Abstract
Research activity and applications of biosensors for measurement of analytes of clinical interest over the last eight years are reviewed. Nanotechnology has been applied to improve performance of biosensors using electrochemical, optical, mechanical and physical modes of transduction, and to allow arrays of biosensors to be constructed for parallel sensing. Biosensors have been proposed for measurement of cancer biomarkers, cardiac biomarkers as well as biomarkers for autoimmune disease, infectious disease and for DNA analysis. Novel applications of biosensors include measurements in alternate sample types, such as saliva. Biosensors based on immobilized whole cells have found new applications, for example to detect the presence of cancer and to monitor the response of cancer cells to chemotherapeutic agents. The number of research reports describing new biosensors for analytes of clinical interest continues to increase; however, movement of biosensors from the research laboratory to the clinical laboratory has been slow. The greatest impact of biosensors will be felt at point-of-care testing locations without laboratory support. Integration of biosensors into reliable, easy-to-use and rugged instrumentation will be required to assure success of biosensor-based systems at the point-of-care.
Keywords: Biosensors, Clinical analysis, Nanotechnology, Point-of-care testing
Highlights
► Nanotechnology improves performance of biosensors using various modes of transduction. ► Research activity into biosensors for clinical analytes outpaces commercialization. ► Novel biosensor designs are focused on point-of-care testing applications.
1. Introduction
In an earlier report [1], biosensors were reviewed from the perspective of fundamental measurement technologies and their applications to clinical analysis, including examples of biosensors available as commercial products. Other recent reviews have covered advances in electrochemical biosensors from both the fundamental science and prospects for commercialization [2], [3], progress and hurdles toward realizing implantable biosensors for clinical application [4] and biosensors for point-of-care testing, particularly those designed for home-use setting [5].
In this report, developments in technologies and applications of biosensors to clinical analysis over the past 8 years will be reviewed. A few recent examples of commercial biosensors for clinical analysis, in particular for point-of-care testing, will be included, but in general, commercialization of biosensors continues to lag behind research output as reflected by the numerous publications and patents appearing in the last 8 years. Many of the biosensor reports found in the literature demonstrate concepts to detect one or a few target analytes, however, biosensors with commercial promise should be based on versatile sensing technologies to support interchangeable recognition elements, the possibility for miniaturization to allow for parallel sensing [6], or biosensors based on technologies presenting performance advantages over existing methods, e.g., improved sensitivity or specificity. The latest work in the area of biosensors and their application to clinical chemistry will be reviewed in light of these requirements.
2. Biosensors based on nanotechnology
Perhaps the most notable trend in biosensor research over the past decade has been application of nanotechnology for construction of biosensors. Besides some analytical performance advantages discussed below, nanotechnology brings to biosensors the possibility for construction of biosensor arrays for high throughput parallel measurements and the possibility for integration of biosensors with microfluidics for construction of lab-on-a-chip devices. Nanotechnology has even been proposed as a method to enhance biocompatibility of biosensors and a possible solution to the problem of fouling of implantable biosensors — a problem which has hindered progress in the area of in-vivo sensing for years [7].
Nanotechnology is defined as the study of synthesis, properties and application of structures and materials having at least one critical dimension on the scale of < 100 nm [8], [9]. In relation to biosensors, a critical dimension is one directly related to the measurement function of the biosensor, such as a dimension that controls the area available for immobilization of a biorecognition element, or a dimension that controls the magnitude of a signal, such as electrode surface area in the case of electrochemical biosensors, or area available for detecting formation of complexes between recognition elements and target analytes in the case of biosensors based on mechanical transducers. Structures such as nanowires, metallic nanoparticles, magnetic nanoparticles, nanopores and carbon nanotubes offer unique electrical, optical and magnetic properties that can be exploited for chemical sensing. The large surface area to volume ratio available on nanostructures for immobilization of labels and biological recognition elements offers the potential for high signal amplification and improved measurement sensitivity. Biosensors based on nanocantilevers measure a mechanical property in response to an affinity reaction and offer the possibility of label free measurements. Recent developments in the field of DNA nanobiosensors have resulted in molecular detection and amplification schemes, capable of outperforming PCR in sensitivity and ease of use [10].
2.1. Carbon nanotubes, metallic nanoparticles and nanowires coupled to electrochemical and optical transducers
The advantages of carbon nanotubes (CNT) as an immobilization matrix for both biocatalytic and affinity biosensors, owing to high surface to volume ratio, fast electron transfer kinetics (advantageous for electrochemical biosensors) and the presence of reactive groups on the surface, have been demonstrated [11], [12]. Applications of CNT to improve sensitivity, response time and other analytical performances for electrochemical biosensors for several analytes of clinical interest have recently been reviewed [13]. Enhanced electron transfer between immobilized proteins, mediators and CNT results in electrochemical biosensors capable of operation at lower overpotentials, where signals from electrochemically active interfering substances are minimized, as was recently demonstrated for an amperometric glucose biosensor [14]. A biosensor for measurement of cholesterol in blood with cholesterol oxidase immobilized on multiwall CNT in a sol–gel matrix showed marked improvements in response time and sensitivity compared to an equivalent biosensor without CNT [15]. A biosensor for measurement of uric acid in serum was realized by immobilization of uricase in a polyaniline/CNT matrix. The sensor exhibited a response time of 8 s which the authors attributed to fast electron transfer properties of CNT [16]. Immunosensors are a particular area where the improvements in sensitivity brought by CNT can be exploited. Immunosensors using field effect transistors (FET) as transducers have been widely used for proof of concept studies because of the inherent sensitivity and signal amplification offered by FET [17]. In one example using prostate specific antigen (PSA) as a model analyte, CNT were deposited on the silicon gate of a FET [18]. Antibodies to PSA were immobilized on the CNT via a succinimidyl linking molecule. In a parallel experiment, indium oxide nanowires were deposited on the gate of a FET and antibodies to PSA immobilized on the nanowire. After incubation of the device with PSA, the nanowire device showed enhanced conductance between the source and drain electrodes, while the CNT device showed reduced conductance. The difference in response can be understood because the nanowire is an n-type semiconductor, while CNT is a p-type semiconductor, leading to different mechanisms of electrical conductance. Based on the signal to noise ratio observed at a PSA concentration of 5 ng/mL, it was predicted that a lower detection limit approaching 0.25 ng/mL, where the signal to noise ratio is approximately 3, could be obtained. In a similar study, an electrochemical immunosensor for PSA was constructed by depositing CNT on a 30 microelectrode array on a silicon substrate and modifying the CNT surface with a succinimidyl linking molecule to which monoclonal antibodies to PSA could be covalently bound [19]. Upon incubation of the array with PSA, the intrinsic oxidation signal of the electroactive amino acid residues present in PSA was detected using differential pulse voltammetry. The detection limit toward PSA was 0.25 ng/mL with a signal to noise ratio of 3. Aptamers have been used in place of antibodies as recognition elements to construct immunosensors by attaching the aptamer to CNT on the gate of a FET. Claimed advantages are improved shelf life of the devices and the potential to realize reversible immunosensors, not practical using antibodies as recognition elements [20]. When used to construct electrochemical DNA biosensors, CNT allow an efficient method to amplify label-free electrochemical detection of DNA hybridization by enhancing charge transfer between surface-anchored DNA sequences and CNT [21], [22]. Increases in sensitivity offered by nanomaterials, when coupled to electrochemical modes of transduction, have resulted in highly sensitive, highly specific sensors attractive for detection of small DNA sequence variations [23].
Similar to CNT, gold nanoparticles (AuNP) have been shown to result in large signal enhancements and lower limits of detection for enzyme-based electrochemical biosensors, electrochemical immunosensors for disease-related protein biomarkers, and for detecting DNA hybridization events [24], [25]. AuNP may be attached to electrode surfaces using self assembled monolayers (SAM) of alkanethiol molecules and the strong S–Au bond, providing a large active surface area for immobilization of biomolecules in close proximity to an electrode surface. AuNP have been used as both immobilization platforms and labels for electrochemical immunosensors [26]. Electrodes with layers of densely packed 5 nm gold nanoparticles were used as a matrix for immobilization of horseradish peroxidase in a sandwich immunoassay for prostate specific antigen with a detection limit of 0.5 pg/mL [27]. Optical properties of AuNP have also been exploited for biosensing. It is known that well dispersed suspensions of AuNP produce a red color, while aggregated AuNP produce a blue color [28]. A simple, aptamer-based colorimetric sensing of thrombin using 13 nm AuNP probes has been described [29]. AuNP, modified with thrombin aptamers, are observed as red because the nanoparticles are in suspension. Upon addition of thrombin, the aptamer undergoes a conformational change, resulting in aggregation of the AuNP and a change to blue. This assay demonstrated a linear range from 0 to 167 nmol/L thrombin with a detection limit of 0.83 nmol/L, as low as the most sensitive currently available methods. In an optical DNA assay, oligonucleotide targets labeled with AuNP instead of fluorophore probes have been shown to enhance sensitivity toward the oligonucleotide target by up to two orders of magnitude [30]. A commercial system for genetic testing using AuNP probes without need for PCR amplification is the Verigene® System (Nanosphere, Northbrook, IL), capable of detecting single nucleotide polymorphisms related to some common genetic disorders, such as thrombophilia, alterations of folate metabolism, cystic fibrosis and hemochromatosis. Application of the Verigene System for detection of gene variants associated with hypercoagulability has recently been reported [31].
2.2. Nanoscale biosensors based on mechanical and physical transducers
Microfabricated cantilevers made of silicon or silicon nitride have been used as transducers in affinity biosensors for label-free monitoring of DNA hybridization, antigen–antibody interactions, and adsorption of bacteria. The devices range from ten to hundreds of μm long, hundreds of nm thick, and may be fabricated in arrays containing ten to thousands of microcantilevers [32]. When fabricated on the nanoscale, detection sensitivity may be improved to the attogram (10− 18 g) level. [33]. Mechanical bending of the cantilever on the order of nanometers is monitored in response to affinity reactions taking place on the surface. Optical read out is the most common scheme for detecting movement of microcantilevers, similar to atomic force microscopy (Fig. 1 ). The sensitivity of cantilever biosensors is limited by non-specific binding; still the technology has been shown equivalent in sensitivity to other label-free quantitative methods, for example, in the picomolar range for detection of oligonucleotides and in the ng/mL range for measurement of antigens [34]. Most commonly, the cantilever sensor is coated with a thin layer of gold followed by a self assembled monolayer of thiol. Immobilization of oligonucleotides and proteins to gold surfaces using thiol chemistry is well known and used in other biosensing applications [32]. Applications for DNA sensors, immunosensors and sensors for detection of pathogens have been reported. Fritz et al. were able to detect a single-base mismatch between two 12-mer oligonucleotides with 10 nmol/L limit of detection [35]. A reference microcantilever with a control (non-complementary) oligonucleotide was included to cancel out effects from non-specific binding and noise. There are some commercial platforms using arrays of cantilever biosensors. CantiChip8 (Cantion, Denmark) is an array of 8 nanocantilevers. The system is an open platform allowing the user to functionalize each cantilever individually. The VeriScan 3000 System (Protiveris, Rockville, MD, USA) uses a 64 nanocantilever array to measure molecular interactions applied to immunoreactions or DNA sensing.
Fig. 1.
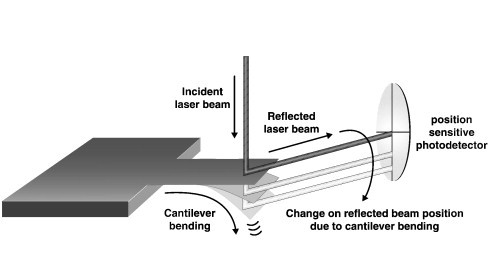
Scheme of the optical read-out method to monitor cantilever bending.
Reprinted from Ref. [32] with permission from Elsevier.
Acoustic biosensors, such as the quartz crystal microbalance (QCM), measure changes in resonant frequency of a piezoelectric crystal in response to changes in surface adsorbed mass resulting from an affinity reaction. Limits of detection on the order of ng/cm2 are possible with microfabricated structures [36], with the possibility to further increase the sensitivity by reducing size of the structures to the nanoscale [37]. A limitation of acoustic biosensors for clinical application is propagation losses due to dampening of the acoustic signal in liquid environments. Recent improvements in sensor design include addition of a waveguide layer to trap acoustic waves along the surface of the piezoelectric substrate to minimize energy losses and to protect the device from corrosion in liquid (surface acoustic wave (SAW), Love wave device) [38]. The surface of the waveguide is often coated with a gold layer (approx. 50 nm thick) to provide a better surface for immobilization of biomolecules, e.g. antibodies. An example of such a device includes a Love wave immunosensor for detection of whole Escherichia Coli bacteria at a detection threshold of 106 bacteria/mL in less than one hour [39].
Nanogap electrode arrays (three dimensional interdigitated electrodes) have been used as transducers for biosensors. Label-free affinity reactions are monitored on the electrode surface by measuring change in impedance across the electrode array. Height and width of the electrodes, as well as width of the gap may be optimized to increase the sensitivity. Antibodies to C-reactive protein (CRP) were immobilized on a gold nanogap electrode array and formation of an immunocomplex with CRP was monitored. Sensor response was linear from 0.1 ng/mL to 1 μg/mL CRP in human serum [40].
Other examples of nanotechnology applied to biosensing are emerging. Pores with nanometer-scale diameters have sizes comparable to single large molecules (proteins, single-stranded DNA). A molecule must be charged to be driven through the pore by an electric field. The contribution of the large molecule to passage of electric (ionic) current through the pore is small compared to mobile small ions, reducing the measured current across the pore [41]. Han et al. have shown that nanopores, 28 nm in diameter formed in silicon nitride, may be used as label-free biosensors using β-human chorionic gonadotropin (β-hCG) as an example. Formation of the immune complex between β-hCG and its antibody results in an increase in current across the nanopore, compared to antibody alone, due to an increase in electrophoretic mobility of the immune complex. A claimed advantage over other label-free biosensors is that immobilization of ligands, which could ultimately change binding kinetics or molecular interactions, is not required. The authors speculate, based on the reported measurement ability of their assay, that antibody concentrations down to 0.5 nmol/L could be measured with a CV of 10% [42].
3. Recent proposals for biosensors to detect clinically important analytes
3.1. Biosensors for cancer biomarkers
Biosensors for detection and monitoring treatment of cancer are emerging. A symposium held June 2005, “Moving Biosensors to Point-of-Care Cancer Diagnostics”, sponsored by the Cancer Diagnostics Program of the National Cancer Institute, highlighted developments in biosensor technologies for detection of important cancer biomarkers [43]. Included are biosensors for proteins, peptides and tumor related metabolites, as well as biosensors to detect DNA mutations in tumor cells to indicate the presence or risk of cancer as well as to monitor therapy [44]. PSA is often used as a model cancer marker for demonstration of biosensor measurement principles. In addition to being used for demonstrating methods to improve sensitivity of new biosensing concepts, the ability to develop a biosensor for PSA offers an attractive possibility to develop a point-of-care testing device to screen for prostate cancer in real time and for forensic testing in near patient locations, without the need for sample transport and delays associated with dedicated central laboratories. Ultrasensitive biosensor assays capable of detecting serum PSA in the low pg/mL region are being sought to enable earlier detection and recurrence of prostate cancer [45]. Known biosensor modes of transduction, with and without labels, are being combined with nanotechnology in order to achieve lower limits of detection for PSA and other clinically relevant cancer biomarkers.
Label-free, direct immunosensors for PSA and other cancer biomarkers using optical, electrochemical and detection of changes in mass as modes of transduction have been reported. A label-free approach for detection of PSA involved using a carbon microelectrode array with conductive polyaniline protrusions [46]. Immobilization of antibodies toward PSA on polyaniline was done using classical avidin–biotin affinity. Formation of the Ag–Ab complex in the presence of PSA increased electron transfer resistance of the conductive polymer. A linear response toward PSA was demonstrated from 1 to 100 pg/mL, with ability to discriminate at 5 pg/mL. A similar, label-free conductometric approach for measurement of tumor marker CA 125 was constructed using polypyrrole nanowires, surface-functionalized with antibody to CA 125 [47]. The nanowire served as a connection between a pair of gold electrodes (Fig. 2 ). A sharp decrease in conductance between the electrodes was measured upon exposure to CA 125 antigen. Lower limit of detection toward CA 125 was estimated at 1 U/mL and the assay time was approximately 5 min. An approach for quantitation of PSA using a piezoresistive microcantilever has been reported [48]. Monoclonal anti-PSA was immobilized on the surface of Au coated microcantilevers, after functionalizing the surface with a self-assembled monolayer using a calixarene derivative. Exposure to PSA resulted in formation of the Ag–Ab complex, bending of the cantilever and a resistance change of the piezoresistive layer. The cantilever surface was treated with silicon nitride to minimize nonspecific binding of undesired proteins, yet quantitation toward PSA was limited to 10 ng/mL. A surface plasmon resonance (SPR) assay for PSA using a commercially available system (Biacore2000™, GE Healthcare) was reported [49]. A SAM of alkanethiol was deposited on a gold SPR substrate, followed by immobilization of single domain antibody fragments, with increased affinity toward PSA, derived from heavy-chain antibodies. A detection limit of 10 ng/mL PSA, marginally acceptable for clinical use, was obtained using this direct assay in serum samples. However, sample dilution was required due to nonspecific adsorption of serum proteins, a problem known to limit the usefulness of SPR biosensors. Detection limit was improved by an order of magnitude using a sandwich assay consisting of a second, biotinylated anti-PSA followed by exposure to gold nanoparticles modified with streptavidin. A recent study, aimed at reducing nonspecific protein adsorption to SPR substrates, indicated that polymers containing the carboxybetaine moiety, grafted to the surface of a SPR substrate coated with SiO2, not only maximized protein immobilization efficiency, but also minimized biofouling from protein adsorption in undiluted plasma or serum. The concept was demonstrated using the cancer biomarker activated leukocyte cell adhesion molecule. Detection of the biomarker down to 64 ng/mL was possible in a direct assay. It was hypothesized that surface hydration formed via ionic solvation of zwitterionic groups is primarily responsible for the ability of the carboxybetaine polymers to prevent protein adsorption [50].
Fig. 2.
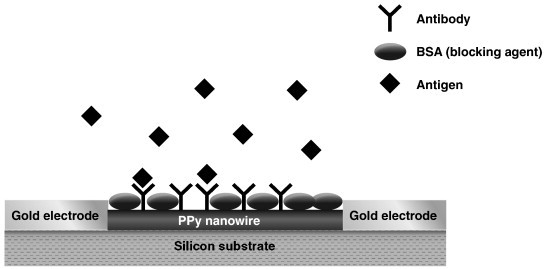
Schematic of a single polypyrrole (PPy) nanowire modified with anti-CA 125 and blocked by BSA to reduce nonspecific binding for detection of tumor marker CA 125.
Reprinted from Ref. [47] with permission from the American Chemical Society.
While label-free, direct immunosensors may offer simplicity in terms of the detection chemistry without need for separation and washing steps, electrochemical or optical labels bring advantages such as higher sensitivity, in general because nonspecific binding of proteins is reduced and the presence of labels serves as a method for signal amplification, especially when combined with nanotechnology. Based on direct electrochemistry of horseradish peroxidase (HRP) labeled immunoreagent, an immunosensor for CA 125 without need for an electrochemical mediator was proposed [51]. CA 125 was immobilized in a sol–gel matrix on a glassy carbon electrode. Based on a competitive assay between analyte CA 125 and HRP-labeled anti-CA 125 in the sample, the immunocomplex between labeled antibody and antigen immobilized on the electrode surface resulted in an electrochemical signal corresponding to direct Fe(III) to Fe(II) reduction within the structure of the HRP label. CA 125 could be measured with a detection limit of 1.29 U/mL. By comparison, the label free, conductometric biosensor for CA 125 using a polypyrrole nanowire, described above, produced a similar limit of detection, pointing to sensitivity advantages brought by nanotechnology. An amperometric biosensor for vascular endothelial growth factor (VEGF), a glycoprotein overexpressed in a number of different human tumors, was recently reported [52]. Anti-VEGF antibodies, labeled with ferrocene monocarboxylic acid, were immobilized on a carbon fiber microelectrode. The ferrocene label is easily oxidized at an applied potential of + 0.52 V vs. Ag/AgCl. Exposure of the sensor to VEGF resulted in a decrease in electrochemical response of ferrocene monocarboxylic acid due to increased spatial blocking. The proposed immunosensor showed a lower detection limit of about 38 pg/mL. Another electrochemical nano-label for signal amplification is the quantum dot (QD). A QD is a nanoparticle containing over 10,000 metal ions. An electrochemical biosensor for detection of PSA using QD labels was constructed using multiple reaction zones on a disposable, immunochromatographic strip [53]. Quantum dots consisting of CdSe as the core and ZnS as the shell were used to label PSA antibodies. PSA in a sample reacted with QD labeled anti-PSA in one zone of the strip, followed by capture of the complex by a second PSA antibody in another zone. Cadmium ions were liberated from the complex, measured voltammetrically, and the resulting signal proportional to PSA concentration. Excellent sensitivity toward PSA was obtained with a detection limit of 0.02 ng/mL. An optical biosensor scheme for the breast cancer marker HER2/neu used fluorescent silica nanospheres as signal amplifiers [54]. Silica nanospheres were encapsulated with the fluorescent dye [Ru(phen)3] and used to label HER2/neu antibodies. A competitive assay between HER2/neu in a sample and HER2/neu immobilized on a glass substrate resulted in an assay with a linear range up to 10 μg/mL and a detection limit approaching 1 ng/mL. Luminescence from the glass substrate was monitored at 590 nm when excited at 460 nm. The biosensor assay was lengthy, requiring up to one hour and involving multiple incubation and washing steps.
3.2. Progress in development of biosensors for DNA analysis
Detection of mutations in a nucleic acid sequence related to development of disease is the basis for research into genetic biosensor technology. Single-nucleotide polymorphism (SNP) is a single base pair variation which can affect how humans develop disease and respond to various therapeutic measures. Proposed biosensors for analysis of DNA are generally based on the process of hybridization, matching one strand of DNA with its complementary strand, and the ability for SNP detection. The hybridization event on the active surface of a biosensor may be detected with or without the use of labels. A commercial biosensor microarray known as the GeneChip™, using fluorescent labels to detect hybridization, was introduced in 1996 by Affymetrix (Santa Clara, CA), a pioneer in the development of commercial biosensor arrays for DNA analysis. Affymetrix continues to expand its applications with a variety of arrays available for analysis of gene expression and whole genome analysis related to cancer, inflammatory disease, infectious disease and diabetes, to guide disease detection and therapy. Another commercially available platform from GeneFluidics (Monterey Park, CA) uses a sensor array chip consisting of 16 nanoscale Au electrodes, modified with thiol SAMs, optimized for biomolecule immobilization [55]. The sensor chip may be used with associated instrumentation as the basis for affinity biosensors, including DNA sensors, using a variety of electrochemical methods. HRP is a preferred label owing to its fast electron transfer kinetics.
Methods for label-free biosensors to detect DNA mutations have been published. A quartz crystal microbalance with piezoelectric sensing has been reported for detection of mutations in the tumor suppressor gene p53, a gene showing a wide spectrum of mutations in human tumors [56]. A biotinylated DNA probe was immobilized on the quartz crystal surface, which had been coated with gold and treated with thiol and streptavidin. Following PCR amplification, the sensor was able to distinguish between complementary and non-complementary oligonucleotides with one base difference at the sub-micromolar level. A new commercial system (QCMagic, Elbatech, Italy) was used to carry out the measurements, with an assay time of approximately 20 min. In a similar study, SAW sensors (Love wave devices) were shown capable of detecting point mutations in the p53 gene and the tumor suppressor gene BRCA1, a gene mutation correlated with breast cancer [57]. The sensor could distinguish between mass changes due to hybridization of short DNA fragments at the sensor surface and longer, non-specifically adsorbed DNA strands which affect viscosity. Single nucleotide mutations could be detected reliably at low nanomolar concentrations based on association and dissociation kinetics of DNA oligonucleotide hybridizations on the sensor chip.
Point mutations in the same BRCA1 gene were detected using a sensitive chronocoulometric method using an electrochemical label and gold nanoparticles for signal amplification [58]. A capture probe DNA is immobilized on a gold electrode. A reporter probe, loaded on AuNP, is capable of hybridizing with one of two sequences on the target DNA, and the other sequence hybridizes with the immobilized capture probe. Each hybridization event on the electrode surface brings multiple reporter probes for each AuNP. Electroactive [Ru(NH3)6]3+ is electrostatically bound to the reporter probes and coulometric signal from reduction of the Ru complex is proportional to concentration of target DNA (Fig. 3 ). The DNA sensor could selectively detect as low as femtomolar concentrations of target DNA in less than 2 h. A variant of a sandwich type DNA assay scheme, using electrochemical detection, involved proximity-dependent surface hybridization of a ferrocene labeled reporter oligonucleotide sequence [59]. In this approach, a thiolated capture oligo probe was immobilized on a gold electrode. When the complementary target DNA was introduced, it was complementary to the region of the capture probe furthest from the surface of the gold. The reporter oligo with the ferrocene label was prepared to be complementary to both one end of the target as well as the sequence of the surface immobilized capture probe that was closest to the gold electrode. Hence, in the presence of the target, the ferrocene tracer ends up being positioned extremely close to the surface of the gold, making it more easily observed electrochemically via a simple differential pulse voltammetric scan, allowing femtomolar detection of the target DNA.
Fig. 3.
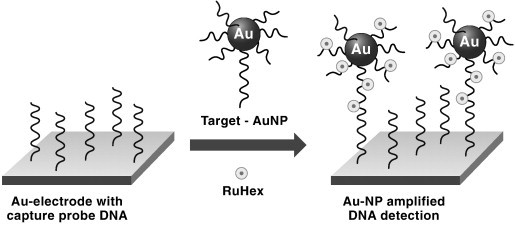
Schematic of a coulometric biosensor for detection of DNA mutations using DNA reporter probes labeled with an electroactive ruthenium complex (RuHex = [Ru(NH3)6]3+) and loaded onto gold nanoparticles.
Reprinted from Ref. [44] with permission from IOP Publishing.
An optical, thin-film biosensor chip for SNP analysis has been constructed on a silicon wafer by depositing a 475 Å layer of silicon nitride as an optical layer [60]. Synthetic oligonucleotide capture probes were covalently attached to the hydrazine-derivatized chip surface. Target DNA sequences, resulting from PCR amplification, were hybridized on the chip in the presence of biotinylated detector probes. An individual SNP was identified by a color change when a biotinylated detector probe perfectly matched a capture oligomer. Color was developed by adding an anti-biotin IgG-HRP conjugate along with an HRP substrate, tetramethyl benzidine, to the chip surface. Low oligonucleotide spotting densities were read as a visual color change, and chips with higher densities were visualized by digital imaging. Although the biosensor assay was complex, requiring separation/washing steps and PCR amplification, the sensitivity was excellent, with as little as 0.1 fmol of target detectable depending on the spot density. Validity of the assay was tested by correctly identifying 3 genetic polymorphisms associated with venous thromboembolism in DNA samples from 100 individuals at risk, compared to a laboratory method.
Detection of DNA methylation at specific transcription sites on tumor suppressor genes is important to identify because several tumor types have a high level of methylation in these regions. A nanowire/FET device, similar to that described above [18] was used to detect DNA methylation of the p16 tumor suppressor without PCR amplification [61]. Methylation of the p16 gene has been associated with development of several types of cancer, particularly lung cancer. Semiconductor nanowires were patterned on the gate of a nano-FET, followed by formation of an alkanethiol SAM on the nanowire and immobilization of anti-5-methylcytosine antibodies. Methylated DNA targets were captured on the FET surface. An increase in conductance was measured with as little as 2.5 × 10− 19 mol DNA.
3.3. Biosensors for markers of autoimmune diseases
Autoimmune diseases are disorders characterized by the presence of autoantibodies which bind proteins, peptides or other structural compounds of the body as target antigens. SPR has proven particularly useful as a mode of transduction for biosensors to detect autoantibodies in serum [62], although some examples of electrochemical biosensors and biosensors based on the quartz crystal microbalance have also been published. SPR is a phenomenon occurring when polarized light is directed from a layer of higher refractive index to one of lower refractive index. Usually, a thin layer of gold (~ 50 nm) is the low RI medium, placed over a prism (higher RI medium). Incident light through the high RI medium is transferred to the surface of the gold layer as packets of electrons called plasmons when the incident light through the prism is equal to a critical angle known as the resonance angle. The plasmons generate an electric field extending approximately 100 nm above the gold surface. Changes in composition of matter within this electric field (from an affinity reaction between sample analyte and immobilized biomolecules on the gold layer), alter the RI between sample and metal layer and appear as changes in the resonance angle. Since the RI change is sensitive to total biomolecule concentration at the gold surface, sensitivity of the SPR technique is limited by nonspecific adsorption at the sensor surface. The problem of nonspecific adsorption often requires sample dilution in cases where high concentrations of protein are present, e.g., serum or plasma. Deposition of certain polymers over the gold surface has been reported to limit nonspecific adsorption at SPR biosensors [50]. SPR offers the possibility for sensitive, label-free detection of autoantibodies in serum to diagnose autoimmune diseases and guide clinical decision making before onset of symptoms, as well as during the course of the disease.
Antiphospholipid syndrome (APS) is an autoimmune disease, the main clinical manifestations of which are venous and arterial thromboses. Lupus erythematosus is a disease associated with APS. Detections of two anti-phospholipid antibodies, anti-cardiolipin (CL) and anti-β2-glycoprotein I (β2-GPI) in serum are laboratory criteria for diagnosis of APS. SPR using the Biacore system has been investigated as a biosensor transduction mode for diagnosis of APS [62]. SAMs of alkanethiols were prepared on gold substrates and CL and β2-GPI attached to the surface using standard coupling chemistries. Analysis of samples from patients with APS showed β2-GPI to be the main antigenic structure in APS. The biosensor device could discriminate between anti-β2-GPI resulting from APS and anti-β2-GPI resulting from infection. Further, because the SPR biosensor monitors affinity reactions in real time, following kinetics of association and dissociation between antigen and antibody allowed discrimination between β2-GPI-specific IgG titers and the IgM isotypes [63].
Detection of antibodies toward native double-stranded DNA (anti-dsDNA) offers significance for diagnosis of lupus erythematosus. A SPR biosensor assay for anti-dsDNA was realized by covalently immobilizing dsDNA to the gold surface of a SPR chip [64]. Using a cut off value of 25 resonance units (1 ru ≈ 1 pg protein/mm2 of the chip surface), the SPR biosensor detected the presence of antibodies in sera from patients with confirmed diagnoses of the disease with 98% specificity at a sensitivity of 83%. The SPR device could be used for multiple sample injections and regeneration cycles in a flow through cell arrangement. A biosensor based on SPR was also described for monitoring the presence of autoantibodies associated with Wegener's Granulomatosis, an autoimmune disease characterized by inflammation and production of granuloma around blood vessels [65]. Antineutrophil cytoplasm autoantibodies toward proteinase 3 (PR3-ANCA) were monitored over time. It was found that the epitope specificities of PR3-ANCA affinity reaction change over the course of the disease and may be responsible for differences in functional properties of autoantibodies in various stages of the disease.
The quartz crystal microbalance (QCM) was proposed as the transducer in a biosensor for detection of autoantibodies in sera from patients with rheumatoid arthritis (RA) [66]. The QCM was coupled with single-walled carbon nanotubes to enhance sensitivity. A carbon nanotube film was formed on a QCM sensing crystal. The antigen, a citrulline-containing peptide, was attached to the surface of the nanotube via polyethylene glycol spacer chains. A decrease in natural resonance of the QCM crystal could be detected upon exposure of the device to sera from RA patients, vs. a control device with immobilized non-citrulline-containing peptide, indicating formation of immune complex on the surface of the QCM crystal. Sensitivity of the device in the femtomole range was shown, superior to the existing ELISA method.
An electrochemical biosensor for autoantibody detection in multiple sclerosis (MS) was reported. A synthetic glycosylated peptide (CSF114(Glc)) was modified at the N-terminus with an electrochemical ferrocenyl derivative and adsorbed on the surface of a gold electrode to form a SAM via the sulfur atom [67]. An electrode modified with the unglycosylated peptide acted as a control. The presence of purified antibodies toward the glycosylated peptide in sera from MS patients produced a positive shift in the oxidation peak for the ferrocene derivative measured by cyclic voltammetry, indicative of inhibition of the electrochemical activity resulting from formation of the immune complex. A detection limit lower than the ELISA method was reported.
3.4. Biosensors for markers of infectious disease
Biosensors offer promise for field diagnosis of infectious diseases in real time and the possibility for early intervention and disease containment [68], [69]. Deployment of hand held or portable point-of-care devices, based on biosensors, in remote field locations could serve such a purpose, especially in developing countries where access to healthcare locations with laboratory support is limited. A 2006 Workshop, co-sponsored by the National Institute of Biomedical Imaging and Bioengineering, the National Heart, Lung, and Blood Institute at the National Institutes of Health and the National Science Foundation, identified delivery of healthcare to developing countries as an area where point-of-care testing devices can fill an unmet need. Biosensors and associated technologies, e.g., lab-on-a-chip fluidics, are expected to be at the heart of such devices [70].
Dengue fever is a disease that has become prevalent in developing countries and represents a major international health concern. Dengue fever has been targeted as a disease where early diagnosis and therapy could be applied to contain spread of the disease. Sensitive detection systems based on biosensors would provide a method, with possible field application, toward this end. Research efforts have been applied in demonstrating measurement concepts using biosensors for identification of dengue virus. The quartz crystal microbalance with immobilized monoclonal antibodies against dengue viral antigens detected those antigens in human serum blended with dengue viral samples and in actual patient samples [71]. The immunochip was capable of detecting dengue E and NS1 proteins with detection limits of 1.727 and 0.740 μg/mL, respectively. However, sample pretreatment and dilution were required to reduce protein interference of the QCM, indicating that improvements are needed before being considered as a viable point-of-care device for field application. The same research group later demonstrated that a molecularly imprinted, thin (70 nm) polymeric film (MIP) toward an epitope of dengue virus NS1 protein could be used as a recognition element on the QCM [72]. Nonspecific interaction between the MIP and proteins was minimized, allowing sample measurements of the dengue virus NS1 protein without sample pretreatment. The binding effect was further enhanced using a monoclonal antibody toward the MIP–NS1 complex on the chip. Signal responses down to 50 ng/mL NS1 protein were demonstrated.
A genomic biosensor was coupled with microfluidics to demonstrate ultrasensitive measurement of RNA sequences specific to the dengue fever virus [73]. Microchannels with defined areas for capture and detection of RNA sequences were created in a polydimethylsiloxane substrate. Two different DNA probes complementary to unique sequences on the target pathogen RNA were used as biorecognition elements. For signal generation and amplification, one probe was coupled to dye encapsulated liposomes and the second probe was coupled to magnetic beads for target immobilization. Following hybridization with target RNA, the liposome-target-bead complex was captured on a magnet. A fluorescence reader was used to measure the quantity of liposomes captured, which is proportional to the concentration of target RNA. A detection limit in the picomolar range was achieved. While excellent sensitivity has been demonstrated in clean samples, much remains to be done to demonstrate performance of this concept in blood and serum to realize the goal of monitoring infectious diseases in the field.
Electrochemical detection of DNA hybridization can be performed by direct oxidation of DNA bases and has been used to identify DNA nucleotides, oligonucleotides and genomic sequences related to dengue virus and other infectious diseases [69]. A conducting polymer such as poly-4-aminophenol on electrode surfaces was found to be efficient for increasing the amplitude of oxidation signals produced by amperometric measurement of purine bases at modified electrodes compared to non-coated electrodes [74], [75]. The benefit of Au nanoparticles coupled to electrochemical sensors for signal amplification was again demonstrated for biosensors related to infectious diseases. Forest-Spring encephalitis is viral, tick-borne encephalitis prevalent in wooded areas of Eastern Europe and Asia. An amperometric biosensor for measurement of antibodies related to Forest-Spring encephalitis was carried out on a screen printed graphite electrode onto which the Forest-Spring encephalitis antigen was immobilized [76]. Antibodies present in human serum resulted in formation of the immune complex, which was recognized by AuNP-labeled Protein A. Signal resulting from Au oxidation voltammogram was proportional to the concentration of antibodies over the interval from 10− 7 to 10− 2 mg/mL, with good agreement to the ELISA method. Electrochemical impedance spectroscopy (EIS) has been applied to detect Hepatitis B antigen. Hepatitis B antibody was immobilized onto a platinum electrode modified with colloidal gold nanoparticles and polyvinyl butyral. Formation of the complex between Hepatitis B surface antigen (HBsAg) in a sample and the immobilized antibody resulted in changes in electron transfer resistance at the electrode surface using a redox probe (ferri/ferro cyanide) measured as an impedance change by EIS. Linear response toward HBsAg was measured over the range of 20–160 ng/mL, with a detection limit of 7.8 ng/mL. The enhanced sensitivity of the biosensor was attributed to the large specific surface area afforded by the Au nanoparticles and the encapsulating effect of polyvinyl butyral [77]. Other examples of EIS in combination with redox probes have been demonstrated capable of measuring formation of immune complexes at electrode surfaces in clean solutions. However, it remains to be seen if the method will be effective in biological fluids owing to changes in electron transfer resistance resulting from nonspecific adsorption at electrode surfaces [67].
Modification of electrochemical sensors with polymeric materials has been used to impart selectivity and sensitivity toward analytes of clinical interest, similar to enhanced oxidation signals toward DNA bases discussed above. A potentiometric immunosensor toward Hepatitis B surface antigen and other clinically important analytes was demonstrated by polymerizing polypyrrole on the surface of a screen printed gold electrode in a format the authors called a “universal transducer system” [78]. Antibodies were adsorbed or immobilized to the polypyrrole layer, followed by capture of the specific analyte from a sample. A second antibody labeled with HRP was complexed to the antigen and exposure to substrate resulted in a change in electrochemical activity of the enzyme conjugate, a change in the redox state of the polypyrrole and a resulting shift in the measured potential. The assay format is a standard ELISA technique; however the potential shift of the sensor resulting from the change in redox state of polypyrrole imparts an enhanced assay sensitivity, the mechanism of which is not well understood. HBsAg was detectable at a concentration of 0.05 IU/mL, corresponding to approximately 50 fM.
3.5. Research toward new biosensors for cardiac markers
Several recent publications highlight the use of biosensors together with nanotechnology to achieve sensitive detection of cardiac biomarkers for early diagnosis of acute myocardial infarction. In addition, biosensors for detection of biomarkers associated with the presence of unstable coronary plaque have appeared. Lin et al. created an array of nanowells by overlaying biogenic nanoporous silica on a gold electrode for measurement of C-reactive protein (CRP) and myeloperoxidase (MPO), two protein biomarkers associated with risk of vulnerable coronary plaque rupture [79]. The nanowells were created by overlaying a suspension of silica derived from the eukaryotic unicellular photosynthetic algae Coscinodiscus wailesii over a gold substrate. Each nanowell was approximately 40 nm in diameter with a gold electrode at the bottom (Fig. 4 ). Within each nanowell, protein detection was carried out by measuring single interactions between antibody and protein antigen. The affinity reaction perturbed the charge distribution in the electrical double layer at the electrode/liquid interface, measured as a change in capacitance by impedance spectroscopy. The measured signal is enhanced due to cumulative signal generation from the array of nanowells. Limit of detection for measurement of CRP and MPO in serum samples was 10 pg/mL, compared to 5 ng/mL on planar gold electrodes. Another electrochemical biosensor for cardiac troponin-T (cTnT) was demonstrated by creating clusters of n-doped silicon nanowires [80], in a biosensor assay similar to that described above for PSA [18]. Antibody to cTnT was attached to surface functionalized nanowires. Increase in conductivity of the nanowires was measured following binding of cTnT antigen to the nanowire surface, consistent with that expected for a n-type semiconductor. Limits of detection toward cTnT down to 30 fg/mL in undiluted serum, and 1 fg/mL in buffer were demonstrated. Nonspecific binding, tested with bovine serum albumin at 109-fold higher concentrations was negligible.
Fig. 4.
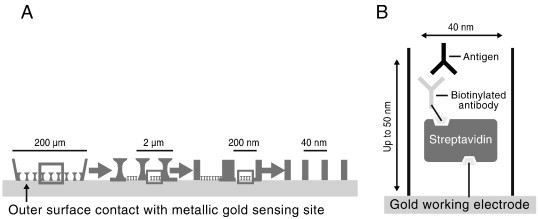
(A) Diagram of a nanowell array created by overlaying biogenic nanoporous silica on a gold substrate for electrochemical detection of C-reactive protein and myeloperoxidase and (B) an expanded view of the immunoreactions taking place in each nanowell.
Reprinted from Ref. [79] with permission from Elsevier.
An acoustic biosensor for CRP was demonstrated using the Resonant Acoustic Profiling (RAP™) biosensor (Akubio Ltd., Cambridge, UK) with comparable sensitivity to a high sensitivity CRP ELISA assay [81]. A high frequency voltage was applied to a piezoelectric crystal to induce oscillation, and its resonance frequency monitored in real time. The device was operated in a differential mode, where antibody toward a specific antigen was bound to the test crystal using standard coupling chemistry and IgG coupled to the control crystal to cancel out background signal. A specific binding reaction at the test crystal produced a reduction in the oscillation frequency. Detection limit for measurement of CRP in serum was found to be 13 ng/mL in the direct capture mode, and 3 ng/mL in the sandwich mode. The authors indicated the device is suitable as a monitor for risk of cardiovascular disease, where a high sensitivity CRP assay with a detection limit less than 300 ng/mL is required.
Utility of surface plasmon resonance as a mode of transduction in biosensors for cardiac troponin-I (cTnI) has been demonstrated. Ability of SPR to monitor association, dissociation and affinity constants in real time for a series of monoclonal antibodies to cTnI allowed optimization of a sandwich immunoassay on a gold coated SPR chip with a limit of detection of 0.25 μg/L, one order of magnitude lower than a direct SPR assay [82]. While the sensitivity of the biosensor based on the sandwich immunoreaction is close to being practical for measuring cTnI in sera from patients with AMI, sensitivity is limited by nonspecific protein adsorption, requiring washing after each incubation step, and an assay time of approximately 40 min. A later study using SPR-based biosensor for cTnI showed minimized nonspecific signal from serum proteins by immobilizing antibody toward cTnI on a self assembled monolayer consisting of N-hydroxysuccinimide activated 16-mercaptohexadecanoic acid [83]. A limit of detection of 0.7 μg/L in undiluted serum was demonstrated for the biosensor operated in a direct assay, similar to cTnI measurements made in saline solution, but still requiring improvements in sensitivity to realize a practical sensor for measurement of cTnI in sera from AMI patients.
Gold nanoparticles were shown to enhance fluorescence from a fluorophore in a fiber optic based immunosensor for cTnI by immobilizing nanoparticles an appropriate distance from the fluorophore by means of a self assembled monolayer. Free electrons from the fluorophore, normally used for self-quenching, were transferred to the strong surface plasmon polariton field of the AuNP [84]. The biosensor was based on a sandwich immunoassay using a fluorescence labeled second antibody toward cTnI. Signal enhancements in the presence of AuNP allowed quantitation of cTnI down to tens of picomolar concentration (approx. 0.2 μg/L). As with most biosensors using optical labels, washing steps were required to remove unbound label.
3.6. Proposed biosensors for other clinically important analytes
Glucose remains the model analyte of choice for demonstration of concepts toward improving biosensor performance, in particular electrochemical biosensors, owing to the widespread use of biosensors for blood glucose in systems for point-of-care testing, ease of working with the enzyme glucose oxidase, and a large literature base documenting known performance problems with state-of-the-art glucose biosensors. Newer enzyme immobilization matrices include sol–gel composite films [85], [86], polymeric microparticles [87] and chitosan [88], to achieve direct electron transfer between bioreactions and electrodes, without the need for nanoparticles. Performance advantages of direct electron transfer include faster response times (~ 10 s). Other recently published improvements include sensor use life of 60 days or longer, improved resistance toward environmental conditions during sensor storage and improved selectivity over typical electrochemical interfering substances (ascorbic acid, acetaminophen, uric acid, etc.). New point-of-care devices for measuring blood glucose continue to appear using disposable, mass produced biosensors requiring reduced blood sample volumes (1 μL or less), providing the ability to sample from sites other than the fingertip [89] and point-of-care devices measuring blood glucose together with other critical care analytes in a disposable sensor cartridge format [90].
An optical array biosensor for simultaneous measurement of markers for renal disease (urea, creatinine, uric acid, glucose) was produced by immobilizing the appropriate enzymes and fluorescent dyes in a sol–gel matrix. For urea and creatinine, actions of the hydrolase enzymes urease and creatinine deiminase produced hydroxide ions which were detected via an immobilized pH-sensitive fluorescent indicator. For glucose and uric acid, actions of the oxidase enzymes glucose oxidase and uricase produced hydrogen peroxide, which was consumed by co-immobilized horseradish peroxidase to give a fluorescent signal at 590 nm as a result of reduction of Amplex red to resorufin. Measurement of analytes in serum was in good agreement with traditional laboratory methods [91]. A hand-held device for measurement of whole blood creatinine in point-of-care environments has recently become available (StatSensor creatinine meter, Nova Biomedical). The device uses a known 3-enzyme cascade to generate hydrogen peroxide from creatinine, and amperometric measurement of peroxide in a disposable strip format. The strip requires a small sample volume (1.2 μL) and minimizes electrochemical interferences by means of a multi-electrode differential approach. A recent evaluation showed blood creatinine results from the StatSensor to correlate well with laboratory plasma measurements (R 2 = 0.9328) but with a negative proportional bias which became less evident in specimens from post-dialysis renal patients. High concentrations of creatine and urea resulted in falsely elevated creatinine measurements [92].
A biosensor for monitoring thrombin generation demonstrated advantages of electrochemical biosensors because they are unaffected by color or turbidity, and may be used for measurements in various sample types, including platelet-poor plasma, platelet-rich plasma and whole blood, even after initiation of clot formation [93]. The assay was developed on a single-use test strip onto which were deposited a palladium microelectrode as the working electrode and an Ag/AgCl combined reference and counter electrode, allowing measurements in < 10 μL of sample. Also deposited on the strip were a reagent mixture consisting of a clotting activating substance (silica) and a substrate for thrombin, an oligopeptide derivative with an amide linkage that mimics the thrombin-cleaved amide linkage in fibrinogen. Upon addition of sample, activation of the coagulation cascade generated thrombin, which cleaved an electroactive group, 4-amino-2-chlorophenol, from the substrate. The electroactive leaving group was measured using chronoamperometry at an applied potential of + 0.300 V. The amount of active thrombin present in the sample was linearly related to the maximum positive slope of the current vs. time curve. Included as proof of concept was the ability to distinguish between normal samples and factor V Leiden samples in platelet-poor plasma and in whole blood. The method holds promise for assessment of thrombin generation as a point-of-care test. A commercially available test for activated clotting time in a single-use, disposable cartridge format (Abbott i-STAT) uses celite as a clotting activator and is based on the same measurement principle. Generation of thrombin is monitored amperometrically based on the ability of thrombin to cleave an electroactive group from a substrate with an amide linkage similar to fibrinogen. The device was evaluated for monitoring heparin therapy during cardiopulmonary bypass surgery [94].
Surface plasmon resonance biosensors have been shown useful in endocrinology, both for quantitative measurement of hormones and fundamental studies related to mechanisms of the endocrine system. Measurement of progesterone in whole blood was demonstrated using a surface sensitive optical wave guide in an optical configuration similar to that used in SPR. A derivative of progesterone was covalently bound to an activated sensor surface. The measurement was carried out as an indirect, competitive immunoassay, requiring a washing step for removal of excess antibodies. Non-specific binding of interfering substances, particularly for measurements in blood, required treatment of the sensor surface with an undisclosed hydrophobic repulsive agent. In this way, progesterone in whole blood could be measured from 0.005 to 10 ng/mL, with a detection limit of 3 pM and a relative standard deviation of 3.5% [95]. A SPR biosensor was also used for simultaneous determination of 4 pituitary hormones, thyroid stimulating hormone, human growth hormone, follicle stimulating hormone and luteinizing hormone, in serum and urine in a competitive assay format. The 4 hormones were covalently attached to the surface of a gold SPR chip, covered with an alkanethiol self-assembled monolayer. The sensors could be regenerated and used for up to 100 cycles by washing with a solution of 2 M NaCl including surfactant. After exposure of the sensors to serum containing analyte and corresponding antibody, the sensor surface was washed with high ionic strength buffer to remove nonspecifically bound protein. In this way, quantitation of the 4 hormones with limits of detection from 1 to 6 ng/mL was realized [96]. Biosensing using SPR has also been used for high throughput screening of chemicals with potential thyroxine (T4) transport disrupting activity by means of their affinities for the thyroid hormone transport protein, T4 binding globulin (TBG). The most potent binding was observed with hydroxylated metabolites of several brominated diphenyl ethers, commonly found in human plasma [97]. In another research study, the Biacore X® SPR biosensor system was used to test the ligand-binding activity of the sex hormone-binding globulin (SHBG) in serum from men with liver cirrhosis. It was shown that high SHBG serum levels associated with liver cirrhosis reduce free testosterone, causing a shift in hormonal balance toward estrogenic compounds accounting for endocrine symptoms of the disease [98]. The same group used the Biacore SPR system to identify formation of high molecular-mass aggregates of SHBG at concentrations above 200 nmol/L in undiluted serum, limiting the upper measurement range of commercially available immunoassay systems [99].
A label-free electrochemical biosensor for measurement of theophylline in serum used a proximity-dependent signal from a ferrocene redox probe, similar to that described earlier for DNA analysis [59]. An RNA aptamer sequence was identified to recognize and selectively bind theophylline. The RNA aptamer was immobilized on a gold electrode via thiol chemistry, and the other terminal position of the aptamer labeled with ferrocene. In the absence of theophylline, the aptamer was in the open conformation and the ferrocene probe was distant from the electrode. In the presence of theophylline, the aptamer folds into a hairpin structure bringing the ferrocene probe very close to the electrode surface, and the signal measured by cyclic voltammetry (Fig. 5 ). The sensor was used to measure therapeutic concentrations of theophylline in serum (20–100 μmol/L) in the presence of 1 mmol/L caffeine, a structurally related compound known to interfere with chromatographic methods and commercial immunoassays for theophylline [100]. Acetylcholinesterase (AChE) inhibitors neostigmine and eserine, known to have applications as therapeutic drugs for neurodegenerative diseases such as myasthenia gravis and Alzheimer's Disease, were detected by SPR using a commercially available system (Spreeta SPR sensor, ICX Nomadics). AChE was immobilized on the gold surface of a SPR sensor through covalent attachment to a SAM of COOH-terminated alkanethiol. Binding of the 2 reversible inhibitors to AChE could be monitored without need to further modify the sensor surface or use other reagents. Measurement of affinity constants, an advantage offered by SPR, showed higher affinity of the sensor for neostigmine, concluding it to be the more potent AChE inhibitor. Due to the small molecular weight of the two compounds, it was further concluded that the SPR sensor was not measuring an increase in mass due to the affinity reaction, rather a conformational change in enzyme structure upon inhibitor binding [101].
Fig. 5.
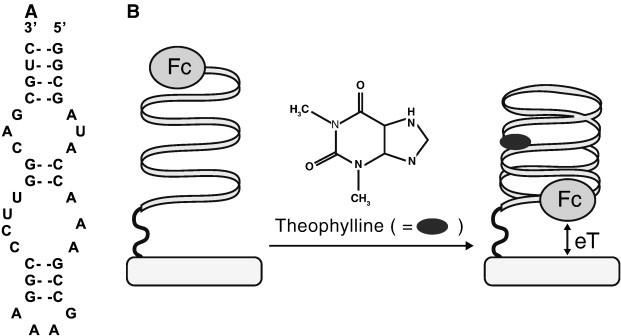
(A) Theophylline-binding aptamer sequence and (B) representation of the electrochemical aptamer-based sensor for theophylline (Fc = ferrocene).
Reprinted from Ref. [100] with permission from the American Chemical Society.
Amyloid-β (Aβ) is suspected to play a role in the pathogenesis of Alzheimer's Disease through formation of insoluble ligands, termed amyloid-derived diffusible ligands (ADDL), leading to neurological dysfunction related to memory. A SPR biosensor, modified with Ag nanoparticles, has been demonstrated for measurement of ADDL in cerebrospinal fluid (CSF) and human brain extract. The SPR biosensor identified 2 ADDL epitopes, with distinct binding constants, which allowed demonstration of a sandwich immunoassay with improved sensitivity compared to a single epitope. Size of the nanoparticles was optimized to extend 35 nm from the surface of the SPR chip for implementation of the sandwich immunoassay. Results indicated detection of approximately 1 pM ADDL in extract from diseased brain was possible while signal from control samples was undetectable. Further, ADDL detected in CSF and correlated with the presence of disease by assaying brain extract, holds promise for development of a diagnostic test for Alzheimer's Disease using CSF while a patient is still alive [102]. A more recent study demonstrated the ability to detect Aβ in serum, along with 3 other potential markers of Alzheimer's Disease (glutamate, acetylcholine and H2O2), using an array biosensor with fluorescent detection. Aβ in the presence of Cu2+ produced H2O2, which in the presence of horseradish peroxidase, reduced the fluorescent dye Amplex red to resorufin. Dye and enzyme were co-immobilized in a sol–gel matrix. Detection limit toward Aβ was 0.63 nM, spiked into serum. It was not stated if this limit of detection is sensitive enough for disease diagnosis in clinically relevant samples; however, the authors speculated that simultaneous detection of multiple markers will provide greater sensitivity for disease detection compared to a single biomarker [103].
4. New directions for clinical biosensors
4.1. Biosensors for alternate sample testing — saliva diagnostics
Biosensors for measurement of physiologically important analytes in samples other than blood, serum or plasma are being applied to selected areas of diagnostic research, outside of traditional clinical chemistry testing. Saliva, in particular, has been targeted as the perfect medium for identification of protein and genetic markers related to disease and physiological status, owing to the ease of collecting, storing and shipping samples in sufficient quantities for analysis. Several studies have demonstrated that saliva can be used to reliably detect HIV, viral hepatitis A, B and C, drugs of abuse and genetic biomarkers related to certain types of cancer. Development of nanoscale and microscale biosensor technologies to detect salivary protein and genetic biomarkers for point-of-care applications has been the focus of research organizations such as the National Institute of Dental and Craniofacial Research [104]. An outcome of this work has been development of a hand-held point-of-care device and disposable biosensor strip for measurement of salivary α-amylase (sAA), a principal salivary protein. Concentration of sAA in saliva has been shown to correlate well with changes in plasma norepinephrine concentrations in response to trauma and stress. The biosensor device was shown to agree with sAA activities measured by a clinical chemistry analyzer from approximately 20–200 kU/L [105]. In a specific application of this device, it was demonstrated that sAA activity increased with mental and physical fatigue. The authors speculated that mental and physical stimuli activate the sympathetic nervous system, which in turn leads to increased production of salivary amylase [106]. The biosensor consisted of a disposable test strip onto which was deposited the substrate 2-chloro-4-nitrophenyl-4-O-β-d-galactopyranosylmaltoside, which is hydrolyzed by amylase to form a yellow colored product. Product formation was monitored kinetically at 430 nm using a LED source in a hand-held reader. Total assay time was 1 min and an algorithm for normalizing individual differences in salivary amylase activity was required.
In the field of dental research, a biosensor for analysis of salivary phosphate has been proposed [107]. Salivary phosphate concentration has been investigated as an indicator of risk for developing dental caries and dental calculus. The biosensor was constructed by immobilizing pyruvate oxidase on a screen-printed electrode consisting of a platinum working electrode and an Ag/AgCl reference/counter electrode. The enzyme consumes phosphate, in the presence of pyruvate and oxygen, to generate H2O2, which is oxidized at a potential of + 0.420 V. Performance of the biosensor for measurement of salivary phosphate was evaluated against a commercial test kit using an optical method. The sensor demonstrated a linear range from 7.5 to 625 μM with a detection limit of 3.6 μM using samples from 50 subjects, with stimulated and unstimulated saliva. Agreement with the commercial kit showed R 2 = 0.9646, slope = 1.17. The authors suggested that higher recovery using the biosensor method was due to negative interference with the optical test kit owing to sample turbidity.
Saliva was used for detection of the virus associated with the outbreak of severe acute respiratory syndrome (SARS) in 2003, using a biosensor based on the quartz crystal microbalance [108]. Horse polyclonal antibodies against the SARS coronavirus were immobilized on a piezoelectric crystal. Saliva samples containing the virus were formed into an aerosol before exposure to the sensor. The SARS virus was specifically adsorbed onto the piezoelectric crystal resulting in a frequency shift. Frequency shifts were linearly dependent on antigen concentration in the range of 0.6–4 μg/mL with an assay time less than 2 min. The biosensor could be used for up to 100 assays without detectable loss of activity. Selectivity was demonstrated by exposure of the sensor to 1 μg/μL of flu-associated virus without interference.
4.2. Novel biosensors based on immobilized whole cells as recognition elements
There has been renewed interest over the past decade in biosensors using intact cells as the recognition element judging from the increasing number of research reports and reviews appearing on this topic [109], [110]. These devices have come to be known as whole cell or microbial biosensors. A claimed advantage of microbial biosensors is that immobilization of whole cells improves the stability of an intracellular biorecognition element, such as an enzyme, because the enzyme is retained in its natural environment. On the other hand, microbial biosensors are limited by low sensitivity and selectivity owing to the fact that a cell wall or membrane serves as a permeability barrier and side reactions catalyzed by other enzymes within the cell may produce unwanted interfering substances. Recent developments in molecular biology offer methods to construct genetically engineered microorganisms, providing a means to alter the selectivity and sensitivity of microbial biosensors. Despite these recent advances, it is important to recognize the pioneering work in this subspecialty of biosensors, which is often not cited in research reports and review articles on microbial biosensors appearing in the last 8 years [111].
An example of how cell biochemistry may be manipulated to optimize production of cells for microbial biosensors was recently reported for constructing a microbial biosensor for urea [112]. The bacterium Brevibacterium ammoniagenes was grown in a nitrogen poor medium to enhance urease expression. The cells were immobilized in a polystyrene sulphonate-polyaniline conducting polymer on the surface of a platinum conductometric electrode. The catalytic action of urease on urea produced ammonia, causing a localized pH increase within the polymer and a simultaneous increase in resistance. The sensor response time was 3 min, however, measurements could be made only in ascending order of analyte concentration and reversal of sensor response following exposure to high urea concentrations required 2 h in analyte-free buffer. In another example of a microbial sensor for l-lactate, cells of the yeast Hansenula polymorpha were genetically engineered for enhanced permeability and over production of mitochondrial flavocytochrome b 2, which catalyzes electron transfer from l-lactate to cytochrome c in yeast mitochondria [113]. Cells were immobilized on the surface of a graphite electrode within an electrodeposited polymer film. Enhanced cell permeability allowed free diffusion of the redox mediator phenazine methosulphate into the cell to couple electrons from l-lactate oxidation to the electrode. The biosensor based on the genetically engineered yeast cells exhibited 20–25 times higher KM compared to native cells, and an increase in the linear range for l-lactate determination to 1.6 mmol/L. These recent examples of microbial biosensors for simple molecules such as urea and lactate are not practical for routine clinical measurements; however, they have been used to demonstrate how manipulation of cell biochemistry can be used to alter analytical characteristics of microbial biosensors.
A more promising application of microbial biosensors is for early detection of cancer and monitoring efficacy of cancer treatment. Tumor angiogenesis occurs during the early phase of cancer, when a tumor progresses from the benign to malignant state. During this phase, tumors secrete various cytokines, such as vascular endothelial growth factor, basic fibroblast growth factor, hepatocyte growth factor and tumor necrosis factor-alpha, which aid in recruiting blood vessels from the existing vasculature. A whole cell biosensor has been reported for detection of these cytokines in sera from cancer patients [114]. Human umbilical vein endothelial cells were immobilized at the surface of a potassium ion selective electrode. The presence of the cells at the sensor surface inhibited response of the sensor to K+. Exposing the cell-based biosensor to individual and combined cytokines at concentrations observed in sera from cancer patients increased cell permeability and enhanced response of the sensor to K+. A dose dependent response of the biosensor to the cytokines showed significantly higher responses in sera from patients with early stages of various types of cancer compared to healthy individuals, and promise as a method for early detection of the disease. Another example of electrochemical biosensors for monitoring response of tumor cells to various drugs for cancer treatment was demonstrated by growing tumor cells directly on silicon-based electrochemical sensors for measuring pH, pO2 and capacitance. Viable cancer cells demonstrate acidification of the surrounding medium and oxygen uptake as a result of cellular metabolism, which could be followed using the pH and pO2 sensors. Capacitance measurement was used to follow morphological changes in cell structure and cell growth. Colon and breast cancer cells and doxorubicin-sensitive and resistant sarcoma cell lines were grown on the silicon chip surface. Drug resistant cells showed expected lowering of pH and increased oxygen consumption as cellular metabolism continued. Drug sensitivity showed reduction in medium acidification and reduced rate in oxygen consumption. An increase in capacitance reflected reduced cell–cell contact due to breakdown in cell structure and less substrate adhesion in response to the drug, correlated with changes observed by microscopy [115].
Finally, microbial biosensors have been proposed as devices for monitoring growth of pathogenic bacteria. A filter paper-based biosensor for semiquantitative detection of bacterial quorum sensing molecules N-acylhomoserine lactones (AHLs) has been described [116]. Quorum sensing molecules are believed to be the mechanism whereby bacteria communicate among themselves to coordinate growth and behavior. The reported device used a genetically engineered E. coli sensing system dried on a paper strip. The presence of AHL resulted in expression of a reporter protein, β-galactosidase. Addition of a chromogenic substrate to the strip resulted in formation of a blue color which could be visually read for semiquantitative determination of AHL. The device was used for detection of AHL in saliva samples from patients with gastrointestinal disorders down to 10− 8 M. The authors proposed the device could be used to detect the presence of low levels of bacteria in point-of-care settings in a noninvasive manner, free from requirements for instrumentation and trained personnel.
5. Conclusions
There remains a great deal of interest in developing biosensors for clinical application evidenced by the increasing number of research reports appearing in the last 8 years. Several reviews, cited herein, have correctly concluded that commercialization of biosensors and their applications to practical clinical analysis have not maintained pace with the large amount of research activity. This report has noted a few isolated examples of newly commercialized clinical biosensors and their applications, especially for near patient testing, and other examples of commercial biosensor platforms designed for research applications. However, with the exception of blood glucose sensing, the prediction of years ago that biosensors would revolutionize clinical analysis has not yet occurred, even though several reported biosensors equal or surpass traditional clinical chemistry methods in important performance metrics such as sensitivity, specificity and time to result.
Similar to blood glucose testing, the impact of biosensor technology will be felt greatest in point-of-care settings, especially in settings without laboratory support. For example, in rural locations or in developing countries, it could be envisioned that biosensors for infectious disease testing will play a role in allowing rapid treatment and containing spread of disease. It will be expected that quantitative performance metrics of these biosensor devices will be comparable to laboratory methods. In addition, successful commercial biosensors for point-of-care testing will be those meeting less quantitative requirements: reagent-free devices with minimal associated instrumentation, assays which are easy to use by non-trained personnel, reliability and freedom from maintenance, ability to accept whole blood (or other sample type) directly without pretreatment and internal, automated quality control. While the present biosensor research literature may seem overburdened by an increasing number of publications, it is important to continue working on a variety of biosensor technologies, with a view to the above requirements, to allow a successful transition from the research laboratory to patient testing locations.
References
- 1.D'Orazio P. Biosensors in clinical chemistry. Clin Chim Acta. 2003;334:41–69. doi: 10.1016/s0009-8981(03)00241-9. [DOI] [PubMed] [Google Scholar]
- 2.Ronkainen N.J., Halsall H.B., Heineman W.R. Electrochemical biosensors. Chem Soc Rev. 2010;39:1747–1763. doi: 10.1039/b714449k. [DOI] [PubMed] [Google Scholar]
- 3.Grieshaber D., MacKenzie R., Voros J., Reimhult E. Electrochemical biosensors — sensor principles and architectures. Sensors. 2008;8:1400–1458. doi: 10.3390/s80314000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li C.M., Dong H., Cao X., Luong J.H., Zhang X. Implantable electrochemical sensors for biomedical and clinical applications: progress, problems, and future possibilities. Curr Med Chem. 2007;14:937–951. [PubMed] [Google Scholar]
- 5.Lee T.M.H. Over-the-counter biosensors: past, present and future. Sensors. 2008;8:5535–5559. doi: 10.3390/s8095535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luong J.H.T., Male K.B., Glennon J.D. Biosensor technology: technology push versus market pull. Biotechnol Adv. 2008;26:492–500. doi: 10.1016/j.biotechadv.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Vaddiraju S., Tomazos I., Burgess D.J., Jain F.C., Papadimitrakopoulos F. Emerging synergy between nanotechnology and implantable biosensors: a review. Biosens Bioelectron. 2010;25:1553–1565. doi: 10.1016/j.bios.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patolsky F., Zheng G., Lieber C.M. Nanowire sensors for medicine and the life sciences. Nanomedicine. 2006;1:51–65. doi: 10.2217/17435889.1.1.51. [DOI] [PubMed] [Google Scholar]
- 9.Yeh J.I., Shi H.B. Nanoelectrodes for biological measurements. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2:176–188. doi: 10.1002/wnan.70. [DOI] [PubMed] [Google Scholar]
- 10.Simmel F.C. Toward biomedical applications for nucleic acid nanodevices. Nanomedicine. 2007;2:817–830. doi: 10.2217/17435889.2.6.817. [DOI] [PubMed] [Google Scholar]
- 11.Rivas G.A., Rubianes M.D., Rodriguez M.C. Carbon nanotubes for electrochemical sensing. Talanta. 2007;74:291–307. doi: 10.1016/j.talanta.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Carbon nanotubes as biosensors: realizing the full potential. May 20, 2010. Nanotechweb.org
- 13.Justino C.I.L., Rocha-Santos T.A., Duarte A.C. Review of analytical figures of merit of sensors and biosensors in clinical applications. TRAC-Trends Anal Chem. 2010;29:1172–1183. [Google Scholar]
- 14.Lozano M.L., Rodriguez M.C., Herrasti P., Galicia L., Rivas G.A. Amperometric response of hydrogen peroxide at carbon nanotube paste electrodes modified with an electrogenerated poly(Fe(III)-5-amino-phenantroline). Analytical applications for glucose biosensing. Electroanalysis. 2010;22:128–134. [Google Scholar]
- 15.Tan X., Li M., Cai P., Luo L., Zou X. An amperometric cholesterol biosensor based on multiwalled carbon nanotubes and organically modified sol–gel/chitosan hybrid composite film. Anal Biochem. 2005;337:111–120. doi: 10.1016/j.ab.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 16.Bhambi M., Sumana G., Malhotra B.D., Pundir C.S. An amperometric uric acid biosensor based on immobilization of uricase onto polyaniline-multiwalled carbon nanotube composite film. Artif Cells Blood Substit Biotechnol. 2010;38:178–185. doi: 10.3109/10731191003716344. [DOI] [PubMed] [Google Scholar]
- 17.Veetil J.V., Ye K. Development of immunosensors using carbon nanotubes. Biotechnol Prog. 2007;23:517–531. doi: 10.1021/bp0602395. [DOI] [PubMed] [Google Scholar]
- 18.Li C., Curreli M., Lin H. Complementary detection of prostate specific antigen using In2O3 nanowires and carbon nanotubes. J Am Chem Soc. 2005;127:12484–12485. doi: 10.1021/ja053761g. [DOI] [PubMed] [Google Scholar]
- 19.Okuno J., Maehashi K., Kerman K., Takamura Y., Matsumoto K., Tamiya E. Label-free immunosensor for prostate specific antigen based on single walled carbon nanotube array-modified microelectrodes. Biosens Bioelectron. 2007;22:2377–2381. doi: 10.1016/j.bios.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 20.Lee J.O., So H.M., Jeon E.K., Chang H., Won K., Kim Y.H. Aptamers as molecular recognition elements for electrical nanobiosensors. Anal Bioanal Chem. 2008;390:1023–1032. doi: 10.1007/s00216-007-1643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X., Bansaruntip S., Nakayama N., Yenilmez E., Chang Y.I., Wang Q. Carbon nanotube DNA sensor and sensing mechanism. Nano Lett. 2006;6:1632–1636. doi: 10.1021/nl060613v. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Kawde A.N., Musameh M. Carbon-nanotube-modified glassy carbon electrodes for amplified label-free electrochemical detection of DNA hybridization. Analyst. 2003;128:912–916. doi: 10.1039/b303282e. [DOI] [PubMed] [Google Scholar]
- 23.Wei F., Lillehoj P.B., Ho C.M. DNA diagnostics: nanotechnology-enhanced electrochemical detection of nucleic acids. Pediatr Res. 2010;67:458–468. doi: 10.1203/PDR.0b013e3181d361c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerman K., Saito M., Yamamura S., Takamura Y., Tamiya E. Nanomaterial-based electrochemical biosensors for medical applications. TRAC-Trends Anal Chem. 2008;27:585–592. [Google Scholar]
- 25.Liu G., Lin Y. Nanomaterial labels in electrochemical immunosensors and immunoassays. Talanta. 2007;74:308–317. doi: 10.1016/j.talanta.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkoci A. Electrochemical biosensing with nanoparticles. FEBS J. 2007;274:310–316. doi: 10.1111/j.1742-4658.2006.05603.x. [DOI] [PubMed] [Google Scholar]
- 27.Rusling J.F., Sotzing G., Papadimitrakopoulosa F. Designing nanomaterial-enhanced electrochemical immunosensors for cancer biomarker proteins. Bioelectrochemistry. 2009;76:189–194. doi: 10.1016/j.bioelechem.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Guo Q., Cui D. Recent advances in nanotechnology applied to biosensors. Sensors. 2009;9:1033–1053. doi: 10.3390/s90201033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei H., Li B., Li J., Wang E., Dong S. Simple and sensitive aptamer-based colorimetric sensing of protein using unmodified gold nanoparticle probes. Chem Commun. 2007:3735–3737. doi: 10.1039/b707642h. [DOI] [PubMed] [Google Scholar]
- 30.Taton T.A., Mirkin C.A., Letsinger R.L. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 31.Lefferts J.A., Jannetto P., Tsongalis G.J. Evaluation of the Nanosphere Verigene (R) System and the Verigene (R) F5/F2/MTHFR nucleic acid tests. Exp Mol Pathol. 2009;87:105–108. doi: 10.1016/j.yexmp.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Carrascosa L.G., Moreno M., Alvarez M., Lechuga L.M. Nanomechanical biosensors: a new sensing tool. TRAC-Trends Anal Chem. 2006;25:196–206. [Google Scholar]
- 33.Ilic B., Craighead H.G., Krylov S., Senaratne W., Ober C., Neuril P. Attogram detection using nanoelectromechanical oscillators. J Appl Phys. 2004;95:3694–3703. [Google Scholar]
- 34.Fritz J. Cantilever biosensors. Analyst. 2008;133:855–863. doi: 10.1039/b718174d. [DOI] [PubMed] [Google Scholar]
- 35.Fritz J., Baller M.K., Lang H.P. Translating biomolecular recognition into nanomechanics. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 36.Erickson D., Mandal S., Yang A.H.J., Cordovez B. Nanobiosensors: optofluidic, electrical and mechanical approaches to biomolecular detection at the nanoscale. Microfluid Nanofluid. 2008;4:33–52. doi: 10.1007/s10404-007-0198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao Y.L., Zhang G.G. Enhancing the sensitivity of SAW sensors with nanostructures. Curr Nanosci. 2006;2:311–318. [Google Scholar]
- 38.Arruda D.L., Wilson W.C., Nguyen C. Microelectrical sensors as emerging platforms for protein biomarker detection in point-of-care diagnostics. Expert Rev Mol Diagn. 2009;9:749–755. doi: 10.1586/erm.09.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moll N., Pascal E., Dinh D.H. A Love wave immunosensor for whole E. coli bacteria detection using an innovative two-step immobilization approach. Biosens Bioelectron. 2007;22:2145–2150. doi: 10.1016/j.bios.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 40.Singh K.V., Whited A.M., Ragineni Y., Barrett T.W., King J., Solanki R. 3D nanogap interdigitated electrode array biosensors. Anal Bioanal Chem. 2010;397:1493–1502. doi: 10.1007/s00216-010-3682-z. [DOI] [PubMed] [Google Scholar]
- 41.Lemay S.G. Nanopore-based biosensors: the interface between ionics and electronics. ACS Nano. 2009;3:775–779. doi: 10.1021/nn900336j. [DOI] [PubMed] [Google Scholar]
- 42.Han A., Creus M., Schurmann G. Label-free detection of single protein molecules and protein–protein interactions using synthetic nanopores. Anal Chem. 2008;80:4651–4658. doi: 10.1021/ac7025207. [DOI] [PubMed] [Google Scholar]
- 43.Rasooly A. Moving biosensors to point-of-care cancer diagnostics. Biosens Bioelectron. 2006;21:1847–1850. doi: 10.1016/j.bios.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Simon E. Biological and chemical sensors for cancer diagnosis. Meas Sci Technol. 2010;21:1–24. [Google Scholar]
- 45.Healy D.A., Hayes C.J., Leonard P., McKenna L., O'Kennedy R. Biosensor developments: application to prostate-specific antigen detection. Trends Biotechnol. 2007;25:125–131. doi: 10.1016/j.tibtech.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Barton A.C., Davis F., Higson S.P.J. Labeless immunosensor assay for prostate specific antigen with picogram per milliliter limits of detection based upon an AC impedance protocol. Anal Chem. 2008;80:6198–6205. doi: 10.1021/ac800491m. [DOI] [PubMed] [Google Scholar]
- 47.Bangar M.A., Shirale D.J., Chen W., Myung N.V., Mulchandani A. Single conducting polymer nanowire chemiresistive label-free immunosensor for cancer biomarker. Anal Chem. 2009;81:2168–2175. doi: 10.1021/ac802319f. [DOI] [PubMed] [Google Scholar]
- 48.Wee K.W., Kang G.Y., Park J. Novel electrical detection of label-free disease marker proteins using piezoresistive self-sensing micro-cantilevers. Biosens Bioelectron. 2005;20:1932–1938. doi: 10.1016/j.bios.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 49.Huang L., Reekmans G., Saerens D. Prostate-specific antigen immunosensing based on mixed self-assembled monolayers, camel antibodies and colloidal gold enhanced sandwich assays. Biosens Bioelectron. 2005;21:483–490. doi: 10.1016/j.bios.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 50.Brault N.D., Gao C., Xue H. Ultra-low fouling and functionalizable zwitterionic coatings grafted onto SiO2 via a biomimetic adhesive group for sensing and detection in complex media. Biosens Bioelectron. 2010;25:2276–2282. doi: 10.1016/j.bios.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Dai Z., Yan F., Chen J., Ju H.X. Reagentless amperometric immunosensors based on direct electrochemistry of horseradish peroxidase for rapid determination of carcinoma antigen-125. Anal Chem. 2003;75:5429–5434. doi: 10.1021/ac034213t. [DOI] [PubMed] [Google Scholar]
- 52.Prabhulkar S., Alwarappan S., Liu G., Li C.Z. Amperometric micro-immunosensor for the detection of tumor biomarker. Biosens Bioelectron. 2009;24:3524–3530. doi: 10.1016/j.bios.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 53.Lin Y.Y., Wang J., Liu G., Wu H., Wai C.M., Lin Y. A nanoparticle label/immunochromatographic electrochemical biosensor for rapid and sensitive detection of prostate-specific antigen. Biosens Bioelectron. 2008;23:1659–1665. doi: 10.1016/j.bios.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 54.Rossi L.M., Shi L., Rosenzweig N., Rosenzweig Z. Fluorescent silica nanospheres for digital counting bioassay of the breast cancer marker HER2/neu. Biosens Bioelectron. 2006;21:1900–1906. doi: 10.1016/j.bios.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Gau V., Ma S.C., Wang H., Tsukuda J., Kibler J., Haaka D.A. Electrochemical molecular analysis without nucleic acid amplification. Methods. 2005;37:73–83. doi: 10.1016/j.ymeth.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dell'Atti D., Tombelli S., Minunni M., Mascini M. Detection of clinically relevant point mutations by a novel piezoelectric biosensor. Biosens Bioelectron. 2006;21:1876–1879. doi: 10.1016/j.bios.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 57.Gronewold T.M.A., Baumgartner A., Quandt E., Famulok M. Discrimination of single mutations in cancer-related gene fragments with a surface acoustic wave sensor. Anal Chem. 2006;78:4865–4871. doi: 10.1021/ac060296c. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J., Song S.P., Wang L.H., Pan D., Fan C.H. A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA. Nat Protoc. 2007;2:2888–2895. doi: 10.1038/nprot.2007.419. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y., Wang Y., Wang H. Electrochemical DNA biosensor based on the proximity-dependent surface hybridization assay. Anal Chem. 2009;81:1982–1987. doi: 10.1021/ac802512d. [DOI] [PubMed] [Google Scholar]
- 60.Zhong X., Reynolds R., Kidd J.R. Single-nucleotide polymorphism genotyping on optical thin-film biosensor chips. Proc Natl Acad Sci USA. 2003;100:11559–11564. doi: 10.1073/pnas.1934783100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maki W.C., Mishra N.N., Cameron E.G., Filanoski B., Rastogi S.K., Maki G.K. Nanowire-transistor based ultra-sensitive DNA methylation detection. Biosens Bioelectron. 2008;23:780–787. doi: 10.1016/j.bios.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 62.Thaler N., Buhl A., Welter H. Biosensor analyses of serum autoantibodies: application to antiphospholipid syndrome and systemic lupus erythematosus. Anal Bioanal Chem. 2009;393:1417–1429. doi: 10.1007/s00216-008-2340-1. [DOI] [PubMed] [Google Scholar]
- 63.Metzger J., von Landenberg P., Kehrel M., Buhl A., Lackner K.J., Luppa P.B. Biosensor analysis of β2-glycoprotein I-reactive autoantibodies: evidence for isotype-specific binding and differentiation of pathogenic from infection-induced antibodies. Clin Chem. 2007;53:1137–1143. doi: 10.1373/clinchem.2006.079632. [DOI] [PubMed] [Google Scholar]
- 64.Buhl A., Metzger J.H., Heegaard N.H.H., von Landenberg P., Fleck M., Luppa P.B. Novel biosensor-based analytic device for the detection of anti-double-stranded DNA antibodies. Clin Chem. 2007;53:334–341. doi: 10.1373/clinchem.2006.077339. [DOI] [PubMed] [Google Scholar]
- 65.Rarok A.A., van der Geld Y.M., Stegeman C.A., Limburg P.C., Kallenberg C.G.M. Diversity of PR3-ANCA epitope specificity in Wegener's Granulomatosis. Analysis using the biosensor technology. J Clin Immunol. 2003;23:460–468. doi: 10.1023/b:joci.0000010422.73892.b5. [DOI] [PubMed] [Google Scholar]
- 66.Drouvalakis K.A., Bangsaruntip S., Hueber W., Kozar L.G., Utz P.J., Dai H. Peptide-coated nanotube-based biosensor for the detection of disease-specific autoantibodies in human serum. Biosens Bioelectron. 2008;23:1413–1421. doi: 10.1016/j.bios.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Real-Fernandez F., Colson A., Bayardon J. Ferrocenyl glycopeptides as electrochemical probes to detect autoantibodies in multiple sclerosis patients' sera. J Pept Sci. 2008;90:488–495. doi: 10.1002/bip.20955. [DOI] [PubMed] [Google Scholar]
- 68.Pejcic B., DeMarco R., Parkinson G. The role of biosensors in the detection of emerging infectious diseases. Analyst. 2006;131:1079–1090. doi: 10.1039/b603402k. [DOI] [PubMed] [Google Scholar]
- 69.Goulart L.R., Vieira C.U., Freschi A.P.P. Biomakers for serum diagnosis of infectious diseases and their potential application in novel sensor platforms. Crit Rev Immunol. 2010;30:201–222. doi: 10.1615/critrevimmunol.v30.i2.70. [DOI] [PubMed] [Google Scholar]
- 70.Price C.P., Kricka L.J. Improving healthcare accessibility through point-of-care technologies. Clin Chem. 2007;53:1665–1675. doi: 10.1373/clinchem.2006.084707. [DOI] [PubMed] [Google Scholar]
- 71.Wu T.Z., Su C.C., Chen L.K., Yang H.H., Tai D.F., Peng K.C. Piezoelectric immunochip for the detection of dengue fever in viremia phase. Biosens Bioelectron. 2005;21:689–695. doi: 10.1016/j.bios.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 72.Tai D.F., Lin C.Y., Wu T.Z., Chen L.K. Recognition of dengue virus protein using epitope-mediated molecularly imprinted film. Anal Chem. 2005;77:5140–5143. doi: 10.1021/ac0504060. [DOI] [PubMed] [Google Scholar]
- 73.Kwakye S., Baeumner A. A microfluidic biosensor based on nucleic acid sequence recognition. Anal Bioanal Chem. 2003;376:1062–1068. doi: 10.1007/s00216-003-2063-2. [DOI] [PubMed] [Google Scholar]
- 74.Brito-Madurro A.G., Ferreira L.F., Vieira S.N., Ariza R.G., Goulart L.R., Madurro J.M. Immobilization of purine bases on a poly-4-aminophenol matrix. J Mater Sci. 2007;42:3238–3243. [Google Scholar]
- 75.Ferreira L.F., Boodts J.F.C., Brito-Madurro A.G., Madurro J.M. Gold electrodes modified with poly (4-aminophenol): incorporation of nitrogenated bases and an oligonucleotide. Polym Int. 2008;57:644–650. [Google Scholar]
- 76.Brainina K., Kozitsina A., Beikin J. Electrochemical immunosensor for Forest-Spring encephalitis based on protein A labeled with colloidal gold. Anal Bioanal Chem. 2003;376:481–485. doi: 10.1007/s00216-003-1912-3. [DOI] [PubMed] [Google Scholar]
- 77.Tang D.P., Yuan R., Chai Y.Q., Dai J.Y., Zhong X., Liu Y. A novel immunosensor based on immobilization of hepatitis B surface antibody on platinum electrode modified colloidal gold and polyvinyl butyral as matrices via electrochemical impedance spectroscopy. Bioelectrochemistry. 2004;65:15–22. doi: 10.1016/j.bioelechem.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 78.Purvis D., Leonardova O., Farmakovsky D., Cherkasov V. An ultrasensitive and stable potentiometric immunosensor. Biosens Bioelectron. 2003;18:1385–1390. doi: 10.1016/s0956-5663(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 79.Lin K.C., Kunduru V., Bothara M., Rege K., Prasad S., Ramakrishna B.L. Biogenic nanoporous silica-based sensor for enhanced electrochemical detection of cardiovascular biomarkers proteins. Biosens Bioelectron. 2010;25:2336–2342. doi: 10.1016/j.bios.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 80.Chua J.H., Chee R.E., Agarwal A., Wong S.M., Zhang G.J. Label-free electrical detection of cardiac biomarker with complementary metal-oxide semiconductor-compatible silicon nanowire sensor arrays. Anal Chem. 2009;81:6266–6271. doi: 10.1021/ac901157x. [DOI] [PubMed] [Google Scholar]
- 81.McBride J.D., Cooper M.A. A high sensitivity assay for the inflammatory marker C-reactive protein employing acoustic biosensing. J Nanobiotechnol. 2008;6:5. doi: 10.1186/1477-3155-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei J., Mu Y., Song D. A novel sandwich immunosensing method for measuring cardiac troponin I in sera. Anal Biochem. 2003;321:209–216. doi: 10.1016/s0003-2697(03)00407-x. [DOI] [PubMed] [Google Scholar]
- 83.Masson J.F., Battaglia T.M., Khairallah P., Beaudoin S., Booksh K.S. Quantitative measurement of cardiac markers in undiluted serum. Anal Chem. 2007;79:612–619. doi: 10.1021/ac061089f. [DOI] [PubMed] [Google Scholar]
- 84.Hong B., Kang K.A. Biocompatible, nanogold-particle fluorescence enhancer for fluorophore mediated, optical immunosensor. Biosens Bioelectron. 2006;21:1333–1338. doi: 10.1016/j.bios.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 85.Tan X.C., Tian Y.X., Cai P.X., Zou X.Y. Glucose biosensor based on glucose oxidase immobilized in sol–gel chitosan/silica hybrid composite film on Prussian blue modified glassy carbon electrode. Anal Bioanal Chem. 2005;381:500–507. doi: 10.1007/s00216-004-2956-8. [DOI] [PubMed] [Google Scholar]
- 86.Liang R.P., Jiang J.L., Qiu J.D. Preparation of GOD/sol–gel silica film on Prussian blue modified electrode for glucose biosensor application. Electroanalysis. 2008;20:2642–2648. [Google Scholar]
- 87.Perez J.P.H., Lopez-Cabarcos E., Lopez-Ruiz B. Encapsulation of glucose oxidase within poly(ethylene glycol) methyl ether methacrylate microparticles for developing an amperometric glucose biosensor. Talanta. 2008;75:1151–1157. doi: 10.1016/j.talanta.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 88.Zhao C., Meng Y., Shao C., Wan Li, Jiao K. Unadulterated glucose biosensor based on direct electrode transfer of glucose oxidase encapsulated chitosan modified glassy carbon electrode. Electroanalysis. 2008;20:520–526. [Google Scholar]
- 89.Wu M.H., Fang M.Y., Jen L.N., Hsiao H.C., Mueller A., Hsu C.T. Clinical evaluation of Bionime Rightest GM310 biosensors with a simplified electrode fabrication for alternative site blood glucose tests. Clin Chem. 2008;54:1689–1695. doi: 10.1373/clinchem.2008.106328. [DOI] [PubMed] [Google Scholar]
- 90.Nichols J., Rodriguez M. Evaluation of glucose on the EPOC™ point of care blood analysis system. Clin Chem. 2009;55:A94–A95. [Google Scholar]
- 91.Tsai H., Doong R. Simultaneous determination of renal clinical analytes in serum suing hydrolase- and oxidase-encapsulated optical array biosensors. Anal Biochem. 2004;334:183–192. doi: 10.1016/j.ab.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 92.Schnabl K.L., Bagherpoor S., Diker P., Cursio C., DuBois J., Yip P.M. Evaluation of the analytical performance of the Nova StatSensor creatinine meter and reagent strip technology for whole blood testing. Clin Biochem. 2010;43:1026–1029. doi: 10.1016/j.clinbiochem.2010.04.055. [DOI] [PubMed] [Google Scholar]
- 93.Thuerlemann C., Haeberli A., Alberio L. Monitoring thrombin generation by electrochemistry: development of an amperometric biosensor screening test for plasma and whole blood. Clin Chem. 2009;55:505–512. doi: 10.1373/clinchem.2008.111963. [DOI] [PubMed] [Google Scholar]
- 94.Paniccia R., Fedi S., Carbonetto F. Evaluation of a new point-of-care celite-activated clotting time analyzer in different clinical settings. Anesthesiology. 2003;99:54–59. doi: 10.1097/00000542-200307000-00012. [DOI] [PubMed] [Google Scholar]
- 95.Ehrentreich-Forster E., Scheller F.W., Bier F.F. Detection of progesterone in whole blood samples. Biosens Bioelectron. 2003;18:375–380. doi: 10.1016/s0956-5663(02)00145-8. [DOI] [PubMed] [Google Scholar]
- 96.Trevino J., Calle A., Rodriguez-Frade J.M., Mellado M., Lechuga L.M. Surface plasmon resonance immunoassay analysis of pituitary hormones in urine and serum samples. Clin Chim Acta. 2009;403:56–62. doi: 10.1016/j.cca.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 97.Marchesini G.R., Meimaridou A., Haasnoot W. Biosensor discovery of thyroxine transport disrupting chemicals. Toxicol Appl Pharmacol. 2008;232:150–160. doi: 10.1016/j.taap.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 98.Luppa P.B., Thaler M., Schulte-Frohlinde E., Schreiegg A., Huber U., Metzger J. Unchanged androgen-binding properties of sex hormone-binding globulin in male patients with liver cirrhosis. Clin Chem Lab Med. 2006;44:967–973. doi: 10.1515/CCLM.2006.186. [DOI] [PubMed] [Google Scholar]
- 99.Thaler M., Metzger J., Schreiegg A. Immunoassay for sex hormone-binding globulin is influenced by high molecular mass aggregates. Clin Chem. 2005;51:401–407. doi: 10.1373/clinchem.2004.034264. [DOI] [PubMed] [Google Scholar]
- 100.Ferapontova E.E., Olsen E.M., Gothelf K.V. An RNA aptamer-based electrochemical biosensor for detection of theophylline in serum. J Am Chem Soc. 2008;130:4256–4258. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- 101.Milkani E., Lambert C.R., McGimpsey W.G. Direct detection of acetylcholinesterase inhibitor binding with an enzyme-based surface plasmon resonance sensor. Anal Biochem. 2011;408:212–219. doi: 10.1016/j.ab.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 102.Haes A.J., Chang L., Klein W.L., Van Duyne R.P. Detection of a biomarker for Alzheimer's Disease from synthetic and clinical samples using a nanoscale optical biosensor. J Am Chem Soc. 2005;127:2264–2271. doi: 10.1021/ja044087q. [DOI] [PubMed] [Google Scholar]
- 103.Doong R., Lee P.S., Anitha K. Simultaneous determination of biomarkers of Alzheimer's Disease using sol–gel-derived optical array biosensor. Biosens Bioelectron. 2010;25:2464–2469. doi: 10.1016/j.bios.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 104.Segal A., Wong D.T. Salivary diagnostics: enhancing disease detection and making medicine better. Eur J Dent Educ. 2008;12:22–29. doi: 10.1111/j.1600-0579.2007.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shetty V., Yamaguchi M. Salivary biosensors for screening trauma-related psychopathology. Oral Maxillofac Surg Clin North Am. 2010;22:269–278. doi: 10.1016/j.coms.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yamaguchi M., Deguchi M., Wakasugi J. Hand-held monitor of sympathetic nervous system using salivary amylase activity and its validation by driver fatigue assessment. Biosens Bioelectron. 2006;21:1007–1014. doi: 10.1016/j.bios.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 107.Kwan R.C.H., Leung H.F., Hon P.Y.T., Cheung H.C.F., Hirota K., Renneberg R. Amperometric biosensor for determining human salivary phosphate. Anal Biochem. 2005;343:263–267. doi: 10.1016/j.ab.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 108.Zuo B., Li S., Guo Z., Zhang J., Chen C. Piezoelectric immunosensor for SARS-associated coronavirus in sputum. Anal Chem. 2004;76:3536–3540. doi: 10.1021/ac035367b. [DOI] [PubMed] [Google Scholar]
- 109.Kintzios S.E. Cell-based biosensors in clinical chemistry. Mini Rev Med Chem. 2007;7:1019–1026. doi: 10.2174/138955707782110141. [DOI] [PubMed] [Google Scholar]
- 110.Su L., Jia W., Hou C., Lei Y. Microbial biosensors: a review. Biosens Bioelectron. 2011;26:1788–1799. doi: 10.1016/j.bios.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 111.Rechnitz G.A., Kobos R.K., Riechel S.J., Gebauer C.R. A bio-selective membrane electrode prepared with living bacterial cells. Anal Chim Acta. 1977;94:357–365. doi: 10.1016/S0003-2670(01)84537-2. [DOI] [PubMed] [Google Scholar]
- 112.Jha S.K., Kanungo M., Nath A., D'Souza S.F. Entrapment of live microbial cells in electropolymerized polyaniline and their use as urea biosensor. Biosens Bioelectron. 2009;24:2637–2642. doi: 10.1016/j.bios.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 113.Smutok O., Dmytruk K., Gonchar M., Sibirny A., Schuhmann W. Permeabilized cells of flavocytochrome b2 over-producing recombinant yeast Hansenula polymorpha as biological recognition element in amperometric lactate biosensors. Biosens Bioelectron. 2007;23:599–605. doi: 10.1016/j.bios.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 114.Ghosh G., Bachas L.G., Anderson K.W. Biosensor incorporating cell barrier architectures on ion selective electrodes for early screening of cancer. Anal Bioanal Chem. 2008;391:2783–2791. doi: 10.1007/s00216-008-2192-8. [DOI] [PubMed] [Google Scholar]
- 115.Otto A.M., Brischwein M., Niendorf A., Henning T., Motrescu E., Wolf B. Microphysiological testing for chemoresistivity of living tumor cells with multiparametric microsensor chips. Cancer Detect Prev. 2003;27:291–296. doi: 10.1016/s0361-090x(03)00093-x. [DOI] [PubMed] [Google Scholar]
- 116.Struss A., Pasini P., Ensor C.M., Raut N., Daunert S. Paper strip whole cell biosensors: a portable test for the semiquantitative detection of bacterial quorum signaling molecules. Anal Chem. 2010;82:4457–4463. doi: 10.1021/ac100231a. [DOI] [PubMed] [Google Scholar]


