Abstract
The PDZ domain is a protein–protein interacting module that plays an important role in the organization of signaling complexes. The recognition of short intrinsically disordered C‐terminal peptide motifs is the archetypical PDZ function, but the functional repertoire of this versatile module also includes recognition of internal peptide sequences, dimerization and phospholipid binding. The PDZ function can be tuned by various means such as allosteric effects, changes of physiological buffer conditions and phosphorylation of PDZ domains and/or ligands, which poses PDZ domains as dynamic regulators of cell signaling. This review is focused on the plasticity of the PDZ interactions.
Keywords: PDZ, Signaling, Scaffolding protein, Phosphoinositide, Lipid binding
1. Introduction
Signal transduction relies on scaffolding proteins that coordinate the assembly of signaling complexes. These molecular scaffolds are composed of modular interaction domains that bring their interacting partners close in space and thereby facilitate the interactions between the proteins [1]. This review is focused on the PDZ domain, which is one of the most widespread protein–protein interacting modules. The name is derived from the three proteins in which it was first identified almost 20 years ago, namely postsynaptic density protein‐95 (PDZ‐95), disks large tumor suppressor (DLG) and zonula occludens‐1 (ZO‐1) [2, 3, 4]. PDZ domains are commonly involved in processes of cell signaling and polarity, and are predominantly found in multi‐cellular organisms, with for example more than 250 human PDZ domains found in over 150 different proteins. The low abundance of PDZ domains in unicellular organisms led to the suggestion that PDZ domains co‐evolved with multi‐cellularity [5].
PDZ domains are over‐and‐above known as protein–protein interacting modules recognizing short peptide stretches at the C‐terminus of their target proteins [6, 7], but their functional repertoire also include recognition of internal peptide motifs [8, 9, 10, 11], hetero‐ and/or homo‐dimerization [12, 13] and interactions with membrane phospholipids [14, 15, 16, 17]. The ligands are commonly transmembrane receptors or ion channels. The interactions tend to be promiscuous, and one PDZ domain can commonly recognize various peptide ligands and the same ligand can be recognized by different PDZ domains [6]. The plasticity of PDZ interactions is further reflected in the ease by which the specificity of the interactions can be changed by mutagenesis of either ligands or PDZ domains [18, 19, 20, 21], which allowed frequent rewiring of PDZ–ligand interactions during evolution [22]. The function of PDZ domains can be tuned by various means such as changes in physiological buffer conditions [23, 24], allosteric changes [25] and phosphorylation [26] posing them as dynamic regulators of cell signaling. This review is focused on the plasticity of PDZ interactions, starting from the structure of the domain and the versatility of the ligand‐binding partners and ending with an overview of the regulatory mechanisms tuning the interactions.
2. Structure of PDZ domains and their integration in scaffolding proteins
PDZ domains consist of 80–90 amino acids and share a common fold of five to six β‐strands (β1–β6) and two α‐helices as shown by the over 300 PDZ structures currently deposited in the PDB data bank (Fig. 1 ). Their average sequence identity is only 30% and although sharing the same core structure, PDZ domains often have variable loop regions and might contain additional secondary structural elements that may affect the structure and function [27]. Indeed, even the archetypical structure of PSD‐95 PDZ3 holds an additional C‐terminal α‐helix that influences the dynamics of the protein [28, 29, 30]. The PDZ structure is robust and tolerates extensive mutagenesis [31] as well as topological changes such as circular permutations (Fig. 1) [32, 33, 34, 35]. The canonical metazoan PDZ domains have a β1–β2–β3–α1–β4–β5–α2–β6 secondary structure arrangements and make part of scaffolding proteins [6]. PDZ domains in bacteria, plant and yeast generally make part of proteases and are circularly permuted as compared to the canonical PDZ domains, typically having a symmetric β2–β3–α1–β4–β5–α2–β6–β1 secondary structure arrangement. In these organisms, PDZ domains are less common, yeast has for example only one PDZ protein holding four PDZ domains [5]. The structure of the PDZ domains of bacterial RseP revealed an alternative connectivity of the secondary structure elements (β3–α1–β4–β5–α2–β6–β1–β2) [36], which is shared with the PDZ tandem of human GRASP55 [37]. In all cases, PDZ domains have their N‐ and C‐termini close in space, which facilitates their integration in pre‐existing protein scaffolds.
Figure 1.
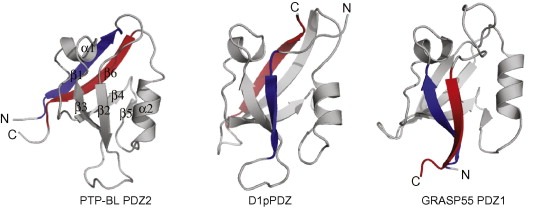
Cartoon representations of canonical and PDZ‐like domains with the N‐ and C‐terminal strands highlighted in blue and red, respectively. PTP‐BL PDZ2 (PDB code http://1GM1, left) with indicated secondary elements (β1–β2–β3–α1–β4–β5–α2–β6) represents a canonical metazoan PDZ domain. The PDZ domain of the D1 C‐terminal‐processing protease (middle, PDB code http://1FC7) and GRASP55 PDZ1 (right, PDB code http://3RLE) illustrate two differentially circularly permuted PDZ structures.
PDZ domains are found in a wide range of proteins, from those that almost exclusively consist of a single PDZ domain such as Tax1‐binding protein 3 (TIP‐1), to proteins that contain several PDZ domains such as multiple PDZ domain protein, and scaffolds combining PDZ domains with other modular domains such as phosphatidylinositol 3,4,5‐triphosphate‐dependent Rac exchanger 2 protein that contains a RhoGef domain, a pleckstrin homology (PH) domain, two DEP domains and a PDZ tandem (i.e. two PDZ domains connected by a short linker). In some cases, two tightly connected PDZ domains form a functional unit, or supramodule, with distinct functional properties from the isolated PDZ domains [38]. For example, PDZ4 in the PDZ4–5 tandem of glutamate receptor‐interacting protein 1 (GRIP1) does not bind to peptides but serve as structural support for the PDZ5 [39]. PDZ domains may also form supramodular units with other domains. For example, a recent study by Pan and co‐workers revealed a supramodular organization of the PDZ3‐SRC homology (SH3)‐guanylate kinase (GUK) domains of ZO‐1, where the PDZ3‐SH3 interface forms a hydrophobic pocket that make additional contacts with the connexin43 peptide [40]. In line with these data, protein engineering experiments have highlighted the ease by which PDZ peptide binding affinity and specificity can be greatly enhanced by the addition of a binding inert domain and optimization of the interface residues [33].
3. Versatility of PDZ interactions
3.1. C‐terminal peptide recognition
The by far most common PDZ interaction is the recognition of C‐terminal ligands with hydrophobic residues at their C‐termini [41]. The carboxylate terminus of the peptide ligand is bound by a conserved carboxylate binding loop (R/K‐XXX‐G‐Φ‐G‐Φ, where X is any amino acid and Φ a hydrophobic residue) and the peptide is added as an additional β‐strand to the grove between β2 and α2, with the last four amino acids in the PDZ‐binding motif being most important for recognition (Fig. 2 ) [21, 42, 43]. The last amino acid of a PDZ‐binding motif is denoted “0”, the penultimate residue “−1” and so on. Deviations from the canonical C‐terminal peptide binding have been reported for example from structural studies on syntenin‐1 PDZ1 [44] and tamalin PDZ [45], for which the peptides were found to be bound perpendicular to the main β‐sheet and only interacting with the PDZ domains through the last two amino acids.
Figure 2.
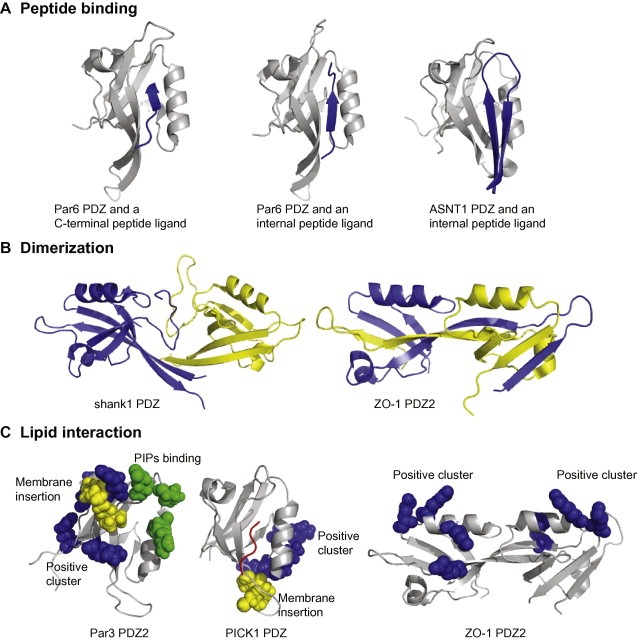
The repertoire of PDZ interactions include peptide binding (A), dimerization (B) and phospholipid interactions (C). (A) Par6 PDZ in complex with a VKESLV‐COO‐ peptide (left; PDB code http://1RZX) representing canonical C‐terminal peptide binding. The structure can be compared with the same protein in complex with a PALS1 internal peptide ligand (middle, PDB code http://1X8S). Note the changes in the β1–β2 loop that allow for the accommodation of the internal peptide ligand. The structure of SNTA1 PDZ in complex with the nNOS internal peptide (right, PDB code http://1QAV) illustrates that internal peptide ligands can be accommodated if they take restricted conformations mimicking free C‐terminal ligands. (B) Different strategies for PDZ–PDZ dimerization as illustrated by the structures of shank1 PDZ (left, PDB code http://1Q3P) and ZO‐1 PDZ2 (right, PDB code http://2RCZ). The constituting monomers are shown in in blue and yellow cartoon representation. (C) PDZ domains employ different means for interacting with PIPs containing lipid membranes. Par3 PDZ2 (PDB code http://2OGP) interacts with negatively charged lipid membranes through a combination of non‐specific electrostatic interactions (Lys491, Arg496, Lys506 and Arg546 indicated in blue spheres), membrane penetration (Leu494, Pro495 and Ile500 in yellow spheres), and a defined PIPs binding site (Glu469, Lys535 and Arg532 in green spheres) [17]. PICK1 PDZ (middle, PDB code http://2PKU) interacts with lipid membranes through a conserved Cys‐Pro‐Cys motif (yellow spheres) and a positive charge cluster (Arg76, Lys79 and Lys81 in blue spheres). A bound peptide is indicated in red. Mutaganic analysis of ZO‐1 PDZ2 (right) indicated a set of basic residues to be of importance for the interactions with PIPs containing liposomes (Arg201, Lys246, Arg251 and Lys253 shown as blue spheres) [86].
PDZ domains were early on divided into specificity classes based on the last amino acids of the C‐terminal target peptide, with the typical class I motif being X[T/S]XΦ‐COO‐, the class II motif being XΦXΦ‐COO‐, and the minor class III motif being X[D/E]XΦ‐COO‐ [41, 46]. Although this classification system is an oversimplification, it is useful to note that the class I binding PDZ domains have a characteristic His in α‐helix 2 that hydrogen bonds with a Ser/Thr at position −2. A more fine‐tuned ligand binding specificity was hinted early on when computer‐aided design of PSD‐95 PDZ3 showed that every position in the target peptide contribute to the binding specificity [19]. Furthermore, it was found that some PDZ domains recognize residues up to position −7; the additional recognition typically being conferred by an extended β2–β3 loop region [47, 48, 49, 50] or an extended α2 [51]. In line with these observations, two large‐scale studies that addressed PDZ–peptide binding specificities on family‐wide scales showed that peptide binding specificity is derived from interactions throughout the binding pocket and that PDZ domains in general are not promiscuous but can be divided in up to 16 different classes based on their ligand binding profile [20, 21].
Experimental and computational data suggest that the binding reaction involves the rate‐limiting formation of a weak encounter complex between the C‐terminus of a disordered peptide and the PDZ domain, followed by formation of native side chain contacts and rearrangement into the final complex [52, 53, 54]. The k on and k off rates are fast and the PDZ–peptide affinities are typically in the low‐to‐mid micro molar range with changes in affinities correlating with changes in k off [53]. PDZ domains exhibit a range of dynamic and entropic behaviors, distinct between PDZ domains but also between the same PDZ domain bound to different ligands, suggesting that the dynamics of the domains contribute to the binding specificity [55, 56, 57, 58]. For example, differences in dynamics were suggested for PDZ domains recognizing class I and class II peptides, such that dynamics of the β1–β2 and the β2–β3 loops are more critical in class I type binding and the dynamic of the β2–β3 loop and α2 is more critical in type II interactions [56]. Double mutant cycle analysis of PSD‐95 PDZ3 and tyrosine‐protein phosphatase non‐receptor type 13 (PTP‐BL) PDZ2 suggested that mutagenic alterations of the dynamic pathways affected the binding and that the whole PDZ domain and not only the binding site may be used to fine tune specificity [59]. However, some PDZ domains such as syntenin‐1 PDZ2 and shank1 PDZ are promiscuous and may interact with ligands of different classes, which is possible owing to a structural flexibility of the binding sites [60, 61].
3.2. Interactions with internal peptide ligands
Although C‐terminal peptide recognition is the hallmark of PDZ domains, some PDZ domains interact also with internal peptide motifs (Fig. 2) [8, 9, 10, 11, 62, 63, 64, 65]. Indeed, a recent high‐throughput yeast two‐hybrid screen found that more than half of the identified PDZ interactions did not require a free C‐terminal consensus sequence, which suggests that binding to internal peptide motifs may be more common than previously appreciated [66]. Although this is an interesting finding that might suggest that a significant percentage of PDZ domains recognize internal peptide motifs, it is unfortunately not clear from the study to what extent the observed interactions are peptide mediated, and to what extent these interactions may engage other parts of the PDZ domains. For binding internal peptide stretches, it is paramount that the PDZ–peptide couple overcomes the problems caused by steric clashes between amino acids extending beyond the 0 position and the carboxylate binding groove. The structures of α‐1‐syntrophin (SNTA1) PDZ in complex with the ca. 30 amino acid residue extension of the nitric oxide synthas (nNOS) PDZ domain, revealed that steric clashes can be avoided by the peptide adopting a β‐strand structure replacing the normally required carboxyl terminus (Fig. 2A) [8]. A recent study on the kinetics of the binding reaction suggested that native contacts form cooperatively after an initial rate‐limiting docking step, and that the β‐hairpin thus folds upon binding [67]. However, the interaction between PDZ domains and internal peptide ligands may also be accomplished by already conformationally constrained cyclic peptides [68]. In addition, internal peptide ligands may be accommodated by structural changes of PDZ domains as shown for the Par6 PDZ domain in complex with an internal region of the InaD‐like protein (PALS1). In this case, there is a conformational change of the carboxylate binding loop upon peptide binding (Fig. 2A) and a conserved lysine in the carboxylate binding loop forms a salt bridge with the aspartic acids side chain in position +1 [69]. Similarly, phage display of Disheveled‐2 (Dvl2) PDZ2 with an internal peptide library revealed that Dvl2 PDZ2 recognizes a continuous stretch of 7–8 amino acids with a conserved aspartate residue mimicking the carboxylate of the C‐terminal ligand. The structural analysis of Dvl2 PDZ2 in complex with different peptide ligands clarified that the protein can interact with both C‐terminal and internal peptide ligands owing to a structural flexibility of its binding site [11, 70].
The recognition of internal peptide ligands allows multiple PDZ domains to interact with the same target protein facilitating the assembly of multi protein signaling complexes. It also opens for the bidentate recognition of a target protein by two PDZ domains that are physically linked or dimerized. For example, INAD PZ3 recognizes an internal peptide stretch of transient receptor potential protein (TRP) and INAD PDZ5 interacts with the TRP C‐terminus, a bidentate interaction that provides INAD with a high affinity for TRP (ca. 0.1 μM) [71]. Another example is given by PSD‐95 that interacts with both an internal region and the C‐terminus of shaker‐type K+ channels [72]. An added value of the bidentate binding may be the possibility to independently regulate the interaction with the distinct PDZ domains.
3.3. PDZ–PDZ interactions
It was early on discovered that some PDZ domains engage in PDZ–PDZ interactions [8, 12, 13]. For example, INAD PDZ3 and PDZ4 can form either homo‐ or hetero oligomers, and the PDZ tandems of NHERF1 and NHERF2 can hetero‐ or homodimerize without affecting peptide binding [12, 13]. However, the extent of PDZ–PDZ dimerizations was not fully appreciated until a recent study by Chang and co‐workers revealed that as much as 30% of PDZ domains engage in PDZ–PDZ interactions with low micromolar affinities [73]. PDZ–PDZ interactions appear more selective than PDZ–peptide interactions and therefore contribute to defining the precise composition of the protein complexes. The prevalence of specific high affinity PDZ–PDZ interactions may suggest that these domains evolved to form multiprotein complexes by the simultaneous interaction with more than one ligand [73].
Structural studies have shown that PDZ–PDZ dimerization can be accomplished by different means (Fig. 2B). For example, shank1 PDZ and GRIP1 PDZ6 form back‐to‐back homo‐dimers that are stabilized by interactions engaging their unusually long β2–β3 loops and N‐terminal β1‐strands, and leave the peptide binding pockets of the constituting domains readily available for engaging in peptide interactions [74, 75]. The second PDZ domain of ZO‐1 forms a peculiar domain swapped dimeric structure, where the constituting PDZ domains retain their overall fold but have exchanged their β1‐ and β2‐strands; the peptide binding sites are in this case located on opposite sides of the dimeric molecule (Fig. 2B) [76, 77]. PDZ–PDZ interactions thus commonly leave the peptide binding sites available and may serve to increase the binding avidity of the interactions. Exceptions to this notion are the SNTA1‐nNOS PDZ and PSD‐95 PDZ2‐nNOS PDZ dimerizations, where the binding of the C‐terminal extension of nNOS blocks the binding site of SNTA1 and PSD‐95 PDZ2, respectively [8, 78].
3.4. PDZ–phospholipid interactions
As PDZ domains are over‐and‐above known as protein–protein interacting modules, it was surprising when the first reports came on PDZ domains as phosphoinositide (PIPs) binders now ten years ago [14]. PIPs are phosphorylated forms of phosphatidylinositol. Only a minor percentage of phosphatidylinositol is phosphorylated on one to three of the hydroxyl groups of the inositol headgroup and the seven biologically relevant PIPs species have a defined cellular localization that is regulated by a system of kinases and phosphatases [79, 80]. PIPs are important regulators of cell polarization and signaling. They can serve as precursors for second messengers, or function as membrane‐bound signaling molecules regulating the localization of signaling complexes. The PDZ tandems of syntenin‐1 and syntenin‐2 represent the paradigm examples of PDZ domains interacting with membrane and nuclear PIPs, respectively [14, 16]. The in vivo importance of the syntenin‐1‐phosphatidylinositol 4.5‐bisphosphate (PIP2) interaction was demonstrated for the recycling of syndecans, cell spreading and directional movements in zebrafish [81, 82]. The syntenin‐2‐PIP2 interaction was suggested to be of importance for targeting the protein to membrane and nuclear pools of PIP2 [16]. Estimates now have it that 20–40% of PDZ domains interacts with phospholipids [15, 17, 83] and in the cases investigated, PDZ–PIPs interactions appear relevant for the function of the PDZ proteins. For example, lipid membrane interactions are crucial for the clustering of AMPA receptors by PICK1 [84]. In other cases, a PIPs binding PDZ domain is linked to another PDZ domain that recruits a phosphatase or kinase and thus target the enzyme to its substrate, as exemplified by Par3 where the second PDZ domain interacts with PIPs and the third PDZ domain interact with the PIP3 phosphatase PTEN [51].
PIPs–PDZ interactions are commonly in the low‐to‐mid micro molar range and often occur in tandem with other lipid binding modules, such as the PDZ domain of PICK1 being linked to a BAR domain [84] and the lipid binding PDZ domain of SNTA1 being connected to a PH domain [85]. Alternatively, PIPs interacting PDZ domains exist as dimers such as ZO‐1 PDZ2 [86] and its Drosophila homologue polychaetoid (Pyd) PDZ2 [87], or form multimers exemplified by of the PDZ domains of bacterial DegP [88]. Combinations of more than one lipid binding module provide proteins with avidity for the lipid membrane making the interactions biologically relevant, although the affinity of each domain may be rather low. As reviewed by Gallardo and co‐authors, PDZ domains employ different means to target PIPs containing lipid membranes such as electrostatic interactions, membrane insertion and basic clusters interacting with the lipid headgroup (Fig. 2C) [15]. The PDZ domains appear in general to lack well‐defined PIPs binding pockets providing stereospecific interactions with distinct PIPs species, which is present in more classic PIPs binding modules such as the PH of PLC‐δ1 [89] and the PX domain of p40(phox) [90], and PDZ domains thus tend to have low stereospecificity [16, 17, 87].
PDZ–PIPs interactions have been mapped to different structural regions in distinct cases and competition as well as synergy between PIPs and peptide binding have been reported, both of which may be involved in regulating the PDZ function [14, 17, 87]. For example, NMR analysis of the PTP‐BAS PDZ2b‐PIP2/PIP3 interaction suggested it to take place in the groove between α2 and β2 and to be competitive with peptide binding [91]. Par3 PDZ2 membrane targeting involves the insertion of a cluster of hydrophobic residues into the membrane and non‐specific electrostatic interactions between a defined positive charge cluster and the negatively charged lipid membrane (Fig. 2) [17]. NMR titration of Par3 PDZ2 with PI3P indicated the strongest chemical shift changes to be located to a polar pocket between the β1–β2 and β5–β6 loops, and the lipid headgroup was modeled into this pocket making contacts with Arg532, Lys535 and Glu469 [17]. The PIPs binding site of Par3 PDZ2 partially overlaps with the peptide binding region and the authors consistently reported on competition between peptide and lipid binding. In line with these data, biochemical analysis of ZO‐1 PDZ2 suggested competition between peptide and lipid binding [14, 86]. The PDZ domain of PICK1 displays a distinct mode of lipid membrane binding as outlined by biochemical and mutational analysis [84]. The PICK1 PDZ interaction with lipid membranes requires a conserved Cys‐Pro‐Cys motif in the β2–β3 loop and a juxtaposed positive charge cluster in the β5–α2 loop [84]. The basic cluster provides non‐specific electrostatic interactions with the lipid membrane and the Cys‐Pro‐Cys motif was suggested to insert into the lipid membrane, although the molecular mechanism is not clear. There is no overlap between the peptide binding site and the lipid binding region of PICK1 PDZ, and peptide binding does not appear to compete with lipid membrane binding.
The first example of synergistic binding between peptide and PIPs ligands was given by a recent study on the Pyd PDZ2 [87], where it was found that preloading Pyd PDZ2 with a LKLPPERLI‐COO‐ peptide conferred a sixfold increase in affinity for PIP2. The results suggest that peptide binding may tune the affinity for the lipid binding partner and that the protein will be efficiently targeted to the membrane when both peptide and PIPs are available. Mutagenic analysis of Pyd PDZ2 revealed that electrostatic interactions along one face of the α2 are crucial for lipid membrane binding [87]. The study on Pyd PDZ2 further highlighted that PDZ–PIPs interactions may be reinforced by additional interactions with other negatively charged phospholipids, such as phosphatidylserine.
Recently, a study by Chen and co‐workers suggested that up to 40% of PDZ domains may interact with lipid membranes and that these domains function as dual‐specificity modules regulating proteins interactions at the membrane [83]. The study further suggested a classification of lipid‐binding PDZ domains based on the localization of their basic clusters, with class A lipid‐binding PDZ domains having topologically distinct peptide and lipid binding sites and acting as coincidence detectors of lipid and peptide signaling. Class B PDZ domains have basic clusters in or near the α2 helix that makes part of the peptide binding site. The class B domains were further subdivided into class B1 having the basic cluster at the C‐terminal end of α2 and potentially acting as coincidence detectors of PIPs and peptide signaling, and class B2 domains having the basic cluster at the N‐terminal end of α2 and displaying competitive peptide and PIPs binding. Future studies of the growing list of lipid binding PDZ domains will elucidate the intriguing interplay between peptide and lipid binding and the biological consequences of these interactions.
4. Regulation of PDZ interactions
PDZ domains are functionally versatile modules that can be dynamically be regulated by various means, as reviewed in the following section.
4.1. Changes of buffer conditions
PDZ interactions can be regulated by changes in physiological buffer conditions [23, 24]. In particular, the salt and pH dependence of the peptide binding of PSD‐95 PDZ3 was found to result from the binding of a chloride to a conserved Arg318 in the carboxylate binding loop and protonation of His372 in α2, respectively [23]. As the His in α2 is the hallmark for class‐1 interacting PDZ domains, it is plausible that pH change may have similar effects on other PDZ domains of the same class.
PDZ function may further br regulated by changes in redox‐potential, with INAD PDZ5 being the best‐characterized example [71, 92, 93]. The peptide binding of INAD PDZ5 is controlled by a light‐dependent redox‐regulated transient change from an open reduced form to an oxidized closed conformation. In the closed conformation, the peptide‐binding pocket is distorted through an intra‐molecular disulfide bond formed between Cys606 in β3 and Cys645 in α2, which confers a 20‐fold decrease in its affinity for a target peptide [71, 92, 93]. INAD PDZ5 is locked in the reduced state by its C‐terminal extension being tucked into a groove at the rear of PDZ4. At lower pH, a His547 in the C‐terminal extension is protonated, which uncouples the PDZ4–PDZ5 supramodule and results in a dramatic decrease in redox potential. INAD assembles a signaling complex essential for signaling in Drosophila photoreceptor cells, and acts as a dynamic regulator of cell signaling through the cycles of conformational changes INAD.
PICK1 PDZ was suggested to be redox‐regulated by the formation of a reversible intermolecular disulfide bond engaging Cys44 in the β2–β3 loop under mildly oxidative conditions [94]. The redox‐regulated dimer formation abolishes the lipid membrane binding and also affects the peptide binding, and the authors suggested that this might be of importance for the regulation of PICK1 mediated trafficking processes. However, it is not clear if PICK1 PDZ is actually redox‐regulated in a cellular context.
4.2. Allosteric changes
PDZ domains often have dynamic properties that can be used for allosteric regulation. Indeed, PDZ domains have been popular model systems for the study of single‐domain allostery as reviewed by Smock and Gierasch [25]. Early work by Ranganathan and co‐workers identified a network of co‐evolving residues stretching from the α1, through the peptide binding site to the α2, and the authors suggested this co‐evolving set of residues to be involved in allosteric communication [95]. Several experimental and computational studies have since then suggested similar networks of connected residues [95, 96, 97, 98]. A recent study by Reynolds et al. demonstrated the ease by which such networks can be used to evolve allosteric switches [97]. In this study, all surface exposed residues in PSD‐95 PDZ3 were probed by saturation mutagenesis. Eleven probed surface positions with no obvious spatial relationship to the binding site or to each other affected the peptide binding. Ten of the eleven residues make part of networks of connected co‐evolving amino acids as suggested by statistical coupling analysis, and the authors proposed that these residues can be used as hot spots for the rapid evolution of allosteric control [97].
Allosteric regulation of PDZ function has also been suggested for human PTP‐BAS/murine PTP‐BL PDZ2. Different studies have indicated the presence of two contiguous interaction pathways in the molecule [57, 59, 95, 99] and the affinity and specificity of the protein was found to be allosterically regulated by the binding of PTP‐BL PDZ1 to a surface on PDZ2 opposite to the PDZ2 peptide binding groove [98]. The PTP‐BL PDZ1–PDZ2 interaction confers a shift of the promiscuous PDZ2 peptide binding specificity towards a more stringent recognition of class 1 peptides [98]. Another example of allosteric regulation is given by the PDZ domain of Par6 that was found to be allosterically regulated by the binding of the small GTPase Cdc42 to a CRIB domain adjacent to the PDZ domain [69, 96]. The Cdc42‐CRIB interaction evokes an allosteric transition of the Par6 PDZ domain from a low‐affinity dynamic state to a more rigid high‐affinity state, with the changes of the dynamics particularly affecting the β1–β2 loop. The details of the allosteric transition were further elucidated in a recent study by Whitney et al., outlining that the conformational switch transposes two adjacent side chains of the conserved carboxylate binding loop [100]. A curiosity in the context is that the interaction between Par6 PDZ and the internal peptide ligand of PALS1 is decoupled from Cdc42 regulation, which might be explained by this interaction deforming the carboxylate binding loop [101].
Finally, allosteric regulation is not limited to the canonical PDZ domains. The binding of specific peptides to PDZ1 of the heat‐shock factor DegP causes a local rearrangement that is allosterically transmitted to the substrate‐binding pocket of the protease domain [102, 103]. Allosteric regulation thus plays an important role in the dynamic regulation of function of several PDZ proteins.
4.3. Autoinhibition
Different studies have reported on autoinhibition of PDZ proteins with structural evidences being available for X11 α, tamalin and NHERF1 [45, 104, 105, 106, 107, 108]. In these cases, the C‐terminal regions folds back and interact with the PDZ domains. The NMR structure of the PDZ tandem of X11α in complex with the C‐terminal tail of X11α provided the first structure of an autoinhibitory PDZ interaction. The intramolecular interaction was supported by analytical gel filtration and was suggested to be regulated by phosphorylation [106]. The structure of the PDZ domain of tamalin bound to the tamalin C‐terminal peptide showed that the peptide can be bound by canonical β‐addition or in a non‐canonical perpendicular fashion [45]. The self‐interaction is of low affinity (630 μM KD) and can be competed by C‐terminal peptides of metabotropic glutamate receptors that interact with tamalin PDZ with higher affinities (ca. 100 μM KD). Finally, the NMR structures of NHERF1 PDZ2 in complex with the NHERF1 C‐terminal ezrin‐binding domain revealed that the ligand takes a helical conformation that is accommodated in the PDZ2 binding pocket and stabilizes the native fold [104, 105, 107]. The self‐interaction can be overcome by competition with a cognate peptide ligand binding to the PDZ domain and/or ezrin interacting with the ezrin‐binding domain, and can further be regulated by phosphorylation of the C‐terminal domain.
PDZ domains may thus be negatively regulated by intramolecular interactions with their C‐terminal tails [45, 104, 105, 106, 107, 108], but there are also studies suggesting regulatory effects exerted by N‐terminal stretches. For example, the PDZ domains of syntenin‐1 appear to be negatively regulated by the N‐terminal domain, although the exact molecular details are not known [109, 110].
4.4. Phosphoregulation
A commonly reported regulatory mechanism of PDZ interactions is phosphorylation of Ser/Thr/Tyr residues in PDZ‐binding motives. Phosphorylation often occurs at the −2 position of a type 1 binding motif and interferes with PDZ–peptide interactions. For example, phosphorylation of Ser411 at the −2 position of the β2‐adrenergic receptor by G‐protein‐coupled‐receptor kinase‐5 disrupts the interaction with NHERF1 and controls the recycling of the receptor [111]. Other residues in PDZ‐binding motifs may be phosphorylated with similar effects, as shown for syndecan‐1 where Tyr −1 phosphorylation confers a loss of interaction with syntenin‐1 [112] and connexin‐43 for which phosphorylation of Ser −9 or −10 disrupts the interaction with ZO‐1 PDZ2 [77, 113, 114]. However, there are also reports on enhanced binding upon phosphorylation of PDZ‐binding motifs, or in the proximity of such motifs, and some PDZ domains thus interact with phosphopeptides. For example, phosphorylation significantly increases the interaction between the PDZ domain of LIM domain‐binding protein 3 and C‐terminal peptides derived from the calsarcin/myozenin and myotilin family [115] and phosphorylation of ephrinB ligands at Ser −9 triggers binding to GRIP [116].
Interestingly, phosphorylation of PDZ binding motifs may have a switch‐like function on PDZ function, simultaneously turning “on” the interaction with one domain while switching of the interaction with other domains. In the case of X11 α, such a behavior is manifested in a single protein, as a Tyr −1 phosphorylation of its C‐terminus confers a switch from the autoinhibited closed conformation where the C‐terminal interacts with the PDZ1 to an open conformation where the C‐terminal is recognized by the PDZ2 [106]. The prevalence of phosphorylation switches was highlighted in a recent study by Akiva et al. [26]. They scanned the available proteomic data of PDZ‐interactions in mouse [20] for cases where single phosphomimetic amino acid substitutions (Ser/Thr/Tyr to Asp/Glu) conferred inverse affinities for two PDZ domains. The study identified 81 potential phosphorylation switches, which suggests this to be a common regulatory mechanism [26]. Indeed, it was early on observed that phosphorylation of PDZ binding motifs may have differential effects on PDZ interactions, as exemplified by the report on phosphorylation of the C‐terminus of B class ephrins that was shown to confer loss of interaction with syntenin‐1 but have little effect on the interaction with PTP‐BAS PDZ5 [117]. It should further be noted that also other posttranslational modifications can regulate PDZ interactions. An interesting case is the interaction between PTEN and DLG2 that was reported to be negatively regulated by phosphorylation, but positively regulated by acetylation [118].
PDZ function can also be down‐regulated by phosphorylation of PDZ domains. For example, the peptide binding of DLG1 PDZ1 is diminished by Ca2+/calmodulin‐dependent protein kinase II phosphorylation of a Ser232 adjacent to the carboxylate‐binding loop [119], and phosphorylation of Ser77 in α2 of NHERF1 PDZ1 confers a loss of peptide binding [120]. In addition, the function of PICK1 PDZ has been proposed to be down‐regulated by phosphorylation of either Ser77 or Thr82. The phosphomimetic mutation of Thr82 at the N‐terminal end of α2 disrupts the interaction between PICK1 PDZ and GluR2 [121] and phosphorylation of Ser77 in the β5‐α2 loop affects the clustering of eYFP‐PICK1 in COS7 cells, although no effect was observed on peptide binding [122]. Interestingly, Thr82 and Ser77 are in vicinity of the positive charge cluster that has been proposed to be of importance for the interaction between PICK1 PDZ and lipid membranes [84] and it is possible that their phosphorylations influence the lipid binding, although no experimental data are available to clarify this point.
Finally, PDZ function can also be regulated by phosphorylation outside the canonical domain boarders, as previously discussed for the INAD PDZ4–PDZ5 supramodule [71]. The phosphorylation of Tyr397 in the C‐terminal non‐canonical α3 of PDS‐95 PDZ3 provides an interesting example of how the dynamics of PDZ domains can be phosphoregulated. The dynamics of PDZ3 of PSD‐95 is dampened by the packing of α3 against the PDZ domain, which enhances its peptide binding affinity by a altering the dynamic properties of the domain [29, 30]. Phosphorylation of Tyr397 in α3 initiates a rapid equilibrium of docked and undocked conformations, with the undocked conformation having reduced ligand binding affinity [123].
4.5. Alternative splicing
PDZ interaction are often regulated by alternative splicing of PDZ proteins or their interacting proteins, as extensively reviewed by Sierralta and Mendoza [124]. Typically, PDZ domains or PDZ binding motifs are deleted from different isoform resulting in variable subcellular localizations and functions [124, 125]. For example, the protein usherin exists in multiple isoforms in murine inner ear of which one is transmembrane and harbors a PDZ‐binding motif that interacts with the PDZ protein whirlin [126]. Whirlin in turn exist as a long isoform containing three PDZ domains of which the first two interact with usherin and a short isoform that only holds one PDZ domain that does not interact with usherin [127, 128].
Alternative splicing within PDZ domains is considerably less reported. However, PTP‐BAS PDZ2 exist as two functionally distinct isoforms a and b, with the difference between the two isoform being the insertion of five amino acids in the β2–β3 loop. The extended loop region folds back onto β2–β3, which leads to a decreased peptide binding affinity. Interestingly, the extended loop region does not interfere with the modest PIPs binding but confers instead a slight increase in PIPs affinity and the alternative splicing may thus tune the preference for the distinct ligands [91].
4.6. Competition and viral hijacking
The competition between distinct PDZ proteins can be used for dynamic regulation of cell signaling. For example, the small protein TIP‐1 that is almost entirely composed of a PDZ domain competes with CASK and other PDZ containing scaffolds with type binding I specificity to negatively regulate their function [129]. The competition between TIP‐1 and CASK for the potassium channel Kir2.3 leads to uncoupling of Kir2.3 from its basolateral membrane‐anchoring complex and confers a shift towards endosomal targeting of the channel. Another case involves the expression and the activity of CFTR in colon cancer cells that is positively regulated by interactions with NHERF1 but negatively regulated by interactions with shank2 [130]. Finally, MAGI‐3 competes with NHERF2 for binding to LPA receptor type 2 and phospholipase C‐β3, with reciprocal effects such that NHERF2 promotes migration and invasion of colon cancer cells, while MAGI‐3 inhibits these processes [131].
Another form of competition for PDZ interaction is the viral hijacking of PDZ proteins, a topic that was extensively reviewed recently [132, 133]. The PDZ‐binding motifs in a cell are often sub‐optimized to allow for rapid changes of cell signaling and viral proteins can therefore rewire cell signaling by presenting C‐terminal motifs of higher affinities. The first examples of viral hijacking of PDZ protein were given by the three oncoproteins adenovirus type 9 E4‐ORF1, human T‐lymphotropoic virus type 1 Tac and high‐risk human papillomavirus E6 and has since then emerged as a common theme among viruses [132, 134, 135, 136]. More recent reports include the hijacking of polarity protein PALS1 by the SARS Coronavirus E protein, which alters tight junction formation and epithelial morphogenesis [137], and a study by Accardi and co‐workers reporting on cooperation between E6 and E7 from human papillomavirus type 16 in the targeting of NHERF1 [138].
5. Concluding remarks
The PDZ domain is a highly versatile interaction module with the main function of recognizing C‐terminal peptide ligands, but with an extended functionality of interacting with alternative ligands such as internal peptide stretches, other PDZ domains and/or membrane lipids. The plasticity of PDZ–ligand interactions has made it one of the most common modular domains in multicellular organisms [5, 6]. The last two decades have seen an intense research on PDZ domains with numerous experimental and computational studies tackling the fine details of the function of isolated PDZ domains and/or the biology of PDZ proteins. It has become clear that at least a subset of PDZ proteins act as dynamic regulators of cell signaling rather than static docking sites. In a cellular context, PDZ interactions depend on several factors such as the intrinsic specificity of the domain, the availability of ligands, the cooperative effects with other interaction partners and the dynamic regulation of the PDZ protein and its ligands. For a better understanding of the biology of PDZ proteins it will be crucial to gain a better understanding of the synergy and/or competition between different binding partners and the regulatory mechanisms governing these interactions. In particular, it will be interesting to follow future studies regarding the interplay between peptide and lipid signaling, the importance of PDZ–PDZ interactions in the organization of multi‐protein scaffolds and the biological importance of the suggested phosphorylation switches. In addition, it will be interesting to follow the efforts being made towards the goal of developing highly specific inhibitors of PDZ proteins through the use of bidentate inhibitors targeting PDZ tandems rather than isolated PDZ domains [139, 140, 141].
Acknowledgements
The author wants to express her gratitude for all support from the members of the groups of professor Pascale Zimmermann at K. U. Leuven and of professors Philip M. Kim and Sachdev Sidhu at University of Toronto. The author apologizes for all papers not included in this review due to space restrictions and the extent of the literature.
and Ivarsson Ylva(2012), Plasticity of PDZ domains in ligand recognition and signaling, FEBS Letters, 586, doi: 10.1016/j.febslet.2012.04.015
References
- 1. Good M.C., Zalatan J.G., Lim W.A., Scaffold proteins: hubs for controlling the flow of cellular information. Science, 332, (2011), 680– 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cho K.O., Hunt C.A., Kennedy M.B., The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron, 9, (1992), 929– 942. [DOI] [PubMed] [Google Scholar]
- 3. Woods D.F., Bryant P.J., ZO-1, DlgA and PSD-95/SAP90: homologous proteins in tight, septate and synaptic cell junctions. Mech. Dev., 44, (1993), 85– 89. [DOI] [PubMed] [Google Scholar]
- 4. Kim E., Niethammer M., Rothschild A., Jan Y.N., Sheng M., Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature, 378, (1995), 85– 88. [DOI] [PubMed] [Google Scholar]
- 5. Ponting C.P., Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci., 6, (1997), 464– 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harris B.Z., Lim W.A., Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci., 114, (2001), 3219– 3231. [DOI] [PubMed] [Google Scholar]
- 7. Jemth P., Gianni S., PDZ domains: folding and binding. Biochemistry, 46, (2007), 8701– 8708. [DOI] [PubMed] [Google Scholar]
- 8. Hillier B.J., Christopherson K.S., Prehoda K.E., Bredt D.S., Lim W.A., Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS–syntrophin complex. Science, 284, (1999), 812– 815. [PubMed] [Google Scholar]
- 9. Hurd T.W., Gao L., Roh M.H., Macara I.G., Margolis B., Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol., 5, (2003), 137– 142. [DOI] [PubMed] [Google Scholar]
- 10. London T.B., Lee H.J., Shao Y., Zheng J., Interaction between the internal motif KTXXXI of Idax and mDvl PDZ domain. Biochem. Biophys. Res. Commun., 322, (2004), 326– 332. [DOI] [PubMed] [Google Scholar]
- 11. Wong H.C., Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol. Cell, 12, (2003), 1251– 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu X.Z., Choudhury A., Li X., Montell C., Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J. Cell Biol., 142, (1998), 545– 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lau A.G., Hall R.A., Oligomerization of NHERF-1 and NHERF-2 PDZ domains: differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry, 40, (2001), 8572– 8580. [DOI] [PubMed] [Google Scholar]
- 14. Zimmermann P., Meerschaert K., Reekmans G., Leenaerts I., Small J.V., Vandekerckhove J., David G., Gettemans J., PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol. Cell, 9, (2002), 1215– 1225. [DOI] [PubMed] [Google Scholar]
- 15. Gallardo R., Ivarsson Y., Schymkowitz J., Rousseau F., Zimmermann P., Structural diversity of PDZ–lipid interactions. Chembiochem, 11, (2010), 456– 467. [DOI] [PubMed] [Google Scholar]
- 16. Mortier E., Wuytens G., Leenaerts I., Hannes F., Heung M.Y., Degeest G., David G., Zimmermann P., Nuclear speckles and nucleoli targeting by PIP2–PDZ domain interactions. EMBO J., 24, (2005), 2556– 2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu H., Feng W., Chen J., Chan L.N., Huang S., Zhang M., PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol. Cell, 28, (2007), 886– 898. [DOI] [PubMed] [Google Scholar]
- 18. Ernst A., Gfeller D., Kan Z., Seshagiri S., Kim P.M., Bader G.D., Sidhu S.S., Coevolution of PDZ domain-ligand interactions analyzed by high-throughput phage display and deep sequencing. Mol. Biosyst., 6, (2010), 1782– 1790. [DOI] [PubMed] [Google Scholar]
- 19. Reina J., Lacroix E., Hobson S.D., Fernandez-Ballester G., Rybin V., Schwab M.S., Serrano L., Gonzalez C., Computer-aided design of a PDZ domain to recognize new target sequences. Nat. Struct. Biol., 9, (2002), 621– 627. [DOI] [PubMed] [Google Scholar]
- 20. Stiffler M.A., Chen J.R., Grantcharova V.P., Lei Y., Fuchs D., Allen J.E., Zaslavskaia L.A., MacBeath G., PDZ domain binding selectivity is optimized across the mouse proteome. Science, 317, (2007), 364– 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tonikian R., A specificity map for the PDZ domain family. PLoS Biol., 6, (2008), e239– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J., Kim I., Yang J.S., Shin Y.E., Hwang J., Park S., Choi Y.S., Kim S., Rewiring of PDZ domain–ligand interaction network contributed to eukaryotic evolution. PLoS Genet., 8, (2012), e1002510– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chi C.N., Engstrom A., Gianni S., Larsson M., Jemth P., Two conserved residues govern the salt and pH dependencies of the binding reaction of a PDZ domain. J. Biol. Chem., 281, (2006), 36811– 36818. [DOI] [PubMed] [Google Scholar]
- 24. Harris B.Z., Lau F.W., Fujii N., Guy R.K., Lim W.A., Role of electrostatic interactions in PDZ domain ligand recognition. Biochemistry, 42, (2003), 2797– 2805. [DOI] [PubMed] [Google Scholar]
- 25. Smock R.G., Gierasch L.M., Sending signals dynamically. Science, 324, (2009), 198– 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akiva E., Friedlander G., Itzhaki Z., Margalit H., A dynamic view of domain-motif interactions. PLoS Comput. Biol., 8, (2012), e1002341– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang C.K., Pan L., Chen J., Zhang M., Extensions of PDZ domains as important structural and functional elements. Protein Cell, 1, (2010), 737– 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morais Cabral J.H., Crystal structure of a PDZ domain. Nature, 382, (1996), 649– 652. [DOI] [PubMed] [Google Scholar]
- 29. Petit C.M., Zhang J., Sapienza P.J., Fuentes E.J., Lee A.L., Hidden dynamic allostery in a PDZ domain. Proc. Natl. Acad. Sci. U.S.A., 106, (2009), 18249– 18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mostarda S., Gfeller D., Rao F., Beyond the binding site: The role of the beta2–beta3 loop and extra-domain structures in PDZ domains. PLoS Comput. Biol., 8, (2012), e1002429– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ernst A., Sazinsky S.L., Hui S., Currell B., Dharsee M., Seshagiri S., Bader G.D., Sidhu S.S., Rapid evolution of functional complexity in a domain family. Sci. Signal, 2, (2009), ra50– [DOI] [PubMed] [Google Scholar]
- 32. Gianni S., Ivarsson Y., De Simone A., Travaglini-Allocatelli C., Brunori M., Vendruscolo M., Structural characterization of a misfolded intermediate populated during the folding process of a PDZ domain. Nat. Struct. Mol. Biol., 17, (2010), 1431– 1437. [DOI] [PubMed] [Google Scholar]
- 33. Huang J., Koide A., Makabe K., Koide S., Design of protein function leaps by directed domain interface evolution. Proc. Natl. Acad. Sci. U.S.A., 105, (2008), 6578– 6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ivarsson Y., Travaglini-Allocatelli C., Brunori M., Gianni S., Engineered symmetric connectivity of secondary structure elements highlights malleability of protein folding pathways. J. Am. Chem. Soc., 131, (2009), 11727– 11733. [DOI] [PubMed] [Google Scholar]
- 35. Ivarsson Y., Travaglini-Allocatelli C., Morea V., Brunori M., Gianni S., The folding pathway of an engineered circularly permuted PDZ domain. Protein Eng. Des. Sel., 21, (2008), 155– 160. [DOI] [PubMed] [Google Scholar]
- 36. Inaba K., Suzuki M., Maegawa K., Akiyama S., Ito K., Akiyama Y., A pair of circularly permutated PDZ domains control RseP, the S2P family intramembrane protease of Escherichia coli . J. Biol. Chem., 283, (2008), 35042– 35052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Truschel S.T., Sengupta D., Foote A., Heroux A., Macbeth M.R., Linstedt A.D., Structure of the membrane-tethering GRASP domain reveals a unique PDZ ligand interaction that mediates Golgi biogenesis. J. Biol. Chem., 286, (2011), 20125– 20129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feng W., Zhang M., Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat. Rev. Neurosci., 10, (2009), 87– 99. [DOI] [PubMed] [Google Scholar]
- 39. Feng W., Shi Y., Li M., Zhang M., Tandem PDZ repeats in glutamate receptor-interacting proteins have a novel mode of PDZ domain-mediated target binding. Nat. Struct. Biol., 10, (2003), 972– 978. [DOI] [PubMed] [Google Scholar]
- 40. Pan L., Chen J., Yu J., Yu H., Zhang M., The structure of the PDZ3-SH3-GuK tandem of ZO-1 protein suggests a supramodular organization of the membrane-associated guanylate kinase (MAGUK) family scaffold protein core. J. Biol. Chem., 286, (2011), 40069– 40074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Songyang Z., Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science, 275, (1997), 73– 77. [DOI] [PubMed] [Google Scholar]
- 42. Daniels D.L., Cohen A.R., Anderson J.M., Brunger A.T., Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nat. Struct. Biol., 5, (1998), 317– 325. [DOI] [PubMed] [Google Scholar]
- 43. Doyle D.A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R., Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell, 85, (1996), 1067– 1076. [DOI] [PubMed] [Google Scholar]
- 44. Grembecka J., Cierpicki T., Devedjiev Y., Derewenda U., Kang B.S., Bushweller J.H., Derewenda Z.S., The binding of the PDZ tandem of syntenin to target proteins. Biochemistry, 45, (2006), 3674– 3683. [DOI] [PubMed] [Google Scholar]
- 45. Sugi T., Oyama T., Muto T., Nakanishi S., Morikawa K., Jingami H., Crystal structures of autoinhibitory PDZ domain of Tamalin: implications for metabotropic glutamate receptor trafficking regulation. EMBO J., 26, (2007), 2192– 2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stricker N.L., PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat. Biotechnol., 15, (1997), 336– 342. [DOI] [PubMed] [Google Scholar]
- 47. Birrane G., Chung J., Ladias J.A., Novel mode of ligand recognition by the Erbin PDZ domain. J. Biol. Chem., 278, (2003), 1399– 1402. [DOI] [PubMed] [Google Scholar]
- 48. Kozlov G., Banville D., Gehring K., Ekiel I., Solution structure of the PDZ2 domain from cytosolic human phosphatase hPTP1E complexed with a peptide reveals contribution of the beta2–beta3 loop to PDZ domain–ligand interactions. J. Mol. Biol., 320, (2002), 813– 820. [DOI] [PubMed] [Google Scholar]
- 49. Balana B., Maslennikov I., Kwiatkowski W., Stern K.M., Bahima L., Choe S., Slesinger P.A., Mechanism underlying selective regulation of G protein-gated inwardly rectifying potassium channels by the psychostimulant-sensitive sorting nexin 27. Proc. Natl. Acad. Sci. U.S.A., 108, (2011), 5831– 5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luck K., Fournane S., Kieffer B., Masson M., Nomine Y., Trave G., Putting into practice domain-linear motif interaction predictions for exploration of protein networks. PLoS One, 6, (2011), e25376– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Feng W., Wu H., Chan L.N., Zhang M., Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J. Biol. Chem., 283, (2008), 23440– 23449. [DOI] [PubMed] [Google Scholar]
- 52. Staneva I., Wallin S., All-atom Monte Carlo approach to protein-peptide binding. J. Mol. Biol., 393, (2009), 1118– 1128. [DOI] [PubMed] [Google Scholar]
- 53. Haq S.R., Side-chain interactions form late and cooperatively in the binding reaction between disordered peptides and PDZ domains. J. Am. Chem. Soc., 134, (2012), 599– 605. [DOI] [PubMed] [Google Scholar]
- 54. Chi C.N., Bach A., Engstrom A., Wang H., Stromgaard K., Gianni S., Jemth P., A sequential binding mechanism in a PDZ domain. Biochemistry, 48, (2009), 7089– 7097. [DOI] [PubMed] [Google Scholar]
- 55. Basdevant N., Weinstein H., Ceruso M., Thermodynamic basis for promiscuity and selectivity in protein–protein interactions: PDZ domains, a case study. J. Am. Chem. Soc., 128, (2006), 12766– 12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gerek Z.N., Keskin O., Ozkan S.B., Identification of specificity and promiscuity of PDZ domain interactions through their dynamic behavior. Proteins, 77, (2009), 796– 811. [DOI] [PubMed] [Google Scholar]
- 57. Kong Y., Karplus M., Signaling pathways of PDZ2 domain: a molecular dynamics interaction correlation analysis. Proteins, 74, (2009), 145– 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ho B.K., Agard D.A., Conserved tertiary couplings stabilize elements in the PDZ fold, leading to characteristic patterns of domain conformational flexibility. Protein Sci., 19, (2010), 398– 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gianni S., Haq S.R., Montemiglio L.C., Jurgens M.C., Engstrom A., Chi C.N., Brunori M., Jemth P., Sequence-specific long range networks in PSD-95/discs large/ZO-1 (PDZ) domains tune their binding selectivity. J. Biol. Chem., 286, (2011), 27167– 27175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kang B.S., Cooper D.R., Devedjiev Y., Derewenda U., Derewenda Z.S., Molecular roots of degenerate specificity in syntenin’s PDZ2 domain: reassessment of the PDZ recognition paradigm. Structure, 11, (2003), 845– 853. [DOI] [PubMed] [Google Scholar]
- 61. Lee J.H., Park H., Park S.J., Kim H.J., Eom S.H., The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands. Biochem. Biophys. Res. Commun., 407, (2011), 207– 212. [DOI] [PubMed] [Google Scholar]
- 62. Uemura T., Mori H., Mishina M., Direct interaction of GluRdelta2 with Shank scaffold proteins in cerebellar Purkinje cells. Mol. Cell. Neurosci., 26, (2004), 330– 341. [DOI] [PubMed] [Google Scholar]
- 63. Zhang Y., Appleton B.A., Wu P., Wiesmann C., Sidhu S.S., Structural and functional analysis of the ligand specificity of the HtrA2/Omi PDZ domain. Protein Sci., 16, (2007), 1738– 1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ellencrona K., Syed A., Johansson M., Flavivirus NS5 associates with host-cell proteins zonula occludens-1 (ZO-1) and regulating synaptic membrane exocytosis-2 (RIMS2) via an internal PDZ binding mechanism. Biol. Chem., 390, (2009), 319– 323. [DOI] [PubMed] [Google Scholar]
- 65. Sengupta D., Linstedt A.D., Mitotic inhibition of GRASP65 organelle tethering involves Polo-like kinase 1 (PLK1) phosphorylation proximate to an internal PDZ ligand. J. Biol. Chem., 285, (2010), 39994– 40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lenfant N., Polanowska J., Bamps S., Omi S., Borg J.P., Reboul J., A genome-wide study of PDZ-domain interactions in C. elegans reveals a high frequency of non-canonical binding. BMC Genomics, 11, (2010), 671– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Karlsson O.A., Chi C.N., Engstrom A., Jemth P., The transition state of coupled folding and binding for a flexible beta-finger. J. Mol. Biol., 417, (2012), 253– 261. [DOI] [PubMed] [Google Scholar]
- 68. Gee S.H., Sekely S.A., Lombardo C., Kurakin A., Froehner S.C., Kay B.K., Cyclic peptides as non-carboxyl-terminal ligands of syntrophin PDZ domains. J. Biol. Chem., 273, (1998), 21980– 21987. [DOI] [PubMed] [Google Scholar]
- 69. Peterson F.C., Penkert R.R., Volkman B.F., Prehoda K.E., Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol. Cell, 13, (2004), 665– 676. [DOI] [PubMed] [Google Scholar]
- 70. Zhang Y., Appleton B.A., Wiesmann C., Lau T., Costa M., Hannoush R.N., Sidhu S.S., Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat. Chem. Biol., 5, (2009), 217– 219. [DOI] [PubMed] [Google Scholar]
- 71. Liu W., Wen W., Wei Z., Yu J., Ye F., Liu C.H., Hardie R.C., Zhang M., The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye. Cell, 145, (2011), 1088– 1101. [DOI] [PubMed] [Google Scholar]
- 72. Eldstrom J., Doerksen K.W., Steele D.F., Fedida D., N-terminal PDZ-binding domain in Kv1 potassium channels. FEBS Lett., 531, (2002), 529– 537. [DOI] [PubMed] [Google Scholar]
- 73. Chang B.H., Gujral T.S., Karp E.S., BuKhalid R., Grantcharova V.P., MacBeath G., A systematic family-wide investigation reveals that ∼30% of mammalian PDZ domains engage in PDZ–PDZ interactions. Chem. Biol., 18, (2011), 1143– 1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Im Y.J., Lee J.H., Park S.H., Park S.J., Rho S.H., Kang G.B., Kim E., Eom S.H., Crystal structure of the Shank PDZ–ligand complex reveals a class I PDZ interaction and a novel PDZ–PDZ dimerization. J. Biol. Chem., 278, (2003), 48099– 48104. [DOI] [PubMed] [Google Scholar]
- 75. Im Y.J., Park S.H., Rho S.H., Lee J.H., Kang G.B., Sheng M., Kim E., Eom S.H., Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization. J. Biol. Chem., 278, (2003), 8501– 8507. [DOI] [PubMed] [Google Scholar]
- 76. Fanning A.S., Lye M.F., Anderson J.M., Lavie A., Domain swapping within PDZ2 is responsible for dimerization of ZO proteins. J. Biol. Chem., 282, (2007), 37710– 37716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen J., Pan L., Wei Z., Zhao Y., Zhang M., Domain-swapped dimerization of ZO-1 PDZ2 generates specific and regulatory connexin43-binding sites. EMBO J., 27, (2008), 2113– 2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Christopherson K.S., Hillier B.J., Lim W.A., Bredt D.S., PSD-95 assembles a ternary complex with the N-methyl-D-aspartic acid receptor and a bivalent neuronal NO synthase PDZ domain. J. Biol. Chem., 274, (1999), 27467– 27473. [DOI] [PubMed] [Google Scholar]
- 79. Balla T., Inositol-lipid binding motifs: signal integrators through protein–lipid and protein–protein interactions. J. Cell Sci., 118, (2005), 2093– 2104. [DOI] [PubMed] [Google Scholar]
- 80. Lemmon M.A., Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol., 9, (2008), 99– 111. [DOI] [PubMed] [Google Scholar]
- 81. Lambaerts K., Syntenin, a syndecan adaptor and an Arf6 phosphatidylinositol 4,5-bisphosphate effector, is essential for epiboly and gastrulation cell movements in zebrafish. J. Cell Sci., (2012), 1129– 1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zimmermann P., Syndecan recycling [corrected] is controlled by syntenin-PIP2 interaction and Arf6. Dev. Cell, 9, (2005), 377– 388. [DOI] [PubMed] [Google Scholar]
- 83. Chen Y., Genome-wide functional annotation of dual-specificity protein- and lipid-binding modules that regulate protein interactions. Mol. Cell, (2012), [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pan L., Wu H., Shen C., Shi Y., Jin W., Xia J., Zhang M., Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes. EMBO J., 26, (2007), 4576– 4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yan J., Wen W., Xu W., Long J.F., Adams M.E., Froehner S.C., Zhang M., Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin. EMBO J., 24, (2005), 3985– 3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Meerschaert K., The PDZ2 domain of zonula occludens-1 and -2 is a phosphoinositide binding domain. Cell. Mol. Life Sci., 66, (2009), 3951– 3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ivarsson Y., Wawrzyniak A.M., Wuytens G., Kosloff M., Vermeiren E., Raport M., Zimmermann P., Cooperative phosphoinositide and peptide binding by PSD-95/discs large/ZO-1 (PDZ) domain of polychaetoid, Drosophila zonulin. J. Biol. Chem., 286, (2011), 44669– 44678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Krojer T., Sawa J., Schafer E., Saibil H.R., Ehrmann M., Clausen T., Structural basis for the regulated protease and chaperone function of DegP. Nature, 453, (2008), 885– 890. [DOI] [PubMed] [Google Scholar]
- 89. Ferguson K.M., Lemmon M.A., Schlessinger J., Sigler P.B., Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell, 83, (1995), 1037– 1046. [DOI] [PubMed] [Google Scholar]
- 90. Bravo J., The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell, 8, (2001), 829– 839. [DOI] [PubMed] [Google Scholar]
- 91. Kachel N., Erdmann K.S., Kremer W., Wolff P., Gronwald W., Heumann R., Kalbitzer H.R., Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): splicing-induced modulation of ligand specificity. J. Mol. Biol., 334, (2003), 143– 155. [DOI] [PubMed] [Google Scholar]
- 92. Mishra P., Socolich M., Wall M.A., Graves J., Wang Z., Ranganathan R., Dynamic scaffolding in a G protein-coupled signaling system. Cell, 131, (2007), 80– 92. [DOI] [PubMed] [Google Scholar]
- 93. Montell C., Dynamic regulation of the INAD signaling scaffold becomes crystal clear. Cell, 131, (2007), 19– 21. [DOI] [PubMed] [Google Scholar]
- 94. Shi Y., Yu J., Jia Y., Pan L., Shen C., Xia J., Zhang M., Redox-regulated lipid membrane binding of the PICK1 PDZ domain. Biochemistry, 49, (2010), 4432– 4439. [DOI] [PubMed] [Google Scholar]
- 95. Lockless S.W., Ranganathan R., Evolutionarily conserved pathways of energetic connectivity in protein families. Science, 286, (1999), 295– 299. [DOI] [PubMed] [Google Scholar]
- 96. Garrard S.M., Capaldo C.T., Gao L., Rosen M.K., Macara I.G., Tomchick D.R., Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO J., 22, (2003), 1125– 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Reynolds K.A., McLaughlin R.N., Ranganathan R., Hot spots for allosteric regulation on protein surfaces. Cell, 147, (2011), 1564– 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. van den Berk L.C., Landi E., Walma T., Vuister G.W., Dente L., Hendriks W.J., An allosteric intramolecular PDZ–PDZ interaction modulates PTP-BL PDZ2 binding specificity. Biochemistry, 46, (2007), 13629– 13637. [DOI] [PubMed] [Google Scholar]
- 99. Dhulesia A., Gsponer J., Vendruscolo M., Mapping of two networks of residues that exhibit structural and dynamical changes upon binding in a PDZ domain protein. J. Am. Chem. Soc., 130, (2008), 8931– 8939. [DOI] [PubMed] [Google Scholar]
- 100. Whitney D.S., Peterson F.C., Volkman B.F., A conformational switch in the CRIB-PDZ module of Par-6. Structure, 19, (2011), 1711– 1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Penkert R.R., DiVittorio H.M., Prehoda K.E., Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex. Nat. Struct. Mol. Biol., 11, (2004), 1122– 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Krojer T., Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc. Natl. Acad. Sci. U.S.A., 105, (2008), 7702– 7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Merdanovic M., Determinants of structural and functional plasticity of a widely conserved protease chaperone complex. Nat. Struct. Mol. Biol., 17, (2010), 837– 843. [DOI] [PubMed] [Google Scholar]
- 104. Bhattacharya S., Dai Z., Li J., Baxter S., Callaway D.J., Cowburn D., Bu Z., A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation. J. Biol. Chem., 285, (2010), 9981– 9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Cheng H., Li J., Fazlieva R., Dai Z., Bu Z., Roder H., Autoinhibitory interactions between the PDZ2 and C-terminal domains in the scaffolding protein NHERF1. Structure, 17, (2009), 660– 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Long J.F., Feng W., Wang R., Chan L.N., Ip F.C., Xia J., Ip N.Y., Zhang M., Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem. Nat. Struct. Mol. Biol., 12, (2005), 722– 728. [DOI] [PubMed] [Google Scholar]
- 107. Morales F.C., Takahashi Y., Momin S., Adams H., Chen X., Georgescu M.M., NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands. Mol. Cell. Biol., 27, (2007), 2527– 2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. LaLonde D.P., Bretscher A., The scaffold protein PDZK1 undergoes a head-to-tail intramolecular association that negatively regulates its interaction with EBP50. Biochemistry, 48, (2009), 2261– 2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zimmermann P., Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol. Biol. Cell, 12, (2001), 339– 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Beekman J.M., Coffer P.J., The ins and outs of syntenin, a multifunctional intracellular adaptor protein. J. Cell Sci., 121, (2008), 1349– 1355. [DOI] [PubMed] [Google Scholar]
- 111. Cao T.T., Deacon H.W., Reczek D., Bretscher A., von Zastrow M., A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature, 401, (1999), 286– 290. [DOI] [PubMed] [Google Scholar]
- 112. Sulka B., Lortat-Jacob H., Terreux R., Letourneur F., Rousselle P., Tyrosine dephosphorylation of the syndecan-1 PDZ binding domain regulates syntenin-1 recruitment. J. Biol. Chem., 284, (2009), 10659– 10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Xiao F., Weng J., Fan K., Wang W., Detailed regulatory mechanism of the interaction between ZO-1 PDZ2 and connexin43 revealed by MD simulations. PLoS One, 6, (2011), e21527– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Park D.J., Wallick C.J., Martyn K.D., Lau A.F., Jin C., Warn-Cramer B.J., Akt phosphorylates Connexin43 on Ser373, a “mode-1” binding site for 14-3-3. Cell Commun. Adhes., 14, (2007), 211– 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. von Nandelstadh P., A class III PDZ binding motif in the myotilin and FATZ families binds enigma family proteins: a common link for Z-disc myopathies. Mol. Cell. Biol., 29, (2009), 822– 834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Essmann C.L., Martinez E., Geiger J.C., Zimmer M., Traut M.H., Stein V., Klein R., Acker-Palmer A., Serine phosphorylation of ephrinB2 regulates trafficking of synaptic AMPA receptors. Nat. Neurosci., 11, (2008), 1035– 1043. [DOI] [PubMed] [Google Scholar]
- 117. Lin D., Gish G.D., Songyang Z., Pawson T., The carboxyl terminus of B class ephrins constitutes a PDZ domain binding motif. J. Biol. Chem., 274, (1999), 3726– 3733. [DOI] [PubMed] [Google Scholar]
- 118. Ikenoue T., Inoki K., Zhao B., Guan K.L., PTEN acetylation modulates its interaction with PDZ domain. Cancer Res., 68, (2008), 6908– 6912. [DOI] [PubMed] [Google Scholar]
- 119. Gardoni F., Mauceri D., Fiorentini C., Bellone C., Missale C., Cattabeni F., Di Luca M., CaMKII-dependent phosphorylation regulates SAP97/NR2A interaction. J. Biol. Chem., 278, (2003), 44745– 44752. [DOI] [PubMed] [Google Scholar]
- 120. Weinman E.J., Parathyroid hormone inhibits renal phosphate transport by phosphorylation of serine 77 of sodium-hydrogen exchanger regulatory factor-1. J. Clin. Invest., 117, (2007), 3412– 3420. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121. Shao X., Threonine 82 at the PDZ domain of PICK1 is critical for AMPA receptor interaction and localization. Neurochem. Int., 56, (2010), 962– 970. [DOI] [PubMed] [Google Scholar]
- 122. Ammendrup-Johnsen I., Thorsen T.S., Gether U., Madsen K.L., Serine 77 in the PDZ domain of PICK1 is a protein kinase Calpha phosphorylation site regulated by lipid membrane binding. Biochemistry, 51, (2012), 586– 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhang J., Petit C.M., King D.S., Lee A.L., Phosphorylation of a PDZ domain extension modulates binding affinity and interdomain interactions in postsynaptic density-95 (PSD-95) protein, a membrane-associated guanylate kinase (MAGUK). J. Biol. Chem., 286, (2011), 41776– 41785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Sierralta J., Mendoza C., PDZ-containing proteins: alternative splicing as a source of functional diversity. Brain Res. Brain Res. Rev., 47, (2004), 105– 115. [DOI] [PubMed] [Google Scholar]
- 125. Garcia-Mata R., Burridge K., Catching a GEF by its tail. Trends Cell Biol., 17, (2007), 36– 43. [DOI] [PubMed] [Google Scholar]
- 126. Adato A., Usherin, the defective protein in Usher syndrome type IIA, is likely to be a component of interstereocilia ankle links in the inner ear sensory cells. Hum. Mol. Genet., 14, (2005), 3921– 3932. [DOI] [PubMed] [Google Scholar]
- 127. Yang J., Ablation of whirlin long isoform disrupts the USH2 protein complex and causes vision and hearing loss. PLoS Genet., 6, (2010), e1000955– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Mburu P., Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat. Genet., 34, (2003), 421– 428. [DOI] [PubMed] [Google Scholar]
- 129. Alewine C., Olsen O., Wade J.B., Welling P.A., TIP-1 has PDZ scaffold antagonist activity. Mol. Biol. Cell, 17, (2006), 4200– 4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lee J.H., Richter W., Namkung W., Kim K.H., Kim E., Conti M., Lee M.G., Dynamic regulation of cystic fibrosis transmembrane conductance regulator by competitive interactions of molecular adaptors. J. Biol. Chem., 282, (2007), 10414– 10422. [DOI] [PubMed] [Google Scholar]
- 131. Lee S.J., Ritter S.L., Zhang H., Shim H., Hall R.A., Yun C.C., MAGI-3 competes with NHERF-2 to negatively regulate LPA2 receptor signaling in colon cancer cells. Gastroenterology, 140, (2011), 924– 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Javier R.T., Rice A.P., Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J. Virol., 85, (2011), 11544– 11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Davey N.E., Trave G., Gibson T.J., How viruses hijack cell regulation. Trends Biochem. Sci., 36, (2010), 159– 169. [DOI] [PubMed] [Google Scholar]
- 134. Glaunsinger B.A., Lee S.S., Thomas M., Banks L., Javier R., Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene, 19, (2000), 5270– 5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lee S.S., Weiss R.S., Javier R.T., Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. U.S.A., 94, (1997), 6670– 6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kiyono T., Hiraiwa A., Fujita M., Hayashi Y., Akiyama T., Ishibashi M., Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. U.S.A., 94, (1997), 11612– 11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Teoh K.T., The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell, 21, (2010), 3838– 3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Accardi R., E6 and E7 from human papillomavirus type 16 cooperate to target the PDZ protein Na/H exchange regulatory factor 1. J. Virol., 85, (2011), 8208– 8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Chi C.N., Bach A., Gottschalk M., Kristensen A.S., Stromgaard K., Jemth P., Deciphering the kinetic binding mechanism of dimeric ligands using a potent plasma-stable dimeric inhibitor of postsynaptic density protein-95 as an example. J. Biol. Chem., 285, (2010), 28252– 28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bach A., Chi C.N., Pang G.F., Olsen L., Kristensen A.S., Jemth P., Stromgaard K., Design and synthesis of highly potent and plasma-stable dimeric inhibitors of the PSD-95-NMDA receptor interaction. Angew. Chem. Int. Ed. Engl., 48, (2009), 9685– 9689. [DOI] [PubMed] [Google Scholar]
- 141. Bach A., A high-affinity, dimeric inhibitor of PSD-95 bivalently interacts with PDZ1-2 and protects against ischemic brain damage. Proc. Natl. Acad. Sci. U.S.A., 109, (2012), 3317– 3322. [DOI] [PMC free article] [PubMed] [Google Scholar]


