineers involved in research and development of biodiagnostic devices and techniques usually employ the term biosensor to describe them, while the medical community and people in general know them preferentially as rapid [diagnostic] tests. Rapid tests are usually seen as simple and inexpensive devices for qualitative or even quantitative biodetection, and able to be used not only by non-specifically skilled healthcare professionals within clinical settings, but also by the end-users (patients) themselves, in domestic and in-the-field contexts. The term biosensor is usually applied to prototypes of new bioanalytical devices, frequently at and in-the-bench development stage (proof-of-concept demonstration) only. The concept has also been used to describe new biosensing techniques and methodologies, even though no commitment exists towards further development of a fully automated and (desirably) portable diagnostic device. As such, as any bulky analytical technique for laboratory analysis, biosensors have been commonly tailored to be highly sensitive, specific, fast and response-proportional [29] (this last requirement, however, is losing importance as ongoing advances in signal processing technology proceed). Although very common in the literature, specific characterization of biosensors based on high sensitivity and selectivity is a questionable issue since, in general, all analytical techniques and devices ultimately envisage this goal. Only few biosensor schemes (research level) proceed towards prototype and device production (development level) and, not surprisingly, the concept of biosensor has been restricted mainly to the laboratory, hence belonging to a scientifically limited domain. Many obstacles arise when attempting to go further, i.e., to mass-production and commercialization. In fact, the still limited availability of commercialized biosensors may be due mainly to a lack in the appropriate technology for cost-effective manufacture than a lack of fundamental (scientific) knowledge [3]. Moreover, the ‘real world’ of biosensor markets fort Health, especially for in-the-field diagnosis in developing countries, poses hard challenges, especially the demand for high-throughput testing and related mass-production. Thus, the common high-performing standard paradigm of conventional laboratory-based analytical devices, usually relying in high sensitivity, selectivity and reproducibility, tends to shift, in the point-of-care, towards the pursuit for simpler, cheaper and faster diagnostic tests. These favorable and desirable characteristics of rapid diagnostic tests for field applicability are frequently achieved at the expense of decreased analytical accuracy (although maintaining reliability, at least at a certain level). Ideally, these tests must be produced in the form of portable (miniaturized), automated (self-powered) readout devices coupled to disposable peripherals (e.g., test strips) where the biorecognition event occurs. Disposability is necessary to avoid contamination when dealing with infectious agents, which guarantees the commercial viability of cheap microfabricated devices for clinical diagnosis [30]. However, the importance of prior purification and chemical modification steps has been relatively underestimated in the development of such simple and inexpensive tests, which is especially relevant in the case of complex biological samples. In the case of DNA detection in clinical samples, there is still the problem of the usual very low levels of DNA. Human genomic DNA levels in blood, for instance, may be 1014 fold that of a pathogen DNA from a typical blood infection, which may significantly hamper achieving good selectivity levels [31]. Suitable inclusion of these sample preparation steps has thus been undertaken in disposable chip-based and microfluidic (lab-on-a-chip) device formats (see following section for more details). Obviously, these devices do not entirely comply with the exquisite requirements of ideal rapid diagnostic tests, constituting rather a somewhat mid-term between these and heavy laboratory analytical devices. It is expected, however, that simultaneous compliance with point-of-care requirements and analytical reliability may define a desired balance towards the production of improved devices for successful in-the-field applicability. In short, the distinction between biosensors and rapid diagnostic tests is not always straightforward. In this paper, some care was taken to assign the concepts of biosensors and rapid tests to, respectively, in-the-bench and fully developed analytical devices. However, usually it is not easy to assess the exact developmental stage of these devices, the reason why establishing such a distinction was not a major concern ahead.
3.2. Micro- and nanofabrication
On the past few decades, advances in micro- and nanofabrication techniques based on the planar silicon industry of integrated circuits have leveraged the development of disposable (single-use) chip-like biosensors with improved performance (namely in terms of sensitivity and selectivity) and amenable for point-of-care testing as a way to overcome the usual problems of low stability (normal room temperature usual limits biosensor shelf-life up to some months [32]), difficult sterilization (due to labile elements) and modest selectivity (due to interfering substances present in the complex biological samples) of current biosensors. Many of these tests have been produced in the form of simple immunochromatographic strips for inexpensive and easy visual (colorimetric) detection, usually requiring dried-stored reagents and no additional instrumentation. Miniaturization trends have greatly obviated the usual limitations of lab-based techniques, by providing lower consumption of power, sample volumes and costly and hazardous reagents, shorter assay times, lower operating costs and improved portability compared to bulky laboratory methods [33], which is especially important for point-of-care diagnosis in the tropical and developing countries where dengue is endemic. High-throughput microarray technology, in particular, has proven to be very useful for simultaneous testing of dengue virus and other arboviruses circulating in a given region, as well as for screening pathogens responsible for the occurrence of dengue-like symptoms [34]. Nevertheless, the standard technology is still too expensive and complex for biological analysis, not only for point-of-care usage, but even for routine laboratory testing. Further limitations of current microarrays include strong sample concentration dependence, limited resolution at high biomolecule densities and difficult array scaling-down [35]. More recently, lab-on-a-chip devices have emerged as second-generation chips. By suitable integration of modules for sample processing and analysis in a single device, lab-on-a-chip schemes offer enhanced flexibility and discriminatory ability over conventional biosensors and chips, although minimization of the sample preparation procedure is an ultimate goal towards device simplification and easiness-to-use. Very often, lab-on-a-chip devices also include a prior calibration step before each measurement [11]. DNA analysis, in particular, usually requires extensive sample processing. Taking in consideration that DNA is the most easily amplifiable biomolecule, a module for DNA amplification by PCR-based techniques is usually required in microfluidic devices, owing to the extremely small concentrations of DNA in biological fluids. To be used as probes in DNA biosensors, short oligonucleotides yield relatively exact sequence identification, while longer DNA fragments provide higher tolerance for sequence mismatches and hence improved ability to detect diverged strains [34]. PCR-free (i.e., unamplified) DNA detection has been pursued along with DNA chip development, but only with moderate success. Progresses in this area may be extremely useful for accurate biomolecular detection of dengue infections and for unambiguous serotype identification as well. However, current lab-on-a-chip devices, despite being simpler than conventional laboratory analytical apparatus, still require complex and somewhat expensive fabrication procedures onto plastic or glass substrates, thus severely limiting their affordability and availability by poor countries. Future development of new biosensor platforms may eventually benefit from materials with improved analytical performance (e.g., paper-based materials), especially in terms of enhanced sensitivity [36], as well as simplicity and cost-effectiveness to allow wide utilization in developing countries, as those affected by dengue disease.
4. Biosensors and rapid tests for dengue molecular diagnosis
The sub-sections ahead refer to an overview of the available literature about reported methodologies and devices for detection and identification of specific biomolecules (both from clinical and synthetic origin) related to dengue virus. The three first sub-sections concern the three main types of physical transduction mechanisms (electrochemical, optical and piezoelectric), while the fourth compiles the information concerning some performance parameters of these biosensors.
4.1. Piezoelectric
Among the reported dengue biosensors in the literature is an immunochip employing two specific monoclonal antibodies, immobilized into a piezoelectric transducer, for detection of glycoprotein-E and NS1 protein from DENV2 [37]. Multi-antibody coating allows the capture of several different antigens, which increases the overall mass and thus the detection signal. This layout exhibited 100-fold higher sensitivity than conventional EIA and a short response time (30–60 min). In another work, instead of conventional antibody immobilization onto a quartz crystal microbalance (QCM) surface for piezoelectric detection of antigens, a thin film of molecularly imprinted polymers (MIPs) specific for the dengue viral NS1 protein was formed by polymerization of monomers onto a QCM chip surface [38]. In this work, which claims the first application of MIPs for clinical early diagnosis of dengue-infected biological samples, the polymerization process leaded to the formation of aminoacid-like cavities that selectively recognize the epitope site of the NS1 dengue protein (Fig. 1 ). These artificial receptors, apart avoiding the need for synthesizing monoclonal antibodies, provide improved assay specificity by avoiding both contaminant anchoring and non-specific binding of NS1 protein molecules to the imprinted analyte-specific polymer cavities [39]. In addition, clinical samples often do not require dilution or other pretreatments for effective analysis [30]. Antigens from all the four serotypes (confirmed by PCR and/or virus isolation) were successfully detected. This MIP-QCM chip, whose results agreed with those of conventional EIA, was able to diagnose dengue infection in less than 1 h and exhibited good sensitivity and specificity when compared to reported monoclonal antibody-based chips for DENV2 [37], [40]. In the future, the use of subgroup-specific glycoprotein-E cross-reactive epitopes as the template may be useful to distinguish dengue serotypes. More recently, a piezoelectric genosensor for detection of a DENV2 oligonucleotide sequence obtained by RT-PCR was developed, through specific hybridization with the complementary oligucleotide immobilized onto a QCM surface [41]. Amplification of the detection signal was achieved through interconnected oligonucleotide-attached gold nanoparticles (GNPs) hybridized on a stepwise, layer-by-layer process with the gold surface-grafted oligonucleotide duplex. Since each primary binding event is amplified through successive hybridization events between complementary oligonucleotides (probe and target), not also the detection sensitivity, but also the specificity of DNA hybridization, are greatly increased. DENV2-specific probes were tested against all the four serotypes, but only responded positively to the homologous genomic region of DENV2. This QCM method was able to detect DENV2-specific RNA plaque titers as low as 2 PFU ml−1 and exhibited a linear concentration range up to 2 × 106 PFU ml−1. Moreover, sensitivity and specificity comparable to those of real-time PCR methods were claimed. In addition, the method is label-free (thus avoiding the usual drawback of signal lost by gradual leakage of the label) and does not require expensive equipment, certainly valuable features for in-the-field applications. In spite of all these methods, piezoelectric biosensing in liquid phase is prone to complex interactions between interfacial parameters [42] and to innumerous environmental interferences over the detection signal (including small air fluctuations), the reason why it is only hardly applicable outside laboratory facilities, in highly uncontrolled conditions.
Fig. 1.
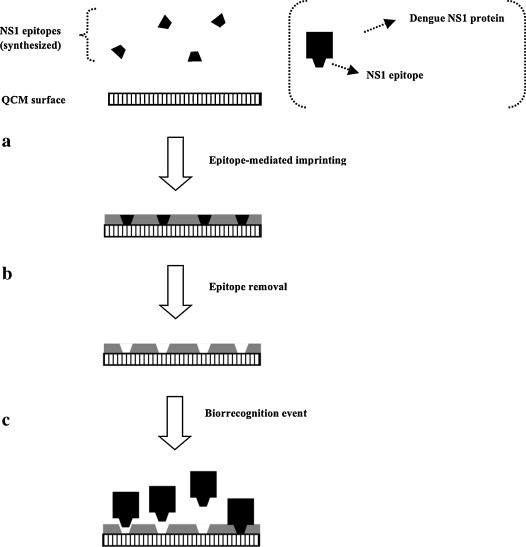
Scheme of a MIP-QCM biosensor for specific detection of the NS1 dengue protein antigen. (a) Synthesized NS1 dengue protein specific epitopes are used as templates for oriented imprinting within a suitable MIP matrix. (b) After matrix polymerization and concomitant imprinting, epitopes are removed, leaving exposed imprinted cavities in the polymer surface. (c) Whole NS1 dengue protein molecules can now be specifically recognized and bound from a clinical sample.
4.2. Optical
Recently, self-assembled monolayer (SAM)-based covalent immobilization of a biorreceptor conjugate of a dengue antigen and bovine serum albumin (BSA) was performed onto a gold chip surface [43]. In this immunoassay for detection of specific IgM dengue antibodies in serum through indirect competitive inhibition, BSA, in a typical application, was employed as a blocking reagent for minimization of non-specific binding of interferents that typically exist in clinical samples, thus improving detection selectivity. The work also employed a surface plasmon resonance (SPR), by measuring an increase in the resonance angle of the surface-deposited sample in the presence of dengue virus. The SPR technique has been useful as a way to circumvent the typical signal resolution of optical fibers and other photonic devices. Unlike the traditional immunoassay formats, SPR allows real-time and label-free analysis. Nevertheless, miniaturization of optical and electronic components of SPR-based devices is still a difficult issue aiming point-of-care applications [44]. Instead of the common methods that use an oligonucleotide probe with a covalently linked label (e.g., a fluorophore), a cheap and simple dengue biosensor employing a liposome-labeled universal reporter-probe (to bind any of each dengue serotype-related targets) and four sequence-specific probes (one for each serotype) was reported [45], where the liposomes allowed signal amplification. Rapid electrochemiluminescent detection (within 15 min) and serotype-discrimination after dengue genome amplification were obtained after nucleic-acid sequence-based amplification (NASBA), a PCR-based technique for thermal-cycling amplification of RNA. A significant advantage of electrochemiluminescence over fluorescence is that it is not affected by a background optical signal coming from the biological sample, which makes the technique very useful for detecting low levels of biomolecules. The sandwich technique integrates a liposome-coupled reporter-probe and a membrane-immobilized biotinylated capture-probe, such that the amount of tagged liposomes is proportional to that of sample RNA. Unlike fluorescence, chemiluminescence does not require an excitation optical source and is not subjected to quenching phenomena. Moreover, it is also highly sensitive and relatively low costly [46]. In parallel, liposome-based signal amplification provides an excellent signal-to-noise (S/N) ratio since liposomes carry many fluorophore molecules. Unfortunately, NASBA requires careful handling and storing of molecular targets, since RNA is inherently much more liable than DNA. Although this results in decreased risk of contamination, it also limits the use of RNA amplification-based techniques for in-the-field applications, including epidemiological studies [47]. In the study, ‘cold probes’ (unlabeled oligonucleotide probes) were used against serotypes 1 and 4 to prevent false-positive signals due to cross-reactions from these serotypes. Given the fact that DENV1, 2 and 4 were correctly identified, whereas DENV3 exhibited significant cross-reactivity with serotypes 1 and 4, designing a ‘cold probe’ for DENV3 would be a valuable contribution for more accurate DENV3 identification. The performance of three available MAC-EIA schemes (chemiluminescent, colorimetric and commercial Panbio®) [48] was compared with that of a pioneering chemiluminescent optical fiber immunosensor for detection of DENV2 IgM antibodies in serum samples. The immunosensor exhibited good specificity and excellent sensitivity, with a lower detection limit than the three MAC-EIA formats and sensitivity at least 10–100 higher than that of the respective colorimetric test, which is of particular interest for diagnosis of asymptomatic dengue disease. Rapidity was also higher with the chemiluminescent immunosensor than with chemiluminescent MAC-EIA. However, difficulties in surface binding might explain the poorer reproducibility of the optical fiber sensor compared with that of MAC-EIA. Microsphere-based immunoassays with covalent binding between an antigen or antibody to microspheres (microbeads) have been considered promising alternatives to enzymatically labeled MAC-EIA owing to their rapidity and ability for multiplexed serological analysis, as well as to the possibility of testing optically opaque samples [34]. Magnetic particles allow improved detection specificity by magnetically controlled removal of nonspecifically bound magnetic beads (magnetic washing) [49], thus avoiding the elimination of the time-consuming washing step of unspecifically bound molecules. A biosensor fabricated through standard complementary metal oxide semiconductor (CMOS) manufacturing was developed, in a format analogous to EIA, for detection of dengue IgG in clinical serum samples via magnetic labels [21]. The sensitivity of this disposable prototype, in the form of an array sensor chip, was similar to that of EIA. This platform, built by inexpensive and straightforward mass production technologies, was claimed to be easily applied to existing EIA protocols and reagents. Unfortunately, magnetic washing and the intrinsic variability of commercial magnetic beads may decrease the sensitivity of detection. An innovative microfluidic immunoassay system for simultaneous detection of dengue IgM and IgG operates under flow-injection analysis (FIA) mode and contains a sample pre-treatment module to enhance sensitivity [18]. The system uses magnetic beads attached to capture antibody/dengue virus complexes that specifically recognize serum IgG or IgM. After on-chip magnetic isolation of these sandwich-based bead complexes, specific fluorescent-labeled secondary IgG or IgM dengue antibodies are bound and the whole complexes transported into the detection module. Purification of these complexes from serum samples is responsible for improved detection limit (21 pg) and assay time (within 30 min). A slight variation of the liposome-based electrochemiluminescent setup described above was reported, referring the immobilization of a magnetic sphere/capture probe complex onto a permanent magnet in a detection region, and built in a microfluidic format [50]. Liposomes were filled with dye molecules for detection of a synthetic DENV3-related RNA sequence by fluorescence microscopy, with inherent signal amplification (and hence to increased sensitivity). Based upon these two layouts, an optical biosensor with a polyethersulfone membrane-immobilized universal reporter-probe and dye-filled liposomes was developed for dengue genome detection. This scheme was applied to the specific identification of the four dengue virus serotypes in a single multianalyte assay, instead of four independent assays [51]. The design of the multi-analyte sensor was based on a prior investigation about cross-reactions between the four serotypes. This was undertaken with four independent, serotype-specific single-analyte biosensors (i.e., with single capture zones). Among them, biosensors for serotypes 2 and 4 were highly specific, whereas for serotypes 1 and 3 showed cross-reactivity with DENV4. In this study (the final multi-analyte biosensor) with amplified dengue virus RNA sequences from all serotypes, 92% reliability (correct serotype identification) was obtained. This biosensor was also successful in detecting concurrent infecting serotypes from mixed clinical samples. By taking advantage of this previously known technology for immunochromatographic RNA detection, the same sandwich scheme was further employed in a lab-on-a-chip format comprising sample preparation and detection, with separated modules for cell lysis, RNA purification, NASBA and detection [33]. The system showed to be feasible for detection of messenger, ribosomal and genomic RNA. In this way, liposomes, reporter probes and beads were incubated with amplified target prior to introduction of the mixture into the microchannels where sandwich complexes were then magnetically captured and detected by fluorescence microscopy. Unlike in the previously described biosensors, a nonionic surfactant was used to easily disrupt liposomes; in this way, rapid releasing of dozens of thousands of fluorophore molecules occurs to the bulk solution, thus resulting in a remarkable increase of the fluorescent signal owing to the elimination of fluorophore self-quenching. This large signal amplification for each binding event provides very sensitive, quantitative and fast detection of biological targets. In another work, this setup was successfully applied to accurate RNA identification, after amplification, of all four dengue virus serotypes, within only 20 min [52]. Identification of serotypes 2, 3 and 4 was unambiguous, but DENV1 cross-reacted significantly with DENV4. The assay was previously optimized with a synthetic DENV3 RNA fragment and achieved similar sensitivity to that of standard electrochemiluminescence detection [47]. In addition, the lysed liposome approach was compared to the intact liposome one, with the first achieving a slightly better performance. However, the lysed liposome strategy has an intrinsically more complex design than that of intact liposomes, since no second channel and sample flow switching are needed. Solving this problem may implicate turning back to the intact liposome layout, although with a decreased fluorophore concentration, in order to lower self-quenching, so enhancing the signal for intact liposomes. Unfortunately, fluorescent and other optical transducers usually require expensive and complex microscopes and electronic peripherals. This has hindered cost-effectiveness and miniaturization of these devices towards truly point-of-care diagnostics. Very often, the ultimate limit is single molecule detection at the sub-milliliter level, which dramatically increases the probability for occurrence of false-positive or false-negative results. Therefore, techniques able to directly detect unamplified (intrinsic) analytes or signals from clinical samples while still envisaging single-molecule detection still lack. With this aim, single dengue viral particles were detected in serum samples, through binding to a dengue-specific antibody, with a microfluidic array coupled to detection by fluorescence cross-correlation spectroscopy (FCCS) [53]. The microfluidic array confined the dengue bulk solution into femtoliter volumes, whereas the recently developed FCCS technique [54] was able to detect single binding events between dengue virus particles and specific dengue antibodies. The method is based in cross-correlation between fluorescence spectra fluctuations from the virus and the target, both labeled with fluorophores that exhibit different colors upon excitation. Perfect cross-correlation occurs when the two differently colored reactants bind each other (Fig. 2 ). FCCS derives from fluorescence correlation spectroscopy (FCS), a technique for single molecule detection and simultaneous characterization of molecular interactions in homogeneous solutions [55]. The use of FCCS permits to surpass the two main limitations of auto-correlation analysis provided by FCS. These include limited sensitivity for low concentration species (since the diffusion time of the virus/target complex is larger than that of the free target, and hence the auto-correlation profile of the mixture is almost exclusively derived from unbound target) and dependence on the geometrical size of the reactants, caused by undistinguishable formation of the virus/target complex (true events) or target self-aggregation (false positives). In conclusion, given the reliability of cross-correlation for biosensing of probe-target complex formation, the above mentioned study with FCCS integrated with a high-throughput microfluidic device proved the usefulness of this method for ultra-low detection of single virus particles without any amplification procedure. However, clinical usefulness of this strategy implicates extension of single viral particle detection in femtoliter sample volumes to the microliter/sub-milliliter domain (typical of clinical samples), in order to reach improved sensitivities in the 10−18–10−21 M range [53]. Furthermore, the technique suffers from the basic constraints of optical detection, particularly expensiveness and high complexity of analytical apparatus, and complex data processing.
Fig. 2.
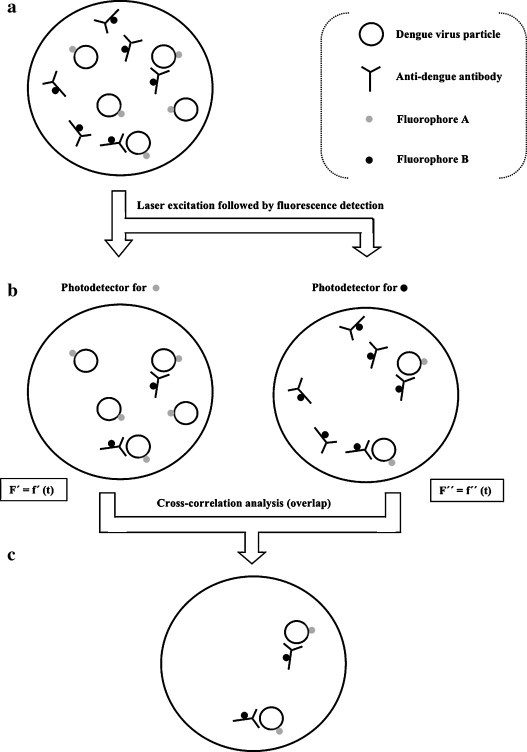
Principle of FCCS applied to the detection of specific interactions between dengue virus and an anti-dengue antibody. (a) Viral particles and antibodies are tagged with fluorophores of different colors and distinct fluorescence spectra (attachment of these dyes does not affect normal virus/antibody interactions). (b) The sample is irradiated at the excitation wavelengths of both dyes (with the same or different laser sources). The emission light beam is split and then two photodetectors (tuned for the emission wavelengths of each one of the dyes) monitor the sample simultaneously. Each photodetector detects both its corresponding particle-labeling dye and the interaction, by measuring fluorescence fluctuations with time. (c) Cross-correlation analysis between the two fluorescence spectra reveals solely the virus/antibody interactions. Cross-correlation analysis allows determining the amount and kinetic parameters of the interacting species.
4.3. Electrochemical
Among reported electrochemical biosensors is a suitable combination between a potentially disposable microfluidic cartridge, an interdigitated ultramicroelectrode array and a minipotentiostat for amperometric detection of NASBA-amplificated DENV3 RNA [56]. In this sandwich hybridization assay, reporter probe molecules were coupled to redox label-entrapping liposomes for signal amplification and capture probes were coupled to superparamagnetic microbeads for magnetic isolation with a magnet in the detection region. Upon capture, lyposomes were lysed for releasing of the redox marker. Compared to a standard lab-bench potentiostat, the integrated minipotentiostat reached a detection limit 10 times lower. This small, portable and inexpensive device is therefore a viable alternative to similar schemes with fluorescent detection [33], [50]. Another work reported a genosensor employing ferrocene as an electroactive hybridization indicator for preferential attachment to single-chain DNA. The resulting unequal ferrocene concentrations near the electrode surface and therefore the distinct electrochemical responses were used for simple electrochemical detection, through cyclic voltammetry, of a dengue-related oligonucleotide synthetic sequence with a chitosan-coated glassy carbon electrode [57]. A more recent electrochemical biosensor employed concanavalin-A immobilized onto a gold electrode through GNPs and polyvinyl butyral [58]. Electrochemical impedance spectroscopy (EIS) is a sensitive and effective technique to probe the interfacial properties of modified electrodes, and was used in this work to compare confirmed dengue fever, DHF and non-infected sera samples. The distinctive patterns of impedimetric responses observed were ascribed to different glycoprotein patterns in the different sera. As with other lectins, the specificity of concanavalin-A glycoprotein for carbohydrate moieties can be used for developing specific biosensors for sugars. Therefore, this biosensor is potentially applicable on the recognition of different serum glycoprotein patterns. In conventional EIS, the toxic redox-couple [Fe(CN)6]4−/3− is usually added to the electrolyte solution for measurement of a faradaic process, although its long-term presence may reduce the bioactivity of a protein layer immobilized onto an electrode surface [59]. This is certainly a favorable point to the utilization of non-faradaic immunosensors without added reagents for point-of-care applications [60]. Accordingly, a recent work describes the development of a non-faradaic, label-free EIS biosensor with thermally inactivated dengue virus, from serotype 2, immobilized onto sol–gel derived barium strontium titanate (BST) thin film pre-coating interdigitated electrodes modified with an organic SAM [59]. In the co-planar symmetric structure, both finger-like electrodes operate as a single working electrode. The BST thin film acts as a middle dielectric layer in the sandwich capacitor, i.e., as an effective sensing medium. In this manner, the BST film provides better coupling for the electrical field on the sensing surface rather than penetrating deep inside the electrolyte. This biosensor rendered signal outputs directly correlated with serum concentrations of dengue antibody, while signal changes upon capture of the antibody are attributed to surface conductivity changes. In fact, an increase in interfacial capacitance can be observed after biomolecule immobilization on the electrode surface. Unlike in conventional electrochemical immunosensors, this sensor uses only deionized water as the electrolyte, and is suitable for integration with external electrical measurement circuits and microfluidic channels. In a similar detection scheme with a field-effect transistor (FET) biosensor, a 69-bp DENV2 RT-PCR amplicon was detected through a specific peptide nucleic-acid (PNA) covalently immobilized onto a Si nanowire surface [61]. In such Si wire-base sensor, PNA probes are responsible for the accumulation of negative charges on the nanowire surface upon specific hybridization with the complementary DNA sequence. Thus, the resulting increase in electrical resistance may be attributed solely to the negative charges of such DNA sequence (Fig. 3 ). The label-free biosensor reached a detection limit below 10 fM within 30 min and sensitivity more than 1000 times higher than the reported method using 32P-labeled probes [62] and more than 10,000 times higher with ethidium bromide staining [63]. The ability to operate in a simple non-sandwich and multiplexed format makes this method suitable for early detection of dengue virus with good sensitivity and specificity. However, FET sensors usually only achieve moderate sensitivity due to significant fluctuations of the interface potential in aqueous environments [64]. Impedance spectroscopy is a time-consuming technique due to the need for recording a full spectrum within a broad frequency region, and hence its use for bioanalytical purposes has been restricted mainly to characterization purposes and as an auxiliary technique [65]. However, impedimetry-based biosensors may find a market where low cost, portability and analysis speed are required and moderate sensitivity is sufficient [66].
Fig. 3.
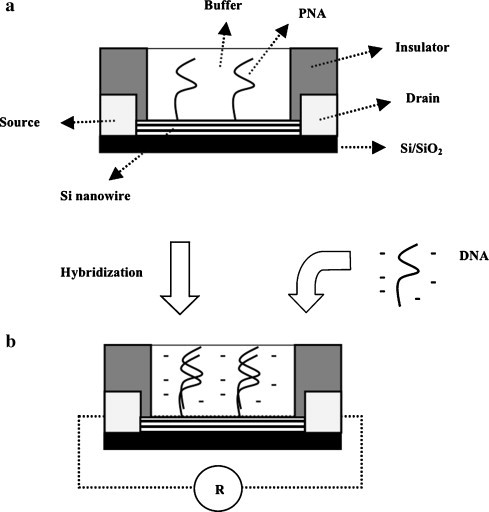
Lateral view scheme of an impedimetric genosensor based on Si nanowire array/PNA. (a) Neutral PNA molecules are grafted on the surface of Si nanowires by single-point attachment. (b) After specific hybridization with the negatively charged complementary single-stranded denatured RT-PCR product of dengue virus genome amplification, accumulation of negative charges (solely derived from the dengue amplicon) near the conducting Si nanowire surface leads to an increase in electrical resistance between the electrodes.
4.4. Performance parameters
Many performance parameters are usually employed to characterize, evaluate and standardize biosensing devices and methodologies. Table 1 presents a compilation of the claimed and reported LODs and assay times among some of the above mentioned biosensors for dengue (and cited in the literature). Despite the significant differences of LODs and even distinct forms of presenting them, significantly higher values can be observed for piezoelectric than for optical or electrochemical biosensors. It can also be observed a slight tendency of most recently reported biosensors for exhibiting lower LODs. However, as for other infectious diseases, attomolar-range LODs for successful detection of dengue viruses in unamplified biological samples must be achieved, although this is still beyond the fundamental limits of common sensing devices.
Table 1.
Reported LODs and assay times for dengue biosensors referenced in the literature (discriminated according to the transduction mechanism and probed analyte).
| Transduction mechanism | Analyte (target) | Limit of detection (LOD) | Assay time (min.) | Reference |
|---|---|---|---|---|
| Piezoelectric | NS1 protein and glycoprotein-E | 0.05 μg mL−1 | 30–60 | [31] |
| NS1 protein | 1–10 μg L−1 | 20–30 | [32] | |
| NS1 protein | 1.727 μg mL−1 | – | [35] | |
| Glycoprotein-E | 0.740 μg mL−1 | |||
| cDNA | 2 PFU mL−1 | – | [36] | |
| Optical | IgM and IgG | 21 pg | 30 | [15] |
| RNA | 1 nM | – | [41] | |
| IgM | 106 dilutions | – | [43] | |
| RNA | 10 pM | – | [45] | |
| RNA | 50 molecules (DENV2) | 25 | [47] | |
| 500 molecules (DENV3–4) | ||||
| 0.08 attomole (DENV1) | ||||
| RNA | 0.125 nM (intact liposomes) | 20 | [48] | |
| 50 pM (lysed liposomes) | ||||
| Viral particle | Single particle | – | [50] | |
| Electrochemical | RNA | 0.01 μM | – | [53] |
| cDNA | 10 fM | 30 | [58] | |
cDNA, complementary DNA.
5. Evaluation of diagnostic tests for dengue
The performance of diagnostic tests strongly depends on local conditions in which they will be used, namely differences in the characteristics of the population or the infectious agent (e.g., infection prevalence, genetic variation of the pathogen or host) and peculiarities of the test (e.g., methodology and local diagnostic practices) [67]. Therefore, the evaluation of diagnostic tests should ideally be carried out in such particular conditions, including multi-centre trials. Obviously, such test conditions will eventually determine the particular in-the-field situations in which the tests will be used. As an example, strong cross-reactivity may be acceptable if there is low prevalence of the disease in such place or setting [27]. The usual lack of proper validation and standardization of many rapid diagnostic tests for both in-the-field and laboratory usage is partially due to the fact that most laboratories use in-house rather than well-standardized assays and techniques, which strongly discourages compared analysis in aggregate. It is thus important that national or reference healthcare entities establish quality assurance programs that guarantee the production of reference materials and protocols for effective quality control of rapid diagnostic tests [34]. In this regard, the Special Programme for Research and Training in Tropical Diseases (TDR) (sponsored by UNICEF, United Nations Development Programme (UNDP), World Bank and WHO) and the Pediatric Dengue Vaccine Initiative (PDVI) have underpinned an extensive collaborative program aimed to evaluate the performance of commercially available diagnostic tests for dengue, especially IgM-based EIA assays and rapid diagnostic tests [27], mainly on the basis of comparison among them. A specific goal was the assessment of the different patterns of immunoglobulin antibody responses, especially when envisaging accurate diagnosis of acute dengue infections. To accomplish this task, the tests were assayed against a panel of well-characterized serum specimens from persons with confirmed dengue infections and from others with confounding infections and conditions. The inclusion criteria were positive identification of all four dengue serotypes, correct identification of primary/secondary infections and discrimination from other flavivirus and dengue-like diseases, high specificity towards systemic disorders and good performance for various geographical conditions. The evaluation comprised four EIA tests and the rapid tests DuoCassette (Panbio Diagnostics), SD Bioline (Standard Diagnostics) and Dengucheck (Zephyr Biomedicals, India), which are all immunochromatographic tests, and the HapalyseM (Pentax Corporation, Japan), a particle agglutination-based test. Three of the EIA kits showed good agreement with reference standards, although, in general, have rendered false-positive results for dengue IgG-positive, rheumatoid factor-positive and dengue IgM-negative/malaria-positive serum samples. In this last case, test usage in settings or regions where both dengue and malaria are endemic must be discarded. The HapalyseM test exhibited sensitivity comparable to the high-performing EIA tests, although the specificity was somewhat inferior to those of the remaining rapid tests. The Dengucheck test exhibited, by far, the lowest sensitivity. The specificity of the lateral flow-based rapid tests was similar to that of EIA tests. Among the tested rapid tests, the three lateral flow tests reached higher classifications than the Hapalyse M or the EIA tests concerning ease-to-use. Even so, to all the four rapid tests, difficult result interpretation (basically due to weak bands in the strip) was claimed (Table 2 ). As a whole, it can be stated that the performances (in terms of sensitivity and specificity) of the rapid tests did not agree with those of reference standard assays as much as those of EIA tests. One major limitation of these evaluation studies is the presumable underestimation of test specificity; in real situations, it is unlikely that all potential causes of false-positive results would be present simultaneously. Moreover, since the four dengue serotypes were not represented in equal proportions in the sample panel (in particular, the small number of DENV4 samples reflected its rarity in most regions), the evaluation of some kits towards primary infections caused by all four serotypes could not be completely assessed. Since most rapid diagnostic tests are targeted to be used by the patients themselves (who, obviously, lack the expertise to properly evaluate and assess the performance of such tests), the Standards for Reporting of Diagnostic Accuracy (STARD) initiative developed a sequenced checklist to guide adequate report of the results from evaluation studies on diagnostics [68]. Similarly, and recognizing the enormous potential threat of dengue disease introduction in Europe, the European Union started, in 2006, the DENFRAME project, aiming to improve the management of dengue disease among human populations, especially in Asia and Latin-America [69]. One of its working programs involves the development, standardization and automation of novel rapid diagnostic test kits for dengue diagnosis. A complete validation program must include a multi-centre evaluation, which ensures accuracy and robustness of the test, as well as reproducibility of the results within and among different assays and lots. Taking into account the seriousness of clinical applications and implications of rapid diagnostic tests, a full evaluation program of these tests must include, apart performance evaluation (validation), the additional steps of quality control and quality assurance [11]. Quality control guarantees correct functioning of the devices with prolonged use, but traditional procedures normally employed for traditional laboratory devices are not feasible for disposable rapid tests. In such cases, randomly selected units from any production batch of a lot must be assayed and compared to infer reproducibility. A final quality assurance program, on the other hand, demonstrates the readiness of the tests to be introduced in routine clinical diagnosis. Institutional evaluation programs, like those mentioned and described above, are thus meritorious attempts for finally establishing some order and comparison criteria among the vast portfolio of rapid diagnostic tests already available – although still lacking standardization – for dengue and other infectious diseases.
Table 2.
Main characteristics of rapid diagnostic tests for dengue and results of the TDR/UNICEF/UNDP/World Bank/WHO/PDVI evaluation program.
| DuoCassette | HapalyseM | SD Bioline | Dengucheck | |
|---|---|---|---|---|
| Probe (antigen) | Recombinant DENV1–4 | DENV1–4 | Recombinant DENV1–4 glycoprotein-E | Recombinant DENV1–4 (undetermined serotype) |
| Target (antibody) | IgM and IgG | IgM | IgM and IgG | IgM and IgG |
| Sensitivity (%)a | 78 | 98 | 61 | 21 |
| Specificity (%)a | 91 | 77 | 90 | 87 |
| Assay time (min.) | 15 | 90 | 15–20 | 15 |
| Requires additional equipment | No | Yes | No | No |
| Requires low-temperature storage | No | Yes | No | No |
Mean values from seven countries (in Latin-America and Southeast Asia).
6. Highlights for the future
This section focuses some promising themes for the future of biosensor development, although no reports or citations have been found on the literature concerning specific applications of many of these themes for the diagnosis of dengue disease.
6.1. New immunochromatographic platforms
New schemes for conventional lateral flow rapid tests, especially in the form of cassette kits, may offer additional advantages over traditional formats, namely the need for smaller sample volumes, no need for dilutions and decreased complexity. As an example, the recently developed cassette colorimetric test from Panbio Diagnostics (Australia) was successfully applied to the diagnosis of acute-phase dengue disease in individuals from a South American-endemic region [9]. The cassette also performed better than conventional EIA, with acute phase serum, in terms of sensitivity. Meanwhile, specificity was similar to those reported for the conventional Panbio lateral flow tests [15]. Thus, the Panbio cassette is effective for early detection of dengue IgM antibodies in acute febrile patients, for use in laboratory settings and by non-skilled individuals as well. Bio-Manguinhos, the technical and scientific unit of the Brazilian Fundação Oswaldo Cruz (FIOCRUZ) public health research institution, signed a technology transfer protocol with Chembio Diagnostic Systems (USA) for the development of new rapid diagnostic tests for leptospirosis, canine leishmaniosis and HIV [70]. The three new products are based on the Chembio Dual Path Platform (DPP®) technology, expected to be extended to other diseases in the future. This novel immunochromatographic technique essentially relies on two intersecting strips instead of a single migration strip: one for the sample and the other for a conjugated marker. Since this scheme provides more effective binding of the analyte to the binding site in the test zone prior to attachment of the conjugated marker, overall sensitivity can be enhanced 10–50 times compared to that of common lateral flow tests, while maintaining the specificity [71]. Moreover, the known lengthiness for biological fluid migration in single-path lateral flow tests may thus be obviated, especially in the case of aggregation/agglutination of larger, supramolecular analytes (e.g., whole cells). Results can be obtained in faster time periods with minimal volumes of different body fluids. Such rapidity comes from the possibility of new adding of buffer solution immediately after color vanishing at the test site, rather than waiting for a pre-determined (and often low accurate) time interval. In addition, this cassette platform contains five test lines, thus allowing multiplexed testing of up to five targets in the same reaction (conventional lateral-flow tests, by possessing only one strip, are not usually able to measure beyond a single analyte). For this, different colored latex particles can conjugate with different antigens or antibodies provided in the conjugated pad or in the buffer solution, thus facilitating reading of results [70], [71]. Therefore, the DPP® may be considered an improved user-friendly test for point-of-care diagnosis, also holding great promise for rapid and multiplexed detection and identification of dengue virus serotypes. Another interesting and promising innovation in the field of immunochromatographic tests is the issue of patterned paper-based substrates as alternative platforms to conventional test strips and to lab-on-a-chip platforms [72]. Paper is a familiar, abundant, cheap, easy-to-use, stable and disposable (easily and safely recyclable by incineration) material; moreover, many standardized printing techniques (e.g., photolithography and inkjet printing) are already available for its functionalization with polymers, biological species and other elements. Mass production of such devices showed to be feasible by coating paper, through inkjet printing, with inks of biomolecule-attached microgel, a suitable material to link biomolecules to paper substrates, thus preserving biomolecule activity after printing [73]. Being usually white, paper is a suitable material for implementing colorimetric tests (namely with enzymes or small-molecule dyes), by providing high contrast with colored substrates. Since color intensity in the detection region is usually a function of the analyte concentration, quantitative measurements are enabled. Other detection methods, however, are also feasible, like electrochemiluminescence and radiolabeling [74]. An enormous advantage of paper-like substrates over solid-support lab-on-a-chip devices and classical porous immunochromatographic substrates is, owing to the fibrous nature of paper, its intrinsic ability for pumping liquid biological samples along imprinted channels by simple capillarity, thus avoiding the need for external pumps and pressure sources [75]. In addition, expensive infrastructures, like clean rooms, are not required for fabrication of sophisticated microdevices through advanced printing techniques [76]. Most of the works with paper-based devices have pointed towards multi-analyte (multiplexed) detection, as a convenient extension of the conventional lateral-flow test abilities, by linking a single patterned inlet with various detection regions [77]. However, some recent works have proposed the opposite approach, i.e., multi-analyte detection by converging multiple inlets into a single outlet and detection region, through controlled transport of liquid samples, by suitable tailoring of the network geometry and fluid flow rate, which is required for autonomous driving of multi-step sequences [78], [79], as seen in Fig. 4 . In this way, more complex processes such as chemical amplification, which requires multiple and sequential reagent delivery and washing steps, can now be extended from the high-performing laboratory bioassays to simple point-of-care devices, as a way to enhance the sensitivity of detection, while maintaining easiness-to-use and pricing comparable to those of conventional lateral-flow tests. Hence, these improved paper networks may circumvent the usual lack of sensitivity faced by many current lateral-flow tests unless more complicated and expensive reagents (e.g., particle nanolabels) or instrumentation (e.g., light sources or detectors) are used [79]. In addition, such complex multi-tasking processes require no more than a single-user activation step, a significant advantage when compared to current lateral-flow tests and even with the Chembio DPP®, which requires an additional user washing step after label binding in the detection region [72]. Ongoing advances in theoretical studies for optimization of engineering design and fluid flow in two-dimensional paper devices [80] will certainly increase the powerfulness and usefulness of these tests for point-of-care diagnosis in resource-limited regions and settings. Alternatively, the performance and flexibility of paper-based analytical devices can also be enhanced by passing from two- to three-dimensional structures. In fact, it is likely that the complexity of three-dimensional paper-based structures may be higher than that of devices based on conventional plastic frames. As an example, the common steps of cell separation and removal and further sample processing may be significantly improved along with liquid wicking throughout the inner layers of the device, until final detection [75]. Despite all these advantages, some concerns must be raised regarding the future of paper-based kits, namely: (a) the ferocious competition of conventional laboratory and lab-on-a-chip analytical devices in terms of sensitivity, accuracy, quantitative outputs and multiplexed ability; (b) the device stability (shelf-life), especially under long-distance transportation and long-time storage at room temperatures; (c) the complexity and current lack of knowledge about fluid flow and biomolecule immobilization and biorecognition onto paper matrices [81]; (d) research dispersion efforts towards the exploitation and application of novel nanostructures and nanoscaled effects in point-of-care tests [82], [83]. Nevertheless, it is expected that these devices may constitute an example of the desirable combination between the simplicity and the inexpensiveness of common diagnostic test strips and the analytical abilities of microfluidic analytical devices.
Fig. 4.
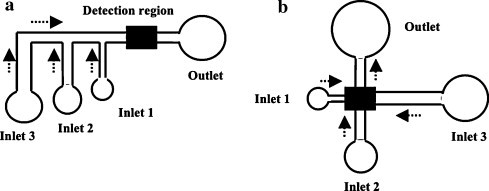
Some possible network geometries in two-dimensional analytical devices with control of fluid transport for sequential delivery of reagents (arrows indicate fluid flow directions). Sub-figure (a) uses multiple staggered inlets to a common way out, whereas sub-figure (b) employs a geometry of non-overlapping flowing channels. The arriving times of fluids 1–3 to the detection region (after simultaneous introduction into the three inlets, in growing volumes, through a single activation step) depend on the engineered length and width of the traveling channels. As such, in (a) and (b), both channel length and width increase from inlet 1 to 3, thus assuring a decreasing flow rate and hence a slowing fluid transport in that sequence.
6.2. New biosensing surfaces
Biosensor surfaces are not only supports for biomolecule immobilization and hence for occurrence of the biorecognition event, but also a part of the signal transduction mechanism itself. Apart the improvements in signal processing, finding suitable supports for immobilization and optimizing such process are of crucial importance for biosensor conception and development. In this regard, new and improved biomaterials have emerged as attractive and suitable platforms for the development of biosensors. In modified carbon pastes, high conductivity, chemical inertness, working potential range, surface area and charge transport speed are common highlights. Other advantages include low electrical resistivity [84], easy surface renewal and possibility of embedding the electroactive compound into the carbon paste matrix (thus avoiding the problem of leakage of the adhered coating). These composites, however, usually render low biosensor stability and reproducibility [85]. Redox hydrogels represent another class of biomaterials for biosensing. They have improved charge transfer ability, chemical inertness and biocompatibility, owing to the high water content [86]. However, the tendency for swelling in water or organic solvents may impair efficient usage for biosensing [30]. In this regard, sol–gel matrices appear as advantageous alternatives for their negligible swelling in solvents. Other pinpoints include operation at room temperature (ideal for labile biomolecules), easy access of the analyte to the probe immobilized within the porous sol–gel network, physical rigidity and chemical inertness, as well as high optical transparency and high photochemical and thermal stability [87]. In general, however, solid–gel matrices exhibit low sensitivity and reproducibility. Undesirable changes in the biochemical properties of entrapped species may occur [88], ultimately leading to biological inactivation. Sol–gel materials have also low conductivity and weak resistance interference, important setbacks in electrochemical biosensors. In this way, sol–gel hybrids have emerged as convenient alternatives, by combining the rigidity of sol–gel materials with the biocompatibility of hydrogels, thus avoiding the unwanted effects of sol–gel cracking, hydrogel swelling and leakage of the immobilized biomolecule [89]. Bilayer lipid membranes (BLMs), usually formed by lipid molecule casting from organic solvents or aqueous vesicle dispersion and subsequent surface-binding [90], constitute another class of biocompatible immobilization matrices. BLMs also allow better electron exchange between redox biomolecules and electrodes. Tighter and more stable attachment of BLMs to solid conductive surfaces has been achieved through self-assembly or by deposition onto conductive polymers [88]. Physicochemical instability of immobilized biomolecules and the resulting lack of reproducibility and unreliability are major concerns in current biosensors (especially affinity-based biosensors, namely immunosensors and genosensors) [30], but the newly developed MIPs may surpass such limitations. MIPs are formed by solvent removal of polymer-entrapped biomolecules, leading to the formation of imprinted nanocavities that become a permanent ‘memory’ of the size and shape of extracted molecules. The imprinted polymer will then be able to selectively rebind the complementary printing molecule [91]. With these tailored surfaces, with predetermined selective molecular recognition properties, the use of conventional and labile immobilized biomolecule-based surfaces can be avoided. However, further improvements in MIP-based biosensors must rely on sensitivity, selectivity and reusability (related with surface regeneration ability) enhancement. Indeed, this last property is one major highlight of ‘smart’ polymers, a new kind of transducing surface amenable for biosensor development. These polymers can undergo large physicochemical structural changes in response to small changes in external conditions (e.g., temperature, light, electromagnetic fields and pH) [30]. In general, when the external physicochemical stimulus reaches a given threshold, sudden collapse of the polymer structure rapidly triggers pronounced conformational changes in attached biorecognition molecules. When a reverse stimulus is applied, both the polymer and the attached biomolecule regenerate their original configurations and bioactivities, making possible the use for multiple cycles of biosensing. It is expected that, by yielding a type of ‘yes/no’ response, ‘smart’ polymer-based biosensors may be used as suitable molecular switches for bioelectronic applications [92].
6.3. Nanobiotechnology
Nanotechnology has provided a brand new range of structures and tools with improved physicochemical characteristics for manipulation of biological systems, the so-called field of nanobiotechnology. In particular, conception and development of biosensors and rapid diagnostic tests has greatly benefited from this advent. Nanomaterials may act not only as immobilization supports, but also as signal amplifiers. Metal nanoparticles (especially GNPs) are probably the best known and widely used nanostructures for biosensing. By providing high S/N and stability, metal nanoparticles can be more advantageous than fluorescence labels [93], being also suitable for incorporation into high-density microchip-based devices. In addition, they are easily synthesizable and functionalized at room temperature, commonly by simple self-assembling [94]. In particular, nanoparticles can be combined with fluorescent tagging and detection in the form of the so-called quantum dots (QDs), a kind of nanoparticles much brighter (due to the high quantum yields) and photostable than conventional fluorophores. Their color directly correlates with particle size, with emission of a single and well-defined wavelength after excitation [95]. The peculiarity of having highly non-overlapping broad absorption spectra and narrow emission spectra, allied to the possibility of exciting QDs of different sizes (and hence different emission peaks) with a single-wavelength excitation source, makes these particles suitable for multiplexed analysis of complex biological systems, by labeling different biological targets with QDs of different sizes [96]. Carbon nanotubes (CNTs) constitute another emerging kind of nanostructure for biosensor construction by their ability to bind, on their surfaces, a large diversity of bioactive biomolecules, through covalent and non-covalent interactions. They belong to the fullerene chemical family, roughly resembling hollow cylinders, with the porous walls composed of carbon networks, and behaving like microcrystals with molecular dimensions [97]. This configuration assures an enlarged area for immobilization of biomolecules. CNT electrical conductivity is comparable to that of copper and much higher than that of polymers [65]. They are also physically robust and chemically inert towards most chemicals [98]. These characteristics favor a peculiar range of improved electronic properties that make CNTs ideal for biosensing. This usually relies on CNT surface functionalization as a way to enhance biocompatibility and improve selective binding to biotargets [30]. Adding CNTs to conducting polymers decreases charge transfer resistance and mass-transfer impedance at a higher extent than other nanomaterials [99]. The manufacture of CNTs, however, is complex, and usually results in high structural variability within the same pool of CNTs, with subsequent lack of reproducibility in electrical properties. Nanotube arrays of high density may not only minimize this setback (due to the statistical average effect), but also lead to multiplexed analysis of many biological targets, namely cheap and simultaneous detection of many specific genes [98]. CNT/GNP composites, produced after functionalization of CNT surfaces, have also been employed for very sensitive and selective detection of biotargets [30]. Conducting polymer nanowires, namely from polyaniline, have also been developed as suitable nanostructures for ultrasensitive biomolecule detection. Apart easy processing, they possess large surface areas, high chemical specificities and tunable conductivities [100]. By their quasi-one dimensional structure, nanowires provide highly sensitive and specific detection and hence high surface/volume ratio. Nevertheless, applications in electrochemical biosensors are limited for the difficulty of adhesion to patterned electrodes [30]. New DNA/protein conjugates have recently emerged as valuable nanoengineered structures for medical diagnostic biosensors, given the wide range of interactions observed between these components in many cellular processes [101]. A remarkable example concerns PNAs which, by possessing a neutral peptidic backbone (unlike true nucleic-acids), allow highly specific and selective binding of a DNA counterpart at low (and relatively varying) ionic strength, being therefore amenable to probe DNA hybridization [102]. An interesting structure for single-molecule DNA detection is the DNA nanopore, usually formed by covalently linking a specific single-chain DNA-probe to the pore of a protein (e.g., Staphylococcus aureus α-hemolysin). The detection principle relies on the production of distinctive patterns of ionic current changes for each different single-chain DNA that crosses the nanochannel, according to its average time interval of binding to the immobilized chain, with direct correlation with the complementarity degree between them. Target-DNA chains can thus be detected with single-base resolution [103]. Some drawbacks of this system are unfeasibility for nucleotide sequencing since DNA sequence nucleotides within the pore at a given instant contribute indistinctively to the overall resistance [104] and also to the effect of using high frequency currents to decrease the experimental noise. Even so, the possibility of being an alternative to high-density probe microarrays is very attractive for a next generation of nanopore-based biosensors [65]. Magnetic nanoparticles (also known as magnetic nanobeads, magnetic nanospheres or simply nanomagnets) are also emerging as interesting structures for biosensing purposes, for their ability to stably bind biological targets. Magnetic particles, in general, can be easily manufactured in a wide range of micro- or nanosized diameters. Basically, magnetic nanoparticles are similar to their microsized counterparts (magnetic microbeads) concerning structure, biological reactivity, biorecognition mechanisms and characteristics. Compared to the typical huge amounts of magnetic nanobeads, even low amounts of magnetic microbeads can be more easily detected, by simple optical microscopy or magnetic detection. However, the higher surface/volume ratio of nanobeads provides much more binding sites for bioprobe and biotarget anchoring, and hence a higher S/N ratio. Maximization of this parameter is extremely important in label-based systems, whose performance (sensitivity, selectivity and reproducibility) is much more dependent on the level of non-specific background signals than on the ability for label detection [105]. Moreover, the use of nanomagnets allows avoiding the size mismatch between microsized magnetic labels and nanosized biological probes and targets. When magnetic particle-bound bioprobes capture the biotarget under exposure to a strong magnetic field, collective alignment of the magnetic moments of magnetic particles occurs, yielding a measurable and specific signal. This signal is given by surface-bound magnetic particles and is proportional to the biotarget concentration. Since the magnetic signal of each functionalized nanomagnet depends on its spatial orientation and unbound magnetic nanoparticle/bioprobe complexes are randomly oriented, there is no net signal from this set of particles, which is responsible for the earlier-mentioned magnetic washing effect. Very often, these magnetic labels are used for separating and pre-concentrating the biotarget rather than for the detection step itself (in which an additional label, usually a metal nanoparticle or a fluorophore, is employed). The detection strategy commonly relies on an initial capture of the target by the probe-functionalized nanomagnet followed by releasing (‘debinding’) for final detection. This may be a significant improvement for the detection, for example, of viral genome RNA, since RNA enrichment due to magnetic confinement also precludes the effect of common interferents and inhibitors of RNA usually existent in thermolysed viral-containing samples [106]. This multi-step process, however, may result in delayed analysis and sample dilution. Moreover, wider applications of magnetic nanoparticles in biosensors will require enhancement of field sensitivity and reduction of noise background [107]. Finally, it is noteworthy the combination of nanomaterials with new biosensing surfaces, like sol–gel materials, for synergic improvement of individual performances, like better conductivity and, in general, to remarkable signal amplification.
6.4. Telemedicine
Another milestone in biosensor development concerns the issue of data processing. In fact, the usage of rapid diagnostic tests by non-experts may lead to misuse and result misinterpretation, a clear example being the very common and improper usage of colorimetric test strips by patients with disturbed visual accuracy. A balanced merging between in-the-field testing with remote data analysis and interpretation by competent clinical staff may be desirable. In this regard, telemedicine emerges as an attractive system for healthcare improvement in developing countries for allowing unskilled persons to provide useful healthcare in remote settings [75]. This will certainly benefit by the unending advances in information technology, especially focused on the development of miniaturized analytical devices with integrated hardware (namely image acquisition through cell phone cameras) and suitable software for storage of results. These can be downloaded to hard copies or directly transferred on-line for a remote healthcare unit central for processing and interpretation by expert personal. Processed results can afterwards be returned to the tester, almost in real-time [108]. Interesting applications of the telemedicine approach have already been developed by coupling to patterned paper-based microfluidic platforms for point-of-care remote multiplexed testing [109]. Since color intensity of an image strongly varies with local lighting and user-dependent conditions, further improvements in local image capturing must be achieved as a way to provide more accurate remote interpretation of results [75].
6.5. Mass spectrometry (MS)
MS is another promising technique for detection and identification of pathogens and other microorganisms, although representing a different paradigm of detection. Coupled to powerful computer software, MS is able to unambiguously detect the presence and quantify unidentified microorganisms in unknown samples by comparing the resulting mass spectra with those included in databases of infectious agents. The system is also able to relate such unknown organisms to those encountered formerly, which could be used for not only straightforward identification of dengue serotypes, but also dengue genotypes during an outbreak. In the form of a commercial packaged integrated platform, this strategy was already applied successfully for inclusion of the human severe acute respiratory syndrome (SARS) virus into the coronavirus family [110] and as a rapid and inexpensive method for global surveillance of emerging influenza virus genotypes [111]. Miniaturization of MS devices still remains a hard challenge, but customized kits comprising modules for genome extraction and amplification by PCR are already available for several infectious agents [34]. It is likely that current rapid diagnostic tests correspond to a time-bridge between conventional laboratory apparatus and future non-invasive sensors for in vivo monitoring [11].
7. Conclusions
In general, rapid tests for point-of-care analysis still suffer from skepticism from the world medical community, for which the questionable validity/usefulness of single-analyte measurements and the interpretation of results by non-clinicians have certainly contributed. In addition, proper validation and standardization of such tests are still required, without which regulatory approvals for testing and commercialization are hardly obtainable. Nevertheless, the suitability of rapid diagnostic tests demonstrated by the validation processes must be complemented with additional figures of merit for implementation in routine diagnosis. Dengue virus is difficult to cultivate and propagate in laboratory, probably due to its wide geographic serotypic and genotypic variability, which has been a major obstacle towards the development and implementation of rapid diagnostic tests for dengue infections. There is still a long way to run from in-the-bench schemes and prototypes to fully developed, commercially attractive biosensing devices, whose wider utilization is still shadowed by well-established and well-performing laboratory assays. Unfortunately, the experimental prerogative for using well-defined, pure synthetic samples has often hindered detailed studies and conclusions about the effect of potentially interfering substances. Cold-chain has supported massive and adequate delivery of rapid diagnostic tests to clinical settings and end-users in remote tropical regions, but is handicapped by frequent irregular power supply in such regions and by the huge costs associated to transportation. Despite all these challenges, rapid diagnostic tests must be considered valuable alternatives rather than competitors to laboratory-based analytical devices and methodologies, even if some loss of sensitivity or selectivity must be accepted in the point-of-care testing. In fact, rapid screening of large amounts of samples may speed up proper decision making by the clinician, while conventional lab-based bioanalytical techniques can be used afterwards for confirmation and better characterization of the infecting pathogen. Reusable diagnostic tests must be avoided when dealing with infectious pathogens, thus explaining the growing interest in the development of chip-based disposable tests. It has been consensual that an efficient rapid diagnostic test for dengue disease should be a multiplexed device for detection and identification of all four dengue serotypes. Future developments and advances in fundamental and engineering sciences will certainly enrich the whole pool of knowledge about rapid diagnostic tests and will also improve micro- and nanofabrication processes for their mass production at an increasing cost-effective basis as well.
Biography

Fernando Sérgio Rodrigues Ribeiro Teles graduated in Biochemistry by the University of Lisbon (Portugal) and concluded a Master in Biotechnology in the Technical University of Lisbon. After an experience, as a teacher of Physics, in the National High-School of the Democratic Republic of São Tomé and Príncipe, his PhD in Biological Sciences (area of Biotechnology) was completed in the Federal University of Pernambuco (Brazil), in the scope of biosensors for dengue. He also teached Biopharmaceuticals in the Portuguese Catholic University. Since 2009, he is a Researcher in the Institute of Hygiene and Tropical Medicine (Portugal), in the field of Science Management.
References
- 1.Castleberry J.S., Mahon C.R. Clin. Lab. Sci. 2003;16:34–38. [PubMed] [Google Scholar]
- 2.Rigau-Pérez J.G., Clark G.G., Gubler D.J., Reiter P., Sanders E.J., Vorndam A.V. Lancet. 1998;352:971–977. doi: 10.1016/s0140-6736(97)12483-7. [DOI] [PubMed] [Google Scholar]
- 3.Teles F.R.R., Prazeres D.M.F., Lima-Filho J.L. Rev. Med. Virol. 2005;15:287–302. doi: 10.1002/rmv.461. [DOI] [PubMed] [Google Scholar]
- 4.Kuno G., Vorndam A.V., Gubler D.J., Gómez I. J. Med. Virol. 1990;32:102–108. doi: 10.1002/jmv.1890320207. [DOI] [PubMed] [Google Scholar]
- 5.The Pan American Health Organization (Regional Office of the WHO) 2010. Emerging and Reemerging Infectious Diseases, Region of the Americas. EID Updates. Available from http://www.paho.org/English/AD/DPC/CD/eid-eer-2008-03-26.htm (accessed September 2010) [Google Scholar]
- 6.Vaughn D.W., Green S., Kalayanarooj S., Innis B.L., Nimmannitya S., Suntayakorn S., Endy T.P., Raengsakulrach B., Rothman A.L., Ennis F.A., Nisalak A. J. Infect. Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 7.Campbell C.A., George A., Salas R.A., Williams S.A., Doon R., Chadee D.D. Acta Trop. 2007;101:153–158. doi: 10.1016/j.actatropica.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Jentes E.S., Robinson J., Johnson B.W., Conde I., Sakouvougui Y., Iverson J., Beecher S., Bah M.A., Diakite F., Coulibaly M., Bausch D.G. Am. J. Trop. Med. Hyg. 2010;83:388–394. doi: 10.4269/ajtmh.2010.09-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez-Veja R.A., Díaz-Quijano F.A., Coronel-Ruiz C., Gómez S.Y., Villar-Centeno L.A. Biomédica. 2009;29:616–624. [PubMed] [Google Scholar]
- 10.Kuno G., Cropp C.B., Wong-Lee J., Gubler D.J. Am. J. Trop. Med. Hyg. 1998;59:757–762. doi: 10.4269/ajtmh.1998.59.757. [DOI] [PubMed] [Google Scholar]
- 11.Aguilera-Herrador E., Cruz-Vera M., Valcárcel M. Analyst. 2010;135:2220–2232. doi: 10.1039/c0an00307g. [DOI] [PubMed] [Google Scholar]
- 12.Baeumner A., Cohen R., Miksic V., Min J. Biosens. Bioelectron. 2003;8:405–419. doi: 10.1016/s0956-5663(02)00162-8. [DOI] [PubMed] [Google Scholar]
- 13.Houng H.-S.H., Chen R.C.-M., Vaughn D.W., Kanesa-thasan N. J. Virol. Methods. 2001;95:19–32. doi: 10.1016/s0166-0934(01)00280-4. [DOI] [PubMed] [Google Scholar]
- 14.Miagostovich M.P., dos Santos F.B., de Araújo E.S.M., Dias J., Schatzmayr H.G., Nogueira R.M.R. Mem. Inst. Oswaldo Cruz. 1997;92:595–600. doi: 10.1590/s0074-02761997000500006. [DOI] [PubMed] [Google Scholar]
- 15.Blacksell S.D., Doust J.A., Newton P.N., Peacock S.J., Day N.P., Dondorp A.M. Trans. R. Soc. Trop. Med. Hyg. 2006;100:775–784. doi: 10.1016/j.trstmh.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Dittmar D., Cleary J., Castro A. J. Clin. Microbiol. 1979;9:498–502. doi: 10.1128/jcm.9.4.498-502.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilling S., Ludolfs D., van An L., Schmitz H. J. Clin. Virol. 2004;31:179–184. doi: 10.1016/j.jcv.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y.-F., Lien K.-Y., Lei H.-Y., Lee G.-B. Biosens. Bioelectron. 2009;25:745–752. doi: 10.1016/j.bios.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groen J., Koraka P., Velzing J., Copra C., Osterhaus A.D. Clin. Diagn. Lab. Immunol. 2000;7:867–871. doi: 10.1128/cdli.7.6.867-871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasquez S., Hafner G., Ruiz D., Calzada N., Guzman M.G. J. Clin. Virol. 2007;39:194–198. doi: 10.1016/j.jcv.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Aytur T., Foley J., Anwar M., Boser B., Harris E., Beatty P.R. J. Immunol. Methods. 2006;314:21–29. doi: 10.1016/j.jim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Shu P.-Y., Yang C.-F., Kao J.-F., Su C.-L., Chang S.-F., Lin C.-C., Yang W.-C., Shih H., Yang S.-Y., Wu P.-F., Wu H.-S., Huang J.-H. Clin. Vac. Immunol. 2009;16:589–591. doi: 10.1128/CVI.00475-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumarasamy V., Wahab A.H., Chua S.K., Hassan Z., Chem Y.K., Mohamad M., Chua K.B. J. Virol. Methods. 2007;140:75–79. doi: 10.1016/j.jviromet.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Lapphra K., Sangcharaswichai A., Chokephaibulkit K., Tiengrim S., Piriyakarnsakul W., Chakorn T., Yoksan S., Wattanamongkolsil L., Thamlikitkul V. Diagn. Microbiol. Infect. Dis. 2008;60:387–391. doi: 10.1016/j.diagmicrobio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Dussart P., Petit L., Labeau B., Bremand L., Leduc A., Moua D., Matheus S., Baril L. PLoS Negl. Trop. Dis. 2008;2:e280. doi: 10.1371/journal.pntd.0000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam S.K., Fong M.Y., Chungue E., Doraisingham S., Igarashi A., Khin M.A., Kyaw Z.T., Nisalak A., Roche C., Vaughn D.W., Vorndam V. Clin. Diagn. Virol. 1996;7:93–98. doi: 10.1016/s0928-0197(96)00257-7. [DOI] [PubMed] [Google Scholar]
- 27.Special Programme for Research and Training in Tropical Diseases (TDR) World Health Organization; Geneva, Switzerland: 2009. Pediatric Dengue Vaccine Initiative (PDVI), Evaluation of Commercially Available Anti-dengue Virus Immunoglobulin M Tests, Diagnostics Evaluation Series 3. [Google Scholar]
- 28.World Health Organization . World Health Organization; Geneva, Switzerland: 1999. Regional Guidelines on Dengue/DHF Prevention and Control, Regional Publication 29/1999. [Google Scholar]
- 29.Fonseca L. In: Engenharia Enzimática. Cabral J.M.S., Aires-Barros M.R., Gama M., editors. Lidel; Lisbon: 2003. pp. 227–244. [Google Scholar]
- 30.Teles F.R.R., Fonseca L.P. Mater. Sci. Eng. C. 2008;28:1530–1543. [Google Scholar]
- 31.Peters R.H.P., van Agtmael M.A., Danner S.A., Savelkoul P.H.M., Vandenbroucke-Grauls C.M.J.E. Lancet Infect. Dis. 2004;4:751–760. doi: 10.1016/S1473-3099(04)01205-8. [DOI] [PubMed] [Google Scholar]
- 32.Cammann K., Lemke U., Rohen A., Sander J., Wilken H., Winter B. Angew. Chem. Int. Ed. Engl. 1991;30:516–539. [Google Scholar]
- 33.Zaytseva N.V., Goral V.N., Montagna R.A., Baeumner A.J. Lab Chip. 2005;5:805–811. doi: 10.1039/b503856a. [DOI] [PubMed] [Google Scholar]
- 34.Special Programme for Research and Training in Tropical Diseases (TDR) new ed. World Health Organization; Geneva, Switzerland: 2009. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. [Google Scholar]
- 35.Bier F.F., Kleinjung F. Fresen. J. Anal. Chem. 2001;371:151–156. doi: 10.1007/s002160101003. [DOI] [PubMed] [Google Scholar]
- 36.Ellerbee A.K., Phillips S.T., Siegel A.C., Mirica K.A., Martinez A.W., Striehl P., Jain N., Prentiss M., Whitesides G.M. Anal. Chem. 2009;81:8447–8452. doi: 10.1021/ac901307q. [DOI] [PubMed] [Google Scholar]
- 37.Su C.-C., Wu T.-Z., Chen L.-K., Yang H.-H., Tai D.-F. Anal. Chim. Acta. 2003;479:117–123. [Google Scholar]
- 38.Tai D.-F., Lin C.-Y., Wu T.-Z., Huang J.-H., Shu P.-Y. Clin. Chem. 2006;5:1486–1491. doi: 10.1373/clinchem.2005.064501. [DOI] [PubMed] [Google Scholar]
- 39.Tai D.-F., Lin C.-Y., Wu T.-Z., Chen L.-K. Anal. Chem. 2005;77:5140–5143. doi: 10.1021/ac0504060. [DOI] [PubMed] [Google Scholar]
- 40.Wu T.Z., Su C.C., Chen L.K., Yang H.H., Tai D.F., Peng K.C. Biosens. Bioelectron. 2005;21:689–695. doi: 10.1016/j.bios.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 41.Chen S.-H., Chuang Y.-C., Lu Y.-C., Lin H.-C., Yang Y.-L., Lin C.-S. Nanotechnology. 2009;20:215501. doi: 10.1088/0957-4484/20/21/215501. 10 pp. [DOI] [PubMed] [Google Scholar]
- 42.Janshoff A., Galla H.-J., Steinem C. Angew. Chem. Int. Ed. Engl. 2000;39:4004–4032. doi: 10.1002/1521-3773(20001117)39:22<4004::aid-anie4004>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Kumbhat S., Sharma K., Gehlot R., Solanki A., Joshi V. J. Pharm. Biomed. Anal. 2010;52:255–259. doi: 10.1016/j.jpba.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Lee W.G., Kim Y.-G., Chung B.G., Demirci U., Khademhosseini A. Adv. Drug Deliv. Rev. 2010;62:449–457. doi: 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baeumner A.J., Schlesinger N.A., Slutzki N.S., Romano J., Lee E.M., Montagna R.A. Anal. Chem. 2002;74:1442–1448. doi: 10.1021/ac015675e. [DOI] [PubMed] [Google Scholar]
- 46.Dodeigne C., Thunus L., Lejeune R. Talanta. 2000;51:415–439. doi: 10.1016/s0039-9140(99)00294-5. [DOI] [PubMed] [Google Scholar]
- 47.Wu S.J., Lee E.M., Putvatana R., Shurtliff R.N., Porter K.R., Suharyono W., Watts D.M., King C.-C., Murphy G.S., Hayes C.G., Romano J.W. J. Clin. Microbiol. 2001;39:2794–2798. doi: 10.1128/JCM.39.8.2794-2798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atias D., Liebes Y., Chalifa-Caspi V., Bremand L., Lobel L., Marks R.S., Dussart P. Sens. Actuators B. 2009;140:206–215. [Google Scholar]
- 49.Baselt D., Lee G.U., Natesan M., Metzger S.W., Sheehan P.E., Colton R.J. Biosens. Bioelectron. 1998;13:731–739. doi: 10.1016/s0956-5663(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 50.Kwakye S., Baeumner A. Anal. Bioanal. Chem. 2003;376:1062–1068. doi: 10.1007/s00216-003-2063-2. [DOI] [PubMed] [Google Scholar]
- 51.Zaytseva N.V., Montagna R.A., Lee E.M., Baeumner A.J. Anal. Bioanal. Chem. 2004;380:46–53. doi: 10.1007/s00216-004-2724-9. [DOI] [PubMed] [Google Scholar]
- 52.Zaytseva N.V., Montagna R.A., Baeumner A.J. Anal. Chem. 2005;77:7520–7527. doi: 10.1021/ac0509206. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y., Bahns J.T., Jin Q., Divan R., Chen L. Anal. Biochem. 2006;356:161–170. doi: 10.1016/j.ab.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 54.Schwille P., Meyer-Almes F.-J., Rigler R. Biophys. J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lagerkvist A.C., Foldes-Papp A., Persson M.A., Rigler R. Protein Sci. 2001;10:1522–1528. doi: 10.1110/ps.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwakye S., Goral V.N., Baeumner A.J. Biosens. Bioelectron. 2006;21:2217–2223. doi: 10.1016/j.bios.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 57.Teles F.R.R., Prazeres D.M.F., Lima-Filho J.L. Sensors. 2007;7:2510–2518. [Google Scholar]
- 58.Oliveira M.D.L., Correia M.T.S., Diniz F.B. Biosens. Bioelectron. 2009;25:728–732. doi: 10.1016/j.bios.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Fang X., Tan O.K., Tse M.S., Ooi E.E. Biosens. Bioelectron. 2010;25:1137–1142. doi: 10.1016/j.bios.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 60.Berggren C., Bjarnason B., Johansson G. Electroanalysis. 2001;13:173–180. [Google Scholar]
- 61.Zhang G.-J., Zhang L., Huang M.J.H., Luo Z.H.H., Tay G.K.I., Lim E.-J.A., Kang T.G., Chen Y. Sens. Actuators B. 2010;146:138–144. [Google Scholar]
- 62.Kulski J.K., Norval M. Arch. Virol. 1985;83:3–15. doi: 10.1007/BF01310960. [DOI] [PubMed] [Google Scholar]
- 63.Puri B., Henchal E.A., Burans J., Porter K.R., Nelson W., Watts D.M., Hayes C.G. Arch. Virol. 1994;134:29–37. doi: 10.1007/BF01379104. [DOI] [PubMed] [Google Scholar]
- 64.Teles F.R.R., Fonseca L.P. Talanta. 2008;77:606–623. [Google Scholar]
- 65.Guan J.-G., Miao Y.-Q., Zhang Q.-J. J. Biosci. Bioeng. 2004;97:219–226. doi: 10.1016/S1389-1723(04)70195-4. [DOI] [PubMed] [Google Scholar]
- 66.Daniels J.S., Pourmand N. Electroanalysis. 2007;19:1239–1257. doi: 10.1002/elan.200603855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.The TDR Diagnostics Evaluation Expert Panel Nat. Rev. Microbiol. 2006:S20–S32. doi: 10.1038/nrmicro1570. [DOI] [PubMed] [Google Scholar]
- 68.Bossuyt P.M., Reitsma J.B., Bruns D.E., Gatsonis C.A., Glasziou P.P., Irwig L.M., Lijmer J.G., Moher D., Rennie D., de Vet H.C.W., for the STARD Group Clin. Chem. 2003;49:1–6. doi: 10.1373/49.1.1. [DOI] [PubMed] [Google Scholar]
- 69.2010. Denframe (Consortium for the Study of Dengue Disease) Available from http://www.denframe.org/index.html (accessed on September 2010) [Google Scholar]
- 70.2010. Bio-Manguinhos, Bio-Manguinhos vai realizar transferência de tecnologia de três novos testes rápidos para diagnóstico com a Chembio. Available from http://www.fiocruz.br/bio/cgi/cgilua.exe/sys/start.htm?infoid=608&sid=227 (accessed on September 2010) [Google Scholar]
- 71.Chembio Diagnostic Systems . 2010. Patented Dual Path Platform (DPP®) Technology. Available from http://www.chembio.com/newtechnologies.html (accessed on September 2010) [Google Scholar]
- 72.Zhao W., van den Berg A. Lab Chip. 2008;8:1988–1991. doi: 10.1039/b814043j. [DOI] [PubMed] [Google Scholar]
- 73.Su S., Ali M.M., Filipe C.D.M., Li Y., Pelton R. Biomacromolecules. 2008;9:935–941. doi: 10.1021/bm7013608. [DOI] [PubMed] [Google Scholar]
- 74.Martinez A.W., Phillips S.T., Whitesides G.M. Anal. Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 75.Zimmermann M., Schmid H., Hunziker P., Delamarche E. Lab Chip. 2007;7:119–125. doi: 10.1039/b609813d. [DOI] [PubMed] [Google Scholar]
- 76.Calvert P. Chem. Mater. 2001;13:3299–3305. [Google Scholar]
- 77.Fenton E.M., Mascarenas M.R., Lopez G.P., Sibbett S.S. Appl. Mater. Interfaces. 2009;1:124–129. doi: 10.1021/am800043z. [DOI] [PubMed] [Google Scholar]
- 78.Fu E., Kauffmann P., Lutz B., Yager P. Sens. Actuators B. 2010;149:325–328. doi: 10.1016/j.snb.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu E., Lutz B., Kauffman P., Yager P. Lab Chip. 2010;10:918–920. doi: 10.1039/b919614e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu E., Ramsey S.A., Kauffman P., Lutz B., Yager P. Microfluid. Nanofluid. 2010 doi: 10.1007/s10404-010-0643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mukhopadhyay R. Anal. Chem. 2008;80:3949. doi: 10.1021/ac801509q. [DOI] [PubMed] [Google Scholar]
- 82.Mijatovic D., Eijkel J.C.T., van den Berg A. Lab Chip. 2005;4:492–500. doi: 10.1039/b416951d. [DOI] [PubMed] [Google Scholar]
- 83.Eijkel J.C.T., van den Berg A. Microfluid. Nanofluid. 2005;1:249–267. [Google Scholar]
- 84.Zhang S., Wright G., Yang Y. Biosens. Bioelectron. 2000;15:273–282. doi: 10.1016/s0956-5663(00)00076-2. [DOI] [PubMed] [Google Scholar]
- 85.Cassidy J.F., Doherty A.P., Vos J.G. In: Principles of Chemical and Biological Sensors. Diamond D., editor. John Wiley & Sons; Toronto: 1998. pp. 73–132. [Google Scholar]
- 86.Rajagopalan R., Atsushi A., Heller A. J. Phys. Chem. 1996;100:3719–3727. [Google Scholar]
- 87.Xu Z., Chen X., Dong S. Trends Anal. Chem. 2006;25:899–908. [Google Scholar]
- 88.Gupta R., Chaudhury N.K. Biosens. Bioelectron. 2007;22:2387–2399. doi: 10.1016/j.bios.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 89.Ionescu R.E., Gondran C., Cosnier S., Gheber L.A., Marks R.S. Talanta. 2005;66:15–20. doi: 10.1016/j.talanta.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 90.Kunitake T., Shimomura M., Kajiyama T., Harada A., Okuyama K., Takayanagi M. Thin Solid Films. 1984;121:L89–L91. [Google Scholar]
- 91.Bossi A., Piletsky S.A., Piletska E.V. Anal. Chem. 2001;73:5281–5286. doi: 10.1021/ac0006526. [DOI] [PubMed] [Google Scholar]
- 92.Hoffman A.S., Stayton P.S. Macromol. Symp. 2004;207:139–151. [Google Scholar]
- 93.Fritzsche W. Rev. Mol. Biotechnol. 2001;82:37–46. doi: 10.1016/s1389-0352(01)00028-9. [DOI] [PubMed] [Google Scholar]
- 94.Tamada K., Nakamura F., Ito M., Li X., Baba A. Plasmonics. 2007;2:185–191. [Google Scholar]
- 95.Hohng S., Ha T. Chem. Phys. Chem. 2005;6:956–960. doi: 10.1002/cphc.200400557. [DOI] [PubMed] [Google Scholar]
- 96.Whitesides G.M., Love J.C. Sci. Am. 2007;17:13. [Google Scholar]
- 97.Gao Y., Wolf L.K., Georgiadis R.M. Nucleic Acids Res. 2006;34:3370–3377. doi: 10.1093/nar/gkl422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gruner G. Sci. Am. 2007;17:48. doi: 10.1038/scientificamerican0507-76. [DOI] [PubMed] [Google Scholar]
- 99.Carrara S., Bavastrello V., Ricci D., Stura E., Nicolini C. Sens. Actuators B. 2005;109:221–226. [Google Scholar]
- 100.Maynor B.W., Filocamo S.F., Grinstaff M.W., Liu J. J. Am. Chem. Soc. 2002;124:522. doi: 10.1021/ja017365j. [DOI] [PubMed] [Google Scholar]
- 101.Odenthal K.J., Gooding J.J. Analyst. 2007;132:603–610. doi: 10.1039/b701816a. [DOI] [PubMed] [Google Scholar]
- 102.Wang J. Nucleic Acids Res. 2000;28:3011–3016. doi: 10.1093/nar/28.16.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Howorka S., Cheley S., Bayley H. Nat. Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 104.Meller A., Nivon L., Branton D. Phys. Rev. Lett. 2001;86:3435–3439. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 105.Mulvaney S.P., Cole C.L., Kniller M.D., Malito M., Tamanaha C.R., Rife J.C., Stanton M.W., Whitman L.J. Biosens. Bioelectron. 2007;23:191–200. doi: 10.1016/j.bios.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 106.Lee W.-C., Lien K.-Y., Lee G.-B., Lei H.-Y. Diagn. Microbiol. Infect. Dis. 2008;60:51–58. doi: 10.1016/j.diagmicrobio.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 107.Freitas P.P., Ferreira H.A., Cardoso F., Cardoso S., Ferreira R., Almeida J., Guedes A., Chu V., Conde J.P., Martins V., Fonseca L., Cabral J.S., Germano J., Sousa L., Piedade M., Silva B., Lemos J.M., Clarke L.A., Amaral M.D. In: Nanotechnology and the Detection of Biomolecular Recognition Using Magnetoresistive Transducers. A Portrait of State-of-the-art Research at Technical University of Lisbon. Pereira M.S., editor. Springer; Dordrecht, The Netherlands: 2007. pp. 3–22. [Google Scholar]
- 108.von Lode P. Clin. Biochem. 2005;38:591–606. doi: 10.1016/j.clinbiochem.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 109.Martinez A.W., Phillips S.T., Carrilho E., Thomas S.W., III, Sindi H., Whitesides G.M. Anal. Chem. 2008;80:3699–3707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sampath R., Hofstadler S.A., Blyn L., Eshoo M., Hall T., Massire C., Levene H., Hannis J., Harrell P.M., Neuman B., Buchmeier M.J., Jiang Y., Ranken R., Drader J., Samant V., Griffey R.H., McNeil J.A., Crooke S.T., Ecker D.J. Emerg. Infect. Dis. 2005;11:373–379. doi: 10.3201/eid1103.040629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sampath R., Russell K.L., Massire C., Eshoo M.W., Harpin V., Blyn L.B., Melton R., Ivy C., Pennella T., Li F., Levene H., Hall T.A., Libby B., Fan N., Walcott D.J., Ranken R., Pear M., Schink A., Gutierrez J., Drader J., Moore D., Metzgar D., Addington L., Rothman R., Gaydos C.A., Yang S., George K.S., Fuschino M.E., Dean A.B., Stallknecht D.E., Goekjian G., Yingst S., Monteville M., Saad M.D., Whitehouse C.A., Baldwin C., Rudnick K.H., Hofstadler S.A., Lemon S.M., Ecker D.J. PLoS One. 2007;2:e489. doi: 10.1371/journal.pone.0000489. [DOI] [PMC free article] [PubMed] [Google Scholar]


