Abstract
Objectives
To determine the viral cause of laryngeal croup by use of highly sensitive methods, and including recently recognized viruses in the analysis.
Study design
One hundred forty-four consecutive children with hoarse voice and inspiratory stridor attending the emergency department were enrolled. Age- and season-matched children presenting with a wheezing illness served as control subjects (n = 76). Nasopharyngeal swabs were analyzed by polymerase chain reaction for rhinovirus and enterovirus, coronavirus, respiratory syncytial virus (RSV), parainfluenza virus (PIV), influenza A and B virus, human bocavirus, human metapneumovirus, adenovirus, and Mycoplasma pneumoniae.
Results
Virus infection was documented in 80% of patients with croup and 71% of control subjects. Children with croup had significantly more positive test results for PIV 1 and 2 (31% vs 4% and 6% vs 0%, respectively) and significantly fewer positive test results for RSV (15% vs 28%) than wheezing children. Rhinoviruses and enteroviruses were present equally in both groups (21% vs 25%). There was no significant difference in the frequency of influenza A virus or human bocavirus. Few subjects with adenovirus or M. pneumoniae were detected.
Conclusion
Acute laryngeal croup is most often associated with PIV, RSV, rhinovirus, and enterovirus. Rhinovirus and enterovirus appeared equally often in croup and in wheezing illness. During late fall, they were found in 39% and 40%, respectively, of the tested samples.
Abbreviations: HBoV, Human bocavirus; PCR, Polymerase chain reaction; PIV, Parainfluenza virus; RSV, Respiratory syncytial virus
The frequency of lower respiratory tract infections with dyspnea is increasing.1, 2 Bronchiolitis is the most common reason for expiratory dyspnea in young children. Their cause has been studied intensively. Respiratory syncytial virus (RSV) is the single most common pathogen associated with wheezing,1, 3 accounting for up to 54% of bronchiolitis episodes in young children.4 Allander et al5 studied children with expiratory wheezing and found viruses by polymerase chain reaction (PCR) in 95% of the cases. Ten different viruses were represented, with RSV (28%), rhinovirus (28%), and enterovirus (27%) being the most common.
Children with laryngeal croup have inspiratory stridor. Based mainly on virus isolation and antigen detection, parainfluenza viruses (PIVs) have been shown to be the most important single agents causing croup, predominantly during late fall and early winter.6, 7 PCR assays for respiratory viruses, however, are more sensitive methods of detection than virus culture and immunologic methods.1, 2, 5, 8, 9 The lack of finding rhinovirus and enterovirus in laryngeal croup studies6 may be due to the virologic methods used, but it also may reflect the real epidemiologic condition.
The purpose of this study was to examine the virologic condition of children with croup, with PCR used to detect the presence of rhinovirus, enterovirus, coronavirus, PIV1-3, RSV, influenza A and B viruses, human metapneumovirus, adenovirus, human bocavirus (HBoV), and Mycoplasma pneumoniae in the nasopharynx of children with laryngeal croup compared with children with acute wheezing illnesses.
Methods
From October 2003 through September 2004 a prospective study was carried out at the 2 pediatric emergency departments of Helsinki University Central Hospital (Hospital for Children and Adolescents and Jorvi Hospital). These hospitals provide emergency care for children in an area with a population of 1 million people. All children who were evaluated at the emergency department with inspiratory stridor and barking cough or hoarse voice were considered to have a laryngeal infection and were enrolled into the study. Season- and age-matched individuals who had wheezing without laryngeal symptoms were selected as the control group.
Ethical approval was obtained from the local Research Ethics committee of Helsinki University Central Hospital. After receiving informed written consent from the parents, a sample of nasopharyngeal mucus was taken via the nostril with a swab (Spectrum Laboratories Inc., Dallas, Texas). The swab was soaked in phosphate-buffered saline solution 0.5 mL in a 2-mL Eppendorf tube, frozen immediately, and stored at −70°C until analyzed. DNA and RNA were later eluted from phosphate-buffered saline solution by QIAamp DNA and RNA Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instruction.
Detection of Microbes
Rhinoviruses and enteroviruses
A multiplex reverse-transcription PCR-hybridization assay for rhinovirus and enterovirus was carried out in 96-well plates as described previously.10, 11, 12 The primers targeted highly conserved sequences in the 5′ noncoding region shared by rhinoviruses and enteroviruses.10 Extracted RNA was reverse-transcribed into cDNA by incubating 5 μL in a total volume of 40 μL with the antisense primer. After RT reaction the cDNA 5 μL was subjected to PCR in 96-well plates with the antisense primer and biotinylated sense primer.
Liquid-phase hybridization assay with lanthanide labeled oligonucleotide probes was done as described previously11 by applying 10 μL of amplified PCR products to the wells of streptavidin-coated microtiter plates and denaturing in 50 mmol/L NaOH. After washing away the detached DNA strand, hybridization with 2 lanthanide probes (Europium for rhinoviruses, Samarium for enteroviruses, Wallac Oy, Turku, Finland), was allowed to take place at 37°C for 30 minutes in Tris-HCl (pH 7.5) 40 mmol/L supplemented with 10 mmol/L ethylenediamine tetraacetic acid in NaCl 1.5 mol/L. Quantification of lanthanide fluorescence was done in a time-resolved manner. Previous work with prototype strains of human rhinoviruses and human enteroviruses11 revealed that although use of these 2 probes provides optimal capture of the vast number of serotypes in both genera of viruses, it is not always possible to permit unequivocal separation of the 2 groups. Therefore, in the following text, positive hybridization result with both probes is referred to as the presence of rhinovirus.
Coronaviruses
RT-PCR was performed with a primer pair that amplifies a 251 bp fragment of the polymerase gene that is well conserved between coronavirus 229E, coronavirus OC43, coronavirus NL-63, and the SARS-coronavirus.13 The amplified product was detected by electrophoresis in 2% agarose gels after ethidium bromide staining. SARS-coronavirus RNA (Frankfurt-1 strain) was included as a positive control in each run. Only samples showing a distinct band of the expected size without any other fragments occurring in the lane were considered positive.
Influenza viruses, RSV, and parainfluenza viruses
Viral RNA was reverse transcribed into cDNA with random hexamer primers (Roche, Mannheim, Germany) and Superscript III reverse transcriptase enzyme (Invitrogen, Carlsbad, Calif) following the manufacturer's procedures. For the detection of influenza A and B viruses and RSV, cDNA was amplified in a real-time multiplex PCR9 with minor modifications. In the case of PIV 1, 2, and 3, cDNA was also amplified in a real-time multiplex PCR.9
Human bocavirus
A fragment of the NS1 gene was amplified by a conventional PCR, as described,14 except with an annealing temperature of 58°C and 40 cycles. All DNA preparations were tested both undiluted and in a dilution of 1:10 for excluding possible inhibitory factors. All results were confirmed by Southern hybridization, with a digoxigenin-labeled probe.
Human metapneumovirus
Viral RNA was amplified in a 1-step real-time RT-PCR.15 Qiagen Onestep RT-PCR kit (Qiagen, Hilden, Germany) was used in the assay and RT-PCR amplification was performed with the following conditions: incubation for 30 minutes at 50°C for reverse transcription, followed by reverse transcriptase enzyme inactivation and DNA polymerase enzyme activation step at 95°C for 15 minutes, followed by 50 PCR cycles consisting of denaturation for 30 seconds at 95°C and annealing and primer extension for 60 seconds at 60°C.
Adenoviruses
Adenoviruses were detected by use of the degenerate primers designed for the amplification of all known adenoviral species.16 Viral DNA was amplified in a real-time PCR with Qiagen Quantitect SYBR Green PCR kit (Qiagen GmbH, Hilden, Germany), following the manufacturer's instructions. After an initial DNA polymerase activation step for 15 minutes, 45 PCR cycles were performed. Each cycle consisted of denaturation for 15 seconds at 94°C, primer annealing for 30 seconds at 55°C and extension for 30 seconds at 72°C.
Mycoplasma pneumoniae
M. pneumoniae was detected by use of a conventional PCR method combined with liquid hybridization assay.17 In this study, the cutoff value of the hybridization assay was determined to be twice the average optical density value of the negative controls.
The patient records were reviewed and the data were analyzed by the SPSS statistical software (SPSS, Chicago, Ill). Significance of differences between mean values was tested by t testing, and ordinal measurements by χ2 test. P value <.05 was considered statistically significant.
Results
Two hundred twenty children were evaluated in the emergency departments and enrolled into this study. A total of 144 children with inspiratory stridor and barking cough or hoarse voice formed the study group of croup children. Seventy-six control subjects had expiratory dyspnea caused by wheezing. There was no difference in sex, but the children with croup were younger than the control subjects (1.5 vs 1.9 years; t test P < .05). Children in both groups had elevated temperature at enrollment. In the emergency department, corticosteroid (Prednisolone, Leiras, Finland) was administered to 88% and inhaled racemic epinephrine (microNEFRIN, Bird comp. Palm Springs, Calif) to 87% of children with croup (Table I).
Table I.
Characteristics of children with croup and with wheezing illness
| Croup | Wheezing | |
|---|---|---|
| Number of patients | 144 | 76 |
| Sex: males/females | 97/47 | 52/24 |
| Age (median) | 1.9 years | 1.5 years⁎ |
| Temperature | 37.7°C | 37.8°C |
| Corticosteroid | 88% | 24%† |
| Inhaled racemic epinephrine | 87% | 25%† |
P < .01 χ2.
P < .001 χ2.
Overall 77% of the samples were found positive by PCR. The children with croup tested positive in 80% of the cases, the children with wheezing in 71% of the cases. The number of dual virus genomes in a single sample was as frequent among subjects with croup or wheezing (Table II).
Table II.
Number of viruses found in 1 sample from children with croup and wheezing illnesses
| Number of virus types | Croup n = 144 | Wheezing n = 76 |
|---|---|---|
| None | 29 (20.1%) | 22 (28.9%) |
| One | 84 (58.3%) | 42 (55.3%) |
| Two | 29 (20.1%) | 12 (15.8%) |
| Three | 2 (1.4%) | 0 (0.0) |
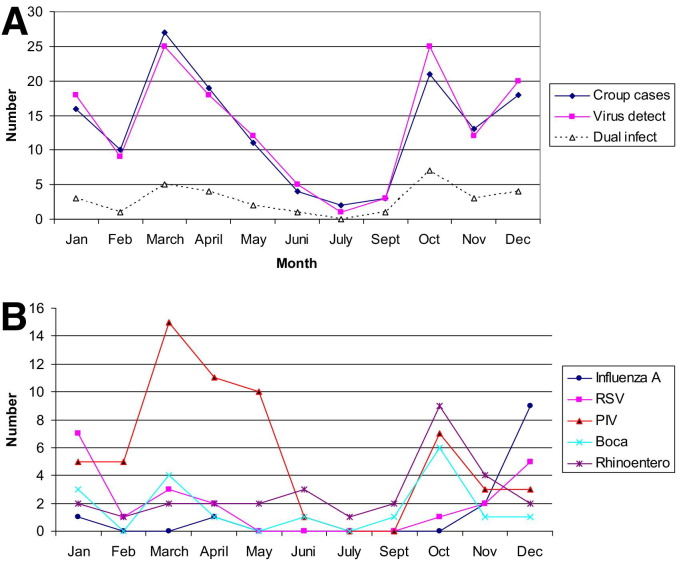
For most of the tested viruses a seasonal difference was observed (Figure). Rhinovirus and enterovirus genome was found more often in samples taken during the fall (September through November; χ2 P < .01), influenza A and RSV were found in samples taken during the winter (December through February; χ2 P < .01), and PIV was found in samples taken during the winter and spring (December through May; χ2 P < .001).
Figure.
Number of patients with laryngeal croup and positive virus findings during 1-year study.
The croup cases appeared in 2 peaks; one from October through January and the other from March through April. During October and November, rhinovirus and enterovirus genomes predominated (Figure, B). Overall, 40% of the samples taken during that period were positive for rhinovirus and enterovirus. This accounts for half of all rhinovirus- and enterovirus-positive cases of croup during the study period. During December, January, and February, several different viruses were associated with the disease. In March, April, and May, PIV was present in more than half of the croup cases (Figure, B).
During the study year, PIV types 1 to 3 genomes were detected in 41% of the children with croup (Table III), compared with children with wheezing; children with croup had significantly more infections with PIV 1 (χ2, P < .001) and PIV 2 (P < .05). For PIV 3 the observed trend was not statistically significant.
Table III.
Virus findings among children with laryngeal croup and acute wheezing illness during the 1-year study
| Croup n = 144 | Wheezing n = 76 | |||
|---|---|---|---|---|
| Parainfluenza virus 1⁎ | 44 | 30.6% | 3 | 3.9% |
| Parainfluenza virus 2† | 7 | 4.9% | 0 | 0.0% |
| Parainfluenza virus 3 | 9 | 6.3% | 1 | 1.3% |
| Respiratory syncytial virus‡ | 21 | 14.6% | 21 | 27.6% |
| Human bocavirus | 18 | 12.5% | 8 | 10.5% |
| Rhinovirus | 17 | 11.8% | 9 | 11.8% |
| Enterovirus | 13 | 9.0% | 10 | 13.2% |
| Influenza A | 13 | 9.0% | 10 | 13.2% |
| Coronavirus | 3 | 2.1% | 1 | 1,3% |
| Adenovirus | 2 | 1.4% | 0 | 0.0% |
| M. pneumoniae | 1 | 0.7% | 1 | 1.3% |
| Human metapneumovirus | 0 | 0.0% | 2 | 2.6% |
| Influenza B | 0 | 0.0% | 0 | 0.0% |
| No. of detected viruses§ | 148 | 66 | ||
| Virus positive | 115 | 79.9% | 54 | 71.1% |
| Virus negative | 29 | 20.1% | 22 | 28.9% |
P < .001 between croup and wheezy bronchitis (χ2 test).
P < .05 between croup and wheezy bronchitis (χ2 test).
P < .01 between croup and wheezy bronchitis (χ2 test).
Some samples harbored more than 1 virus type.
Children with wheezing more often had RSV detected than children with croup (28% vs 15%; χ2, P < .001). Only 2 children tested positive for human metapneumovirus, both of whom had bronchitis (Table III).
Rhinovirus and enterovirus genome was found as often in the nasopharynx of children with croup (30/144) as in those with wheezing (19/76). This holds true for the whole year, as well as for late fall. In October and November, 39% of children with croup and 40% of children with wheezing harbored rhinovirus and enterovirus genome. Among children with croup, half of all rhinovirus- and enterovirus-positive findings appeared during these 2 months; the rest were evenly scattered throughout the year.
Overall, 22% of children with croup and 16% with wheezing harbored more than just 1 pathogen (Table II). Rhinovirus and enterovirus often were found in dual infections. In half (croup 46%, wheezing 57%) of the rhinovirus- and enterovirus-positive samples, no other virus was detected. Thus, rhinovirus and enterovirus genome was the only positive finding in 11% of children with croup and in 15% of children with wheezing (difference not statistically significant).
HBoV appeared equally frequently in both groups (Table III). HBoV tended to be found more often in summer and fall than in winter (Figure). HBoV was frequently found in addition to other pathogens. Children with croup harbored bocavirus in 18 of 144 cases, but it was the sole pathogen in only 5 cases. HBoV was detected in 8 of 76 cases of bronchiolitis but was the sole pathogen in only 4 cases. Of the 35% of children with croup who were hospitalized, virus detected in the nasopharynx did not correlate with frequency of hospitalization.
Discussion
In this study, 80% of the children with croup had a common respiratory virus genome in their nasopharynx, as determined by PCR assays. This figure is higher than those found by less sensitive methods. Denny et al6 studied 951 children with croup and found 360 (34%) virus-positive children by use of virus culture. PIV accounted for about 65% of the positive cases. Thus about 21% of their children with croup had culture-positive PIV type 1 to 3 findings, and RSV was the second most common virus, accounting for about 4% of the culture-positive cases of croup.
In this study, PIV types 1 to 3 were detected in 41% of the children with croup. This is in accordance with earlier reports and confirms the fact that one third of croup cases are caused by PIV.7 We detected PIV in children with croup more frequently in late spring than during any other season. This finding mirrors reported PIV activity in Finland during the spring of 200418 but is at variance with some earlier studies from the United States where PVI 1 infection has been reported to occur in fall.19, 20
Our control subjects with acute wheezing illnesses significantly less often had PIV infections and significantly more often had RSV infections than those with croup. For other virus-positive findings, the groups were similar.
In this study, rhinoviruses could not always be separated from enteroviruses. This was due to the cross-reactivity in the probes used. The close genetic relationship is being recognized also by the taxonomists, and reclassification of enteroviruses and rhinoviruses in a single genus is taking place. In the present study, a virus identification that tested positive for both probes was considered to be rhinovirus.
Rhinoviruses and enteroviruses were the second most common finding among children with croup and bronchitis. The result is similar to that found in bronchiolitis1, 2, 3, 4, 5 but considerably more than that reported earlier6 for acute laryngeal croup. In this study, rhinovirus and enterovirus was the sole positive finding in half of the cases. This likely reflects the real epidemiologic condition and also the fact that modern PCR techniques enhance the detection of rhinoviruses and enteroviruses. However, the diagnostic significance of rhinovirus and enterovirus genome in the nasopharynx of children remains unclear. It has been shown21 that rhinovirus and enterovirus is a common finding in the nasopharynx, especially in the fall. It is often associated with a brief illness21, 22 but is equally often present in the absence of respiratory symptoms.22 The prevalence of rhinovirus and enterovirus genome in this study is quite similar to that reported by Winther et al21 and resembles the result obtained from children with recurrent middle ear infections.23 This warrants more studies to determine the specific role of rhinoviruses and enteroviruses in lower respiratory infections.
Human bocavirus was found in nasopharyngeal aspirates when searching for new viruses by random PCR amplification.24 It has been shown to be one of the most common viruses in respiratory secretions, with a prevalence of 3% to 19%. It typically is found in children hospitalized for lower respiratory tract diseases, and healthy children and adults are generally HBoV-PCR negative.25 This study shows a fairly equal prevalence of HBoV in both groups, and the results are in line with those reported earlier for bronchiolitis. In our study, HBoV-positive specimens frequently were positive for another potential pathogen. This is also in accordance with studies among children with lower respiratory tract infections.5, 26
Acute laryngeal croup in childhood is associated with several different virus types. Analyzed with PCR method, nasopharyngeal swabs from this patient group contained pathogens in 8 of 10 cases. Several viruses were involved, with PIV 1-3, rhinoviruses and enteroviruses, RSV, HBoV, and influenza A virus being the most common ones. In comparison with age- and season-matched control subjects with acute wheezing illnesses, PIV 1 and 2 predominated in children with croup, and RSV was less frequently present. The rest of the nasopharyngeal viral genome detections were similar in both groups. A clear seasonal predilection for most of the tested virus genomes was present.
Footnotes
Supported by Helsinki University Central Hospital Research Funds.
References
- 1.Weigl J.A., Puppe W., Belke O., Neususs J., Bagci F., Schmitt H.J. The descriptive epidemiology of severe lower respiratory tract infections in children in Kiel, Germany. Klin Paediatr. 2005;217:259–267. doi: 10.1055/s-2004-820352. [DOI] [PubMed] [Google Scholar]
- 2.Henrickson K.J., Hoover S., Kehl K.S., Hua W. National disease burden of respiratory viruses detected in children by polymerase chain reaction. Pediatr Infect Dis J. 2004;23:S11–S18. doi: 10.1097/01.inf.0000108188.37237.48. [DOI] [PubMed] [Google Scholar]
- 3.Zamberi S., Zulkifli I., Ilina I. Respiratory viruses detected in hospitalised paediatric patients with respiratory infections. Med J Malaysia. 2003;58:681–687. [PubMed] [Google Scholar]
- 4.Andreoletti L., Lesay M., Deschildre A., Lambert V., Dewilde A., Wattre P. Differential detection of rhinoviruses and enteroviruses RNA sequences associated with classical immunofluorescence assay detection of respiratory virus antigens in nasopharyngeal swabs from infants with bronchiolitis. J Med Virol. 2000;61:341–346. doi: 10.1002/1096-9071(200007)61:3<341::AID-JMV10>3.0.CO;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allander T., Jartti T., Gupta S., Niesters H.G.M., Lehtinen P., Österback R. Human bocavirus and acute wheezing in children. CID. 2007;44:904–910. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny F.W., Murphy T.F., Clyde W.A., Jr, Collier A.M., Henderson F.W. Croup: an 11-year study in a pediatric practice. Pediatrics. 1983;71:871–876. [PubMed] [Google Scholar]
- 7.Henrickson K.J., Kuhn S.M., Savatski L.L. Epidemiology and cost of infection with human parainfluenza virus types 1 and 2 in young children. Clin Infect Dis. 1994;18:770–779. doi: 10.1093/clinids/18.5.770. [DOI] [PubMed] [Google Scholar]
- 8.Osiowy C. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J Clin Microbiol. 1998;36:3149–3154. doi: 10.1128/jcm.36.11.3149-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Templeton K.E., Scheltinga S.A., Beersma M.F., Kroes A.C., Claas E.C. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blomqvist S., Skytta A., Roivainen M., Hovi T. Rapid detection of human rhinoviruses in nasopharyngel aspirates by a microwell reverse transcription-PCR-hybridization assay. J Clin Microbiol. 1999;37:2813–2816. doi: 10.1128/jcm.37.9.2813-2816.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nokso-Koivisto J., Raty R., Blomqvist S., Kleemola M., Syrjanen R., Pitkaranta A. Presence of specific viruses in the middle ear fluids and respiratory secretions of young children with acute otitis media. J Med Virol. 2004;72:241–248. doi: 10.1002/jmv.10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moës E., Vijgen L., Keyaerts E., Zlateva K., Li S., Maes P. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sloots T.P., McErlean P., Speicher D.J., Nissen M.D., Mackay I.M. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. KE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suvilehto J., Roivainen M., Seppanen M., Meri S., Hovi T., Carpen O. Rhinovirus/enterovirus RNA in tonsillar tissue of children with tonsillar disease. J Clin Virol. 2006;35:292–297. doi: 10.1016/j.jcv.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Maertzdorf J., Wang C., Brown J., Quinto J., Chu M., de Graaf M. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol. 2004;42:981–986. doi: 10.1128/JCM.42.3.981-986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allard A., Albinsson B., Wadell G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J Clin Microbiol. 2001;39:498–505. doi: 10.1128/JCM.39.2.498-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Räty R., Ronkko E., Kleemola M. Sample type is crucial to the diagnosis of Mycoplasma pneumoniae pneumonia by PCR. J Med Microbiol. 2005;54:287–291. doi: 10.1099/jmm.0.45888-0. [DOI] [PubMed] [Google Scholar]
- 18.The Infectious Disease Register of the Finnish National Public Health Institute http://www3.ktl.fi/stat/ Accessed May 11, 2006.
- 19.Knott A.M., Long C.E., Hall C.B. Parainfluenza viral infections in pediatric outpatients: seasonal patterns and clinical characteristics. Pediatr Infect Dis J. 1994;13:269–273. doi: 10.1097/00006454-199404000-00005. [DOI] [PubMed] [Google Scholar]
- 20.Marx A., Torok T.J., Holman R.C., Clarke M.J., Anderson L.J. Pediatric hospitalizations for croup (laryngotracheobronchitis): biennial increases associated with human parainfluenza virus 1 epidemics. J Infect Dis. 1997;176:1423–1427. doi: 10.1086/514137. [DOI] [PubMed] [Google Scholar]
- 21.Winther B., Hayden F.G., Hendley J.O. Picoronavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol. 2006;78:644–650. doi: 10.1002/jmv.20588. [DOI] [PubMed] [Google Scholar]
- 22.Winther B., Alper C.M., Mandel E.M., Doyle W.J., Hendley J.O. Temporal relationships between colds, upper respiratory viruses detected by polymerase chain reaction, and otitis media in young children followed through a typical cold season. Pediatrics. 2007;119:1069–1075. doi: 10.1542/peds.2006-3294. [DOI] [PubMed] [Google Scholar]
- 23.Pitkaranta A., Roivainen M., Blomgren K., Peltola J., Kaijalainen T., Raty R. Presence of viral and bacterial pathogens in the nasopharynx of otitis-prone children: A prospective study. Int J Pediatr Otorhinolaryngol. 2006;70:647–654. doi: 10.1016/j.ijporl.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maggi F., Andreoli E., Pifferi M., Meschi S., Rocchi J., Bendinelli M. Human bocavirus in Italian patients with respiratory diseases. J Clin Virol. 2007;38:321–325. doi: 10.1016/j.jcv.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Manning A., Russell V., Eastick K., Leadbetter G.H., Hallam N., Templeton K. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J Infect Dis. 2006;194:1283–1290. doi: 10.1086/508219. [DOI] [PMC free article] [PubMed] [Google Scholar]