Abstract
Proteins fused to the elastin-like polypeptide (ELP) tag can be selectively separated from crude cell extract without chromatography. To avoid the interference of the ELP tag on properties of the target protein, it is necessary to remove the ELP tag from target protein by protease digestion. Therefore, an additional chromatographic purification step is required to remove the proteases, and this is time- and labor-consuming. Here we demonstrate the utility of the ELP-tagged proteases for cleavage of ELP fusion proteins, allowing one-step removal of the cleaved ELP tag and ELP-tagged proteases without chromatography.
It has been an urgent desire to find a simple reliable, and economic method for protein purification during the postgenomic era. For this purpose, a lot of protein purification methods have been explored. Among them, Meyer and Chilkoti reported a nonchromatographic method for purification of recombinant proteins using an elastin-like polypeptide (ELP) 2 tag [1]. The ELP tag is an artificial biopolymer consisting of repeating pentapeptides, Val-Pro-Gly-Xaa-Gly (VPGXG), where Xaa can be any amino acid except Pro [2]. The ELP can undergo a reversible phase transition from soluble forms into aggregates by increasing the salt concentration or heating. The ELP aggregates can be recovered by centrifugation and resolubilized in buffer at a temperature below the transition temperature. Importantly, target proteins fused to the ELP tag still retain the phase transition behavior [1]. This characteristic transition allows the recombinant ELP fusion protein to be isolated from the cell lysate by repeated steps of aggregation, centrifugation, and resolubilization without chromatography. This nonchromatographic method for protein purification was termed as inverse transition cycling (ITC) [1], [3]. Recombinant ELP fusion proteins at different expression levels can be efficiently recovered by the ITC method [4], [5]. In addition to simple separation of recombinant proteins, ultra-high-level recombinant proteins can be produced by fusion with the ELP tag [6]. Due to the attractive advantages of the ELP tag, various proteins of interest have been expressed and purified using the ITC method [7], [8], [9], [10], [11]. For crystal structural and biochemical characterization, however, it is necessary to remove the ELP tag from the target proteins. Cleavage of fusion proteins using proteases is feasible, but an additional purification step is needed to remove the proteases, and this is time-consuming. To solve this problem, self-cleaving ELP tags have been devised for purification of the tag-free recombinant proteins without using proteases [12], [13].
Here we report the application of ELP-tagged proteases for cleavage of the ELP fusion proteins to improve the ELP-tag-based nonchromatographic purification method. The principle using ELP-tagged proteases for the separation of tag-free target proteins is shown in Fig. 1 . The human rhinovirus 3C protease and small ubiquitin-related modifier (SUMO) protease from yeast were used to demonstrate the proof-of-principle for this method. The human rhinovirus 3C protease is a specific protease that recognizes a peptide LEVLFQ/GP and cleaves between the Gln and Gly residues [14]. SUMO protease can recognize the tertiary structure of SUMO moiety instead of a peptide sequence and cleave in the diglycine motif at the junction between SUMO and the target proteins [15], [16]. Both proteases have been expressed and purified from Escherichia coli [16], [17].
Fig.1.
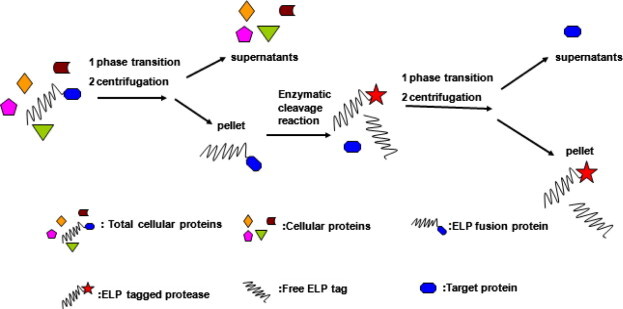
Schematic map of illustrating purification of recombinant proteins using ELP-tagged proteases. The ELP fusion protein was separated by ITC from the lysate and digested by ELP-tagged protease to release the ELP tag from the target protein. After enzymatic cleavage reaction, another round of ITC was carried out to precipitate the cleaved ELP tag and ELP-tagged protease. Thus, the tag-free recombinant protein with high purity was recovered from cell lysate without chromatography.
An ELP gene encoding 90 repeating VPGXG was synthesized as described previously [1] and inserted into the plasmid pET23a (Novagen) to generate the vector pET23a–ELP under control of the T7 promoter. Then the vectors pET23a–ELP–3C protease, pET23a–ELP–GFP (green fluorescent protein), pET23a–ELP–SUMO protease, and pET23a–ELP–SUMO–GFP were constructed (see Supplementary material). For expression of ELP fusion proteins, the E. coli BL21 containing each expression vector was cultured at 37 °C until the OD600 reached 0.8. Isopropyl β-d-1-thiogalactopyranoside was added at a final concentration of 0.5 mM for induction at 20 °C overnight.
Initially, the ELP–3C protease was expressed and purified. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis of the purified products for each step is shown in Fig. 2 A, and the PAGE gel was stained with Coomassie Brilliant Blue G-250 [18]. ELP–3C protease was produced at a high level with an apparent molecular weight of approximately 58 kDa. To aggregate ELP–3C protease, solid NaCl was added into the cleared lysate to a final concentration of 2 M at room temperature to trigger the phase transition of ELP. The ELP–3C protease aggregation was precipitated by centrifugation at 14,000g for 10 min at room temperature. No obvious trace of ELP–3C protease was found in the supernatants (Fig. 2A, lane 3), indicating that ELP–3C protease was fully recovered. The precipitated ELP–3C protease was then dissolved in cooled 50 mM Tris–HCl (pH 8.0) buffer at 4 °C. The insoluble materials were removed by additional centrifugation at 4 °C and 14,000g for 10 min, and the purified ELP–3C protease was recovered from the supernatant. The final yield of ELP–3C protease was approximately 22.5 mg/L after two rounds of ITC.
Fig.2.
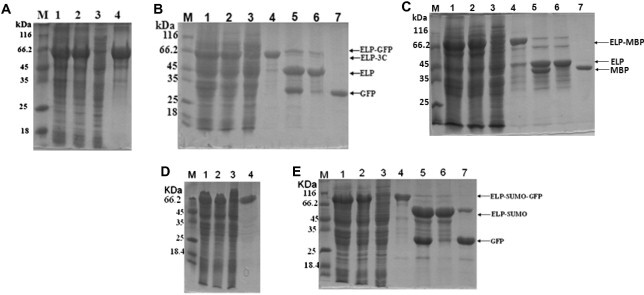
SDS–PAGE analysis of expression and purification of ELP-tagged protease and test proteins. Protein samples are separated by 12% SDS–PAGE and analyzed by Coomassie Brilliant Blue G-250 (20% methanol, 10% phosphoric acid, 10% ammonium sulfate, and 0.12% G-250) staining. (A) Expression and purification of ELP–3C protease. Lane M: molecular weight marker; lane 1: crude extract of cell expressing ELP–3C protease; lane 2: cleared lysate; lane 3: supernatant after precipitation of the ELP–3C protease; lane 4: purified ELP–3C protease. (B) Expression and purification of ELP–GFP and GFP. Lane M: marker; lane 1: crude extract of cell expressing ELP–GFP; lane 2: cleared lysate; lane 3: supernatant after precipitation of ELP–GFP; lane 4: purified ELP–GFP; lane 5: cleavage product of ELP–GFP; lane 6: precipitation of cleaved ELP, ELP–3C protease, and undigested ELP–GFP; lane 7: purified GFP. (C–E) Expression and purification of ELP–MBP (C), ELP-tagged SUMO protease (D), and ELP–SUMO–GFP (E). Gels are as described in panels (A) and (B).
To test the activity of ELP–3C protease and the feasibility using ELP-tagged protease to purify the tag-free recombinant proteins, the ELP–GFP fusion protein was selected as a model protein that contains a 3C protease recognition site between the ELP tag and GFP. The ELP–GFP was purified by ITC as described above. It was shown that highly purified ELP–GFP was recovered after two rounds of ITC (Fig. 2B, lane 4). To release the ELP tag from the GFP, the ELP–GFP was digested using ELP–3C protease at a protease-to-substrate ratio of 1:100 (w/w) in 50 mM Tris–HCl (pH 8.0) buffer (150 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA], and 1 mM dithiothreitol [DTT]) at 5 °C for 16 h. SDS–PAGE analysis showed that the ELP–GFP was efficiently digested by ELP–3C protease. After enzymatic cleavage, NaCl was added into the digestion mixtures to trigger the phase transition of ELP, resulting in aggregation of the cleaved ELP tag and ELP–3C protease. After centrifugation at 14,000g at room temperature for 10 min, the supernatants and pellet were analyzed by SDS–PAGE. Only the GFP band was found in the supernatants (Fig. 2B, lane 7). The cleaved ELP tag, ELP–3C protease, and undigested ELP–GFP were separated from the GFP and kept in the pellet (Fig. 2B, lane 6). The purified GFP that we obtained was approximately 25.6 mg/L. The fluorescence of purified GFP was detected by a fluorescence microscopy. The same method was successfully used to purify another tested protein, maltose binding protein (MBP) (Fig. 2C). Because it is efficient to purify the tag-free recombinant protein using ELP–3C protease, we purified several other proteins with various sizes, including glutathione S-transferase, thioredoxin, and severe acute respiratory syndrome-associated coronavirus (SARS–CoV) nucleocapsid (data not shown).
The use of ELP–3C protease for enzymatic cleavage of fusion protein has several advantages. First, ELP–3C protease is easy to produce in ordinary laboratories because the procedure for preparation of ELP–3C protease is simple, and this can save the cost of buying commercial proteases. Second, ELP-tagged protease as well as the cleaved ELP tag can be removed from the target proteins easily by centrifugation after the triggering phase transition of the ELP, so it saves time, labor, and cost. Third, higher yield of the recombinant proteins can be achieved compared with that of the utility of the self-cleaving ELP tag given that there is no risk of premature cleavage of the fusion protein containing a protease recognition site in vivo. Besides 3C protease, the principle can also be applied to other proteases for various purposes. Unlike 3C protease, SUMO protease from yeast recognizes the tertiary structure of the SUMO tag and releases the target protein with no additional amino acids at the N terminus, which is considered as a robust protease [15], [16]. Therefore, ELP–SUMO protease has also been expressed and isolated by ITC from E. coli (Fig. 2D). The yield of purified ELP–SUMO protease was approximately 11.8 mg/L. ELP–SUMO protease could efficiently digest the ELP–SUMO–GFP to release the GFP from the ELP–SUMO tag (Fig. 2E, lane 5). But a band of ELP–SUMO was observed in addition to the GFP band in the PAGE gel after ITC (Fig. 2E, lane 7), suggesting that the ELP–SUMO was not completely precipitated. To solve this problem, the free ELP protein was added to the digestions to facilitate the aggregation of ELP-SUMO. After triggering the transition of ELP, ELP–SUMO and free ELP were coaggregated and completely separated from GFP by centrifugation (data not shown). The final yield of GFP was approximately 17.1 mg/L. ELP-tagged SUMO protease can be applied to isolate target proteins with a native N terminus.
In summary, we have developed a greatly improved nonchromatographic method for isolation of recombinant proteins using ELP-tagged protease. This method provides an opportunity for efficient and cost-effective purification of a broad range of tag-free proteins for high-throughput analysis of proteins.
Acknowledgments
This work was supported by the European Commission’s Sixth Framework Programme (511060), Development of Intervention Strategies against SARS in a European–Chinese Taskforce (DISSECT), Project 2008AA092603 of the State High-Tech Development Project (863), and Project 2007DFA30840 of the International S&T Cooperation Program from the Ministry of Science and Technology of China.
Footnotes
Abbreviations used: ELP, elastin-like polypeptide; ITC, inverse transition cycling; SUMO, small ubiquitin-related modifier; GFP, green fluorescent protein; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; EDTA, ethylenediaminetetraacetic acid; DTT, dithiothreitol; MBP, maltose binding protein; SARA–CoV, severe acute respiratory syndrome-associated coronavirus.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ab.2011.04.034.
Appendix A. Supplementary data
References
- 1.Meyer D.E., Chilkoti A. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nat. Biotechnol. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 2.Urry D.W. Free energy transduction in polypeptides and proteins based on inverse temperature transitions. Prog. Biophys. Mol. Biol. 1992;57:23–57. doi: 10.1016/0079-6107(92)90003-o. [DOI] [PubMed] [Google Scholar]
- 3.Meyer D.E., Trabbic-Carlson K., Chilkoti A. Protein purification by fusion with an environmentally responsive elastin-like polypeptide: effect of polypeptide length on the purification of thioredoxin. Biotechnol. Prog. 2001;17:720–728. doi: 10.1021/bp010049o. [DOI] [PubMed] [Google Scholar]
- 4.Christensen T., Trabbic-Carlson K., Liu W., Chilkoti A. Purification of recombinant proteins from Escherichia coli at low expression levels by inverse transition cycling. Anal. Biochem. 2007;360:166–168. doi: 10.1016/j.ab.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge X., Filipe C.D. Simultaneous phase transition of ELP tagged molecules and free ELP: an efficient and reversible capture system. Biomacromolecules. 2006;7:2475–2478. doi: 10.1021/bm060507n. [DOI] [PubMed] [Google Scholar]
- 6.Chow D.C., Dreher M.R., Trabbic-Carlson K., Chilkoti A. Ultra-high expression of a thermally responsive recombinant fusion protein in E. coli. Biotechnol. Prog. 2006;22:638–646. doi: 10.1021/bp0503742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trabbic-Carlson K., Liu L., Kim B., Chilkoti A. Expression and purification of recombinant proteins from Escherichia coli: comparison of an elastin-like polypeptide fusion with an oligohistidine fusion. Protein Sci. 2004;13:3274–3284. doi: 10.1110/ps.04931604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimazu M., Mulchandani A., Chen W. Thermally triggered purification and immobilization of elastin–OPH fusions. Biotechnol. Bioeng. 2003;81:74–79. doi: 10.1002/bit.10446. [DOI] [PubMed] [Google Scholar]
- 9.Lin M., Rose-John S., Grotzinger J., Conrad U., Scheller J. Functional expression of a biologically active fragment of soluble gp130 as an ELP-fusion protein in transgenic plants: purification via inverse transition cycling. Biochem. J. 2006;398:577–583. doi: 10.1042/BJ20060544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang H.J., Kim J.H., Chang W.J., Kim E.S., Koo Y.M. Heterologous expression and optimized one-step separation of levansucrase via elastin-like polypeptides tagging system. J. Microbiol. Biotechnol. 2007;17:1751–1757. [PubMed] [Google Scholar]
- 11.Trabbic-Carlson K., Meyer D.E., Liu L., Piervincenzi R., Nath N., LaBean T., Chilkoti A. Effect of protein fusion on the transition temperature of an environmentally responsive elastin-like polypeptide: a role for surface hydrophobicity? Protein Eng. Des. Sel. 2004;17:57–66. doi: 10.1093/protein/gzh006. [DOI] [PubMed] [Google Scholar]
- 12.Banki M.R., Feng L., Wood D.W. Simple bioseparations using self-cleaving elastin-like polypeptide tags. Nat. Methods. 2005;2:659–661. doi: 10.1038/nmeth787. [DOI] [PubMed] [Google Scholar]
- 13.Ge X., Yang D.S., Trabbic-Carlson K., Kim B., Chilkoti A., Filipe C.D. Self-cleavable stimulus responsive tags for protein purification without chromatography. J. Am. Chem. Soc. 2005;127:11228–11229. doi: 10.1021/ja0531125. [DOI] [PubMed] [Google Scholar]
- 14.Cordingley M.G., Callahan P.L., Sardana V.V., Garsky V.M., Colonno R.J. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J. Biol. Chem. 1990;265:9062–9065. [PubMed] [Google Scholar]
- 15.Mossessova E., Lima C.D. Ulp1–SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 16.Malakhov M.P., Mattern M.R., Malakhova O.A., Drinker M., Weeks S.D., Butt T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 17.Leong L.E.-C., Walker P.A., Porter A.G. Efficient expression and purification of a protease from the common cold virus, human rhinovirus type 14. J. Cryst. Growth. 1992;122:246–252. [Google Scholar]
- 18.Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G.M., Carnemolla B., Orecchia P., Zardi L., Righetti P.G. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


