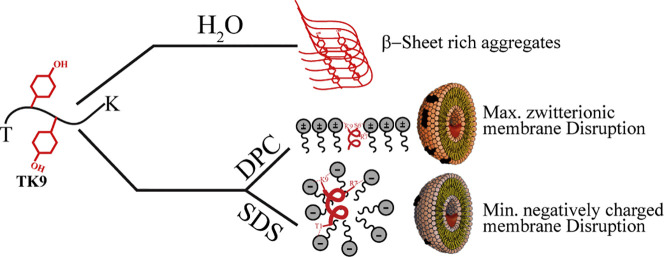
Abstract
In recent years, several studies based on the interaction of self-assembling short peptides derived from viroporins with model membranes, have improved our understanding of the molecular mechanism of corona virus (CoV) infection under physiological conditions. In this study, we have characterized the mechanism of membrane interaction of a short, 9-residue peptide TK9 (T55VYVYSRVK63) that had been derived from the carboxyl terminal of the Severe Acute Respiratory Syndrome (SARS) corona virus (SARS CoV) envelope (E) protein. The peptide has been studied for its physical changes in the presence of both zwitterionic DPC and negatively charged SDS model membrane micelles, respectively, with the help of a battery of biophysical techniques including two-dimensional solution state NMR spectroscopy. Interestingly, in both micellar environments, TK9 adopted an alpha helical conformation; however, the helical propensities were much higher in the case of DPC compared to those of SDS micelle, suggesting that TK9 has more specificity towards eukaryotic cell membrane than the bacterial cell membrane. The orientation of the peptide TK9 also varies in the different micellar environments. The peptide's affinity was further manifested by its pronounced membrane disruption ability towards the mammalian compared to the bacterial membrane mimic. Collectively, the in-depth structural information on the interaction of TK9 with different membrane environments explains the host specificity and membrane orientation owing to subsequent membrane disruption implicated in the viral pathogenesis.
Abbreviations: SARS, Severe Acute Respiratory Syndrome; CoV, corona virus; ssRNA, single-stranded ribonucleic acid; TOCSY, total correlation spectroscopy; NOESY, nuclear overhauser effect spectroscopy; DSS, 4,4-dimethyl-4-silapentane-1-sulfonic acid; SDS, sodium dodecyl sulfate; DPC, dodecylphosphocholine; RMSD, root-mean-square deviation; HSQC, heteronuclear single-quantum correlation; CPMG, Carr-Purcell-Meiboom-Gill; DPH, 1,6-diphenyl-1,3,5-hexatriene; LUVs, large unilamellar vesicles; POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine; POPG, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol
Keywords: NMR, SARS CoV, Micelle, NOESY, Paramagnetic relaxation
Graphical abstract
1. Introduction
Corona viruses (CoV) have been known for decades to be the causative agent of common respiratory problems in both human and domestic fauna [1] and to infect the respiratory as well as the gastrointestinal tract, on a global scale [2]. Among the different strains of corona viruses that are economically important, the SARS-CoV, a human corona virus, have gained much importance in the last few decades as being responsible for the Severe Acute Respiratory Syndrome, typically referred as SARS disease [2], [3]. This deadly virus accounted for a global epidemic with about 8096 respiratory infections resulting in almost 10% (~ 775) mortality cases around the world in the year 2003 [4]. SARS CoVs are a group of ssRNA viruses with a characteristic morphology, due to the spike proteins (S) that form the outer ‘corona’ giving the genera its name [3], [5]. Apart from the spike protein, the overall structure is defined by the presence of the envelope (E), membrane (M) and nucleocapsid (N) proteins. These proteins interact to assist the virus in propagating within host cells. Among these, the E-protein that is expressed in large quantities in infected host cells serves as an essential component of SARS-CoV infection and has been shown to be critical for mediating viral pathogenesis. The E-protein is of particular significance because of its interaction with host membrane and maintaining the membrane curvature and scission necessary for viral propagation [6]. The E-protein consists of short polypeptide chains (~ 75–109 amino acids) with a hydrophobic domain (HD). It can be divided into three subunits: the hydrophilic N-terminal domain (~ 8–12 residues), alpha helical transmembrane domain (~ 21–30 residues) and a flexible C-terminal domain (~ 29–76 residues). The C-terminal domain is specifically essential for viral self-assembly and virus trafficking making a significant contribution to viroporin biology [7], [8]. Therefore, biophysical and structural studies of the C-terminal domain of SARS-CoV E-proteins are of prime importance to understand viroporin biology from the structural point of view [9].
In our previous study, we highlighted the significance of a short stretch of 9 residues (TK9) derived from the host-membrane associating C-terminal end of the E-protein [10]. The study demonstrated that TK9 self-assembles in aqueous solution and more specifically the aromatic residues in the motif ‘VYVY’ accelerate formation of the amyloidogenic deposit [10]. In the present study, we now focus on the structural aspect of the peptide in the presence of the host membrane mimicking micelles and large unilamellar vesicles (LUVs) to understand the biological significance of this amyloidogenic stretch in the viral pathogenesis. Membrane association has often been studied for the initiation of the structural changes, triggering the aggregation of amyloid proteins and peptides in many amyloidogenic diseases [11], [12], [13]. This study, thus, will allow us to correlate the amyloidogenic properties of the peptide in the light of its membrane disrupting activity.
The interaction of TK9 with simplified biological model membranes such as zwitterionic dodecylphosphocholine (DPC) and negatively charged sodium dodecyl sulfate (SDS) was studied using different biophysical and high-resolution solution NMR experiments. From our study, we find that this small, self-assembling domain has a membrane directed activity exhibiting a significant binding affinity and structural changes in the presence of membrane mimics. The peptide adopts an alpha-helical conformation in the presence of both lipid micelles but orients deeper into the DPC lipid layer compared to the SDS micelles. The fluorescence based dye leakage experiment suggests that the membrane disruption by TK9 is more pronounced in the case of eukaryotic cell membrane LUV mimics than the bacterial cell membrane LUVs. This highlights the importance of the critical C-terminal domain in SARS CoV E-proteins in morphogenesis through which the virus propagates [7].
2. Materials and methods
2.1. Reagents
4,4-Dimethyl-4-silapentane-1-sulfonic acid (DSS), deuterium oxide (D2O), deuterated methanol (d4-MeOH), perdeuterated SDS (d35-SDS) and perdeuterated DPC (d38-DPC) were purchased from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA). The lipids (POPG, POPE, and POPC) were purchased from Avanti Polar Lipids (Alabama, USA) and prepared using a lipid extruder from the same manufacturer. Other reagents such as DPH, carboxyfluorescein, etc. were procured from Sigma-Aldrich (St. Louis, MO) unless mentioned otherwise.
2.2. Peptide synthesis
The TK9 peptide was synthesized in a semi-automated peptide synthesizer (Aapptec Endeavor 90) using Fluorenylmethyloxycarbonyl (Fmoc) protected amino acids (Novabiochem) and Rink Amide MBHA resin as described elsewhere [14], [15]. The crude peptide was purified by High-performance liquid chromatography (HPLC) using a SHIMADZU (Japan) instrument with a Phenomenix C18 column (dimension 250 × 10 mm, pore size 100 Å, 5-μm particle size) and linear gradient elution. The ratio of methanol: water was varied linearly from 0 to 100% during the elution and the eluent fractions were lyophilized. Mass spectroscopy and NMR were used to confirm the purity and molecular weight of the eluted peptides.
2.3. Circular dichroism spectroscopy
The circular dichroism (CD) spectra were recorded with a Jasco J-815 spectrometer (Jasco International Co., Ltd. Tokyo, Japan) furnished with a Peltier cell holder and temperature regulator CDF-426L. CD spectra were recorded for TK9 alone (25 μM) and in the presence of various concentrations of SDS and DPC (0–50 mM) at 37 °C. The scan range spanned 190–260 nm, with a 1 nm data interval. Spectra were averaged over 4 consecutive scans for each set of experiments. The raw data were corrected for buffer contribution and changed to molar ellipticity (θ) (deg·cm2·dmol− 1), using the following equation:
| (1) |
where m0 is milli-degrees, M is molecular weight, L represents path length of the cuvette (cm), and C denotes molar concentration.
2.4. Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) experiments were performed using a VP-ITC Micro-Calorimeter at 310 K. DPC and TK9 stock solutions were prepared in 10 mM sodium phosphate buffer at pH 7.2 and degassed before use. The titration involved 20 injections of 25 mM DPC (5 μl per injection) at 180 s intervals, into the sample chamber containing 10 μM TK9. The reaction cell was constantly stirred at 200 rpm. The heat of dilution of DPC in buffer was subtracted from raw data and analyzed using the Micro-Cal Origin 7.0 software in the instrument. The data were fitted to a single site binding model using the nonlinear least square algorithm. The thermodynamic and binding parameters such as equilibrium association constant (Ka), and enthalpy were obtained. Gibb's free energy change (ΔG) was calculated using the relation, ΔG = − RT ln Ka and entropy change (ΔS) was calculated using the equation ΔS = (ΔH − ΔG)/T as described previously [16], [17].
2.5. DPH fluorescence assay
DPH (1,6-diphenyl-1,3,5-hexatriene) fluorescence was measured to elucidate peptide interaction with different micelles i.e. SDS and DPC [18], [19]. Initially, a stock solution of DPH was prepared in acetone and subsequently diluted into the working buffer (10 mM phosphate buffer, pH 7.2) to a final concentration of ~ 18 μM. Next, fixed concentration of micelles, e.g., 5 mM for DPC or 20 mM for SDS was added to the above solution and initial fluorescence intensity (F0) was recorded with an excitation wavelength of 358 nm and emission profile in the range of 400 to 600 nm with a 5-nm slit width. Increasing concentrations of TK9 peptide (0–50 μM) were added separately to each system containing DPH and micelle, following which the fluorescence intensity (F) was measured. Finally, relative DPH fluorescence intensity enhancement (ΔF) was calculated using the following equation:
| (2) |
2.6. Dye leakage release assay
Large unilamellar vesicles (LUVs) were prepared with lipids that mimic the bacterial (Escherichia coli) and the mammalian membrane lipid compositions. For LUVs mimicking the bacterial membrane, POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine) and POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol) were mixed in the ratio of 7:3. The mammalian membrane mimicking LUVs were prepared from POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and 30% cholesterol. Initially, the lipids were mixed in chloroform and then the solutions were subjected to a stream of nitrogen gas, forming a dried film on the wall of the container, and then lyophilized overnight so as to make it void of any residual organic solvent.
The lipid film was then hydrated using 300 μl of a buffer containing the dye, 6-carboxyfluorescein (i.e., 70 mM 6-carboxyfluorescein and 100 mM NaCl in a 10 mM phosphate buffer solution with pH adjusted to 7.4) such that the resultant concentration of the lipids in solution was 10 mg/ml. The solution was then vortexed vigorously, followed by extrusion through a 100 nm polycarbonate Nucleopore filter membrane to obtain dye filled vesicles of an average diameter of 100 nm. The excess dye in the solution, which was not entrapped in the vesicles, was removed by passing the filtered solution through a Centrisep column (equilibrated with extra vesicular buffer). The colored elute from the obtained column contained the dye-filled vesicles separated from the non-encapsulated dye, which was finally used for the leakage assay.
For membrane leakage assay, increasing concentrations (5 to 30 μM) of TK9 were incubated with the 6-carboxyfluorescein dye entrapped LUVs and the fluorescence intensity of the dye was measured. Leakage experiments were achieved on a Hitachi F7000 fluorimeter with excitation wavelengths of 495 nm and emission maxima at 520 nm, keeping 5 nm slit widths for excitation and emission, and a PMT detector voltage of 700 V.
The total fluorescence (I100) was determined by the addition of 5 μl of 10% Triton X-100, with the fluorescence monitored for a minimum of 1 min or until the fluorescence readings had stabilized. The percent leakage was calculated as follows:
| (3) |
where I is the fluorescence intensity of the sample following incubation with peptide, I0 is the fluorescence intensity obtained in the absence of peptide and I100 is the fluorescence intensity obtained after the addition of 5 μl of 10% Triton X-100, which served as the positive control, causing 100% disruption of the vesicles.
2.7. Nuclear magnetic resonance (NMR) experiments
One- and two-dimensional NMR experiments were performed at 298 K and 310 K using a Bruker Avance III 500 MHz spectrometer, equipped with a 5-mm smart probe. Topspin 3.1 (Bruker Biospin Germany) and SPARKY 3.113 (https://www.cgl.ucsf.edu/home/sparky) softwares were used for processing and analysing the NMR data, respectively. Two-dimensional total correlation spectroscopy (TOCSY) and nuclear overhauser effect spectroscopy (NOESY) spectra of TK9 (1 mM) alone and in the presence of perdeuterated d25-SDS (200 mM) and d38-DPC (125 mM) were performed in an aqueous solution containing 10% D2O and at pH ~ 4.5 [20], [21], [22]. All spectra were acquired using states-TPPI mode with 1024 and 512 complex data points along t 2 and t 1 dimensions, respectively. The sweep width and offset due to water were fixed at 12 and at 4.703 ppm with a 2 s relaxation delay. TOCSY spectra were performed using the standard Bruker pulse program (with the MLEV-17 spin lock block) and excitation sculpting being the mode of water suppression. A spin-lock mixing time of 80 ms was used for the TOCSY experiment, whereas 80, 100, 150 and 200 ms mixing times were set for NOESY experiments. Two-dimensional heteronuclear natural abundance 1H–13C HSQC experiment was acquired to estimate the 13CαH chemical shifts of each amino acid residues of TK9 bound to either SDS or DPC at 37 °C with 1024 and 128 data points at t 2 and t 1 dimensions, respectively. The sweep widths were fixed at 12 and 210 ppm with offsets at 4.703 and 100 ppm for 1H and 13C, respectively. The number of scans and dummy scans were 48 and 16, respectively with 2.0 s relaxation delay. Referencing for 1H chemical shifts were achieved using DSS (2,2-dimethyl-2-silapentane-5-sulfonate sodium salt) as an internal standard and indirect referencing with respect to DSS were performed for 13C chemical shifts [23].
The dynamics of each residue of TK9 in the micelle-bound conformation, one-dimensional transverse (R2) and longitudinal (R1) relaxation experiments for protons were performed on a 500 MHz Bruker Avance III spectrometer, as described in our earlier studies [24], [25]. Briefly, the R1 experiments were run with incremental inversion recovery delays starting from 10 ms to 3 s (10, 50, 100, 200, 400, 600, 800, 1000, 1400, 1800, 2400, 3000 ms). Similarly, the measurement for R2 using the Carr-Purcell-Meiboom-Gill (CPMG) refocusing scheme was performed with increasing delays from an initial 0.1 ms to 600 ms (i.e., 0.1, 2, 6, 16, 32, 50, 100, 150, 200, 300, 400, 600 ms). The spectral width was fixed to 10 ppm with 128 scans and 32 dummy scans, respectively. The peak intensities from 1H NMR spectra were plotted against different delays and fitted using exponential equations for estimation of the relaxation rates.
Paramagnetic Relaxation Enhancement (PRE) experiments were carried out by the addition of a fixed concentration (0.5 mM) of paramagnetic quencher, 16-doxyl-stearic acid (16-DSA) dissolved in deuterated methanol (d4-MeOH), into the NMR samples containing peptide and micelles. The sample was allowed to equilibrate for 1 h before the acquisition of 2D TOCSY spectra with the same acquisition parameters as mentioned above [26], [27].
2.8. NMR-derived structure calculation
The three-dimensional NMR structures for TK9 in SDS and DPC micelles were calculated by CYANA 2.1 software suite [28]. From the NOESY spectra of TK9 obtained in the presence of SDS and DPC, upper bound distance constraints were calculated from NOE build-up curves with respect to the NOE intensities of resolved ring protons of Y3 (viz., Y3-2H and Y3-3H). We categorized these distances, obtained from NOE build up curves as strong (2.5 Å), medium (2.6–3.5 Å) and weak (3.6–5.0 Å). A lower bound cutoff of 2.0 Å was used for all distance restraints mentioned above. Apart from that the dihedral angle restraints were calculated from PREDITOR web server with the help of Hα and 13CαH chemical shifts and the predicted torsional angles were further relaxed with ± 30° as upper and lower limits for further structure calculation [29]. Subsequently, several rounds of structure calculation were repeated based on the violation of distance restraints, excluding the non-satisfactory constraints in an iterative manner. Concurrently, the conformational structure was also calculated limiting the conformational space for all the residues, the backbone dihedral angle (phi) was allowed to vary from − 30° to − 120°. This served as a control to reduce the leverage of the torsional angle, obtained from PREDITOR.
Finally, an ensemble of 100 structures, which satisfied most of the experimental distance and dihedral angle constraints, were selected. Among these 100 structures, the top 20 structures with the lowest target function values and RMSD were chosen for further structural analysis. Stereochemical aspects of the final structures were validated using PROCHECK-NMR software package [30].
3. Results and discussion
3.1. TK9 adopts a change in secondary structure in the presence of micelles
TK9 adopted a random coil conformation in aqueous solution, as seen from the characteristic single negative minima at 195 nm in the CD spectra (Fig. 1A). Interestingly, in the presence of micelle, the CD spectra exhibited two negative minima at 208 and 222 nm, indicative of a structural transition from the random coil to an alpha-helical conformation in the micellar environment. In-depth solution state NMR experiments in both micellar systems helped to gain further atomistic resolution understanding of the conformational alternation of TK9 in either case.
Fig. 1.
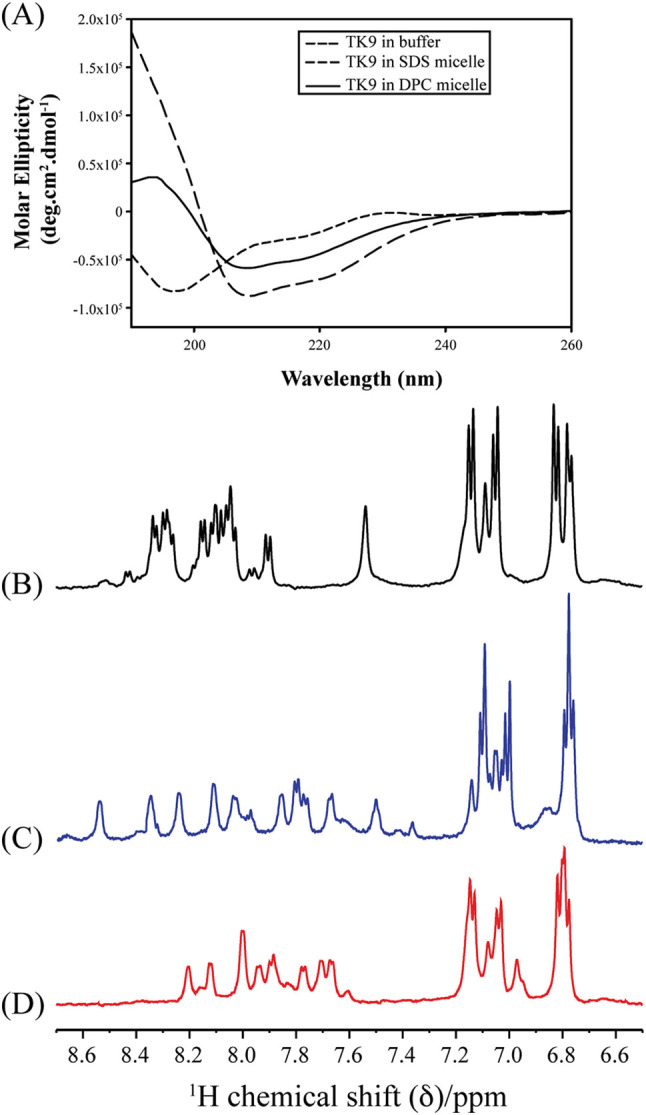
Structural change of TK9 in DPC and SDS micelles. (A) Far UV Circular dichroism (CD) plots of TK9 (25 μM) in aqueous solution (- - -), in DPC micelle (----) (10 mM) and in SDS micelle (- - -) (30 mM). All the experiments were performed at 310 K. (B–D) Selected regions (6.6–8.6 ppm) of 1D 1H NMR spectra of TK9 (1 mM), showing aromatic and amide proton resonances in (B) water (black spectra); in presence of (C) 125 mM perdeuterated DPC (blue spectra); and in (D) 200 mM perdeuterated SDS (red spectra). The experiments were performed using Bruker Avance III 500 MHz spectrometer at 310 K.
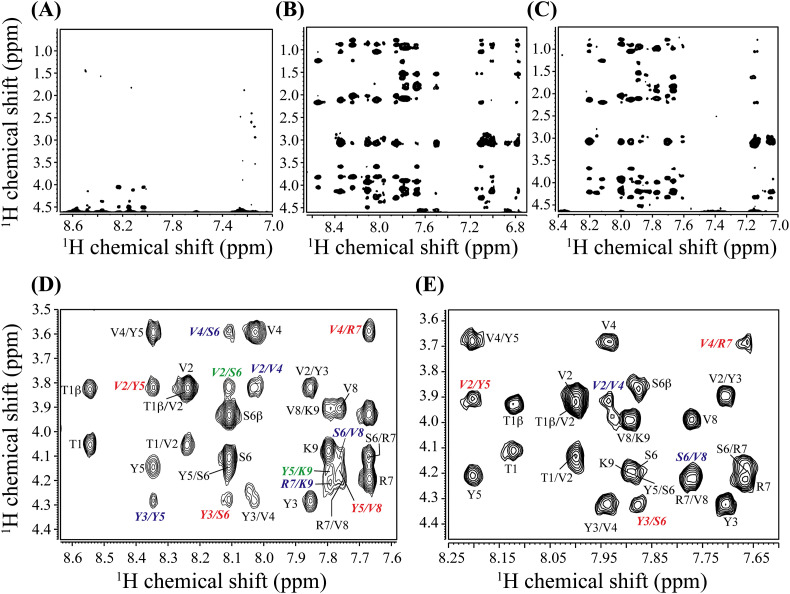
It is already well established that micelles like DPC or SDS are useful for NMR-derived structural elucidation of peptides in solution due to the quick tumbling motion of the peptide-micelle complex in solution [31], [32]. Hence, change in the peptide's conformation in presence of the micelles was further studied using the solution state NMR spectroscopy. Fig. 1B to D shows the amide and aromatic proton region of the one-dimensional 1H NMR spectra of TK9 in aqueous solution, in the presence of DPC and SDS micelle, respectively. TK9 in aqueous solution was less dispersed and showed sharp signals in the amide proton region (Fig. 1B), whereas, in presence of micelles, the spectra were more scattered and, broadened, owing to the well-defined conformational change induced by the micellar environment (Fig. 1C and D). Furthermore, a greater degree of dispersion in the 1D amide proton region in case of DPC micelles when compared to SDS might suggest at the subtle conformational difference among the two micellar environments. Thus to gain atomic resolution understanding of the peptide's conformation, 2D 1H-1H NOESY spectra were acquired in the presence of DPC and SDS micellar environments, which were further assigned and used for the structural elucidation of TK9 in each case as detailed below.
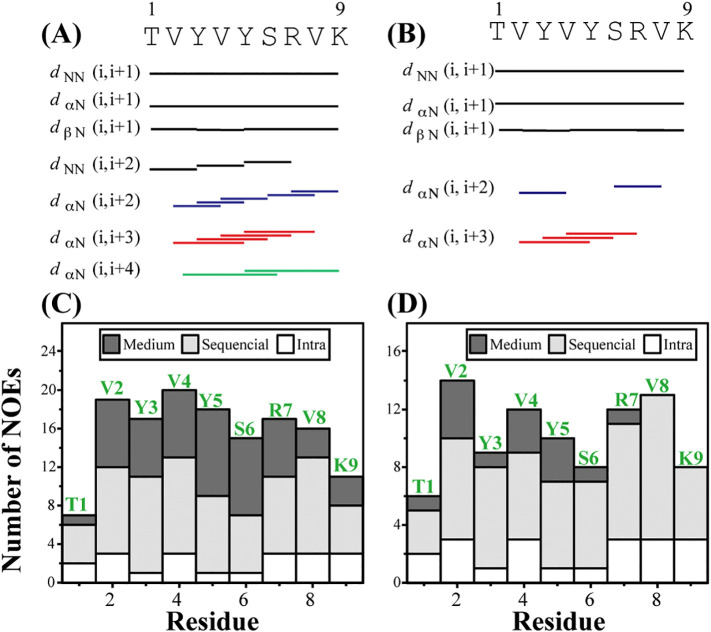
The lack of medium to long range NOEs of TK9 in aqueous solution signifies that the structure is mostly flexible and does not adopt any particular well-defined conformation in aqueous solution (Fig. 2A). On the other hand, the NOESY spectra in DPC (Fig. 2B) and SDS (Fig. 2C) micelle contained a sufficient number of sequential and medium-range NOE cross peaks indicating the existence of a distinct structural fold. The fingerprint region in both micellar environments contained several characteristic, sequential and medium-range CαH/NH NOE contacts as illustrated in Figs. 2D–E, 3 . In addition to the intense sequential CαH/NH (i to i + 1) NOEs, medium range CαH/NH (i to i + 2) NOEs were observed throughout the sequence of TK9 namely between V2/V4, Y3/Y5, V4/S6, S6/V8, R7/K9 in DPC, while in SDS only two CαH/NH (i, i + 2) NOEs such as V2/V4 and S6/V8 were observed (Figs. 2D–E and 3). More specifically, the pattern of CαH/NH (i to i + 3) medium range NOEs were different for the two micelles, e.g., V2/Y5, Y3/S6, V4/R7, and Y5/V8 were observed in DPC micelle, while V2/Y5, Y3/S6, and V4/R7 were predominant in SDS micelle. Also, V2/S6 and Y5/K9 showed CαH/NH (i to i + 4) in DPC micelle and no such NOEs were observed in SDS micelle. Furthermore, a number of medium range NH/NH (i to i + 2) NOE connectivity were found throughout the sequence such as T1/Y3, Y3/Y5, Y5/R7 in DPC while none had been seen in SDS apart from the usual characteristics NH/NH sequential (i to i + 1) NOEs. Therefore, it is evident that the number of medium range NOE connectivities were much higher in DPC micelle in comparison to SDS micelles. These data suggest that there is a structural difference of TK9 in the two different membrane environments. This observation was also well supported by the difference in the aromatic proton region in the NOESY spectra for these two micelles (Fig. S1). In DPC micelle, several aromatic/aliphatic NOE connectivities were observed for TK9, e.g., tyrosine phenyl ring protons (Y3 and Y5) are in close proximity to methylene and methyl protons of neighboring aliphatic residues. In contrast, such NOE contacts were absent in SDS micelle, suggesting the relative orientation of Tyr residues in DPC micelles is much well-ordered and packed within the hydrophobic pockets provided by the side chains of neighboring Val residue in comparison to SDS micelles.
Fig. 2.
Selected 1H-1H NOESY spectra of TK9 in aqueous solution and the presence of micelles. (Upper panel) 2D 1H-1H NOESY spectra (150 ms mixing time) of TK9 (1 mM) in its free state (A); in 125 mM DPC (B) and in 200 mM SDS (C) micellar environment. (Lower panel) Finger print region of NOESY spectra of TK9 in (D) 125 mM DPC; and in (E) 200 mM SDS micelle. The sequential αN (i to i + 1) NOE cross peaks were marked using black color. The medium range αN (i to i + 2/i + 3/i + 4) NOE cross peaks were marked in blue, red and green color, respectively. The experiments were performed using Bruker Avance III 500 MHz spectrometer at 310 K.
Fig. 3.
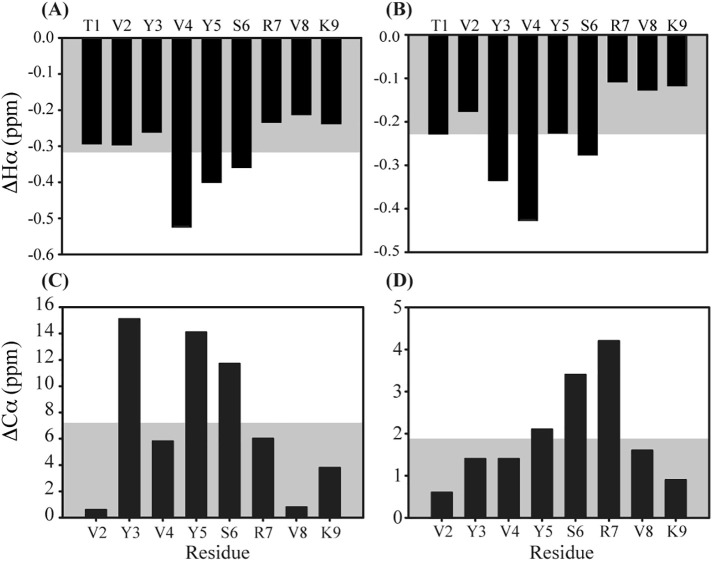
Chemical shift deviations of TK9 in presence of DPC and SDS micelles. (Upper panel) The chemical shift deviation for Hα resonances of each residue of TK9 from the standard random coil values either in (A) DPC micelle or in (B) SDS micelle. (Lower panel) The chemical shift deviation for 13Cα resonances of each residue of TK9 from the standard random coil values either in (C) DPC micelle or in (D) SDS micelle. The grey region indicates the average chemical shift deviation.
The chemical shift deviation of the 13Cα and Hα resonances from the standard random coil value also provides the signature of the secondary structure of the peptide [33], [34]. A helical segment is characterized by negative chemical shift deviation of Hα and positive chemical shift deviation of 13Cα for four consecutive residues while the reverse trend is a signature of the beta sheet structure. In our systems, all the Hα resonances throughout the sequence of the TK9 demonstrate a downfield trend in Hα chemical shift deviation in both the DPC and SDS micelles (Fig. 4A and B). It is also worthy to mention that, the corresponding 13Cα chemical shift deviations for TK9 were all positive in both micelles (Fig. 4C and D). This pattern of chemical shift deviation serves as a signature of the predominance of alpha helical conformation in DPC and SDS micelle. The standard chemical shift values used herein for simplicity were for the aqueous solution. However, a similar trend was obtained for the chemical shift deviations for Hα and 13Cα in case of DMSO and a tri-solvent solution including chloroform, methanol, and water (4:4:1 by v/v). While DMSO best mimics the water-bilayer interface, the latter serves to represent the membrane environment [35]. In either case, a general negative deviation for Hα and a positive deflection was obtained for 13Cα over four consecutive residues (Fig. S2).
Fig. 4.
A summary of NOESY derived NOE statistics of TK9 in micelles. Bar diagram showing the sequential, medium range and long-range NOE connectivities of TK9 in (A) DPC and (B) SDS micelles. The width of the bars, obtained from NOESY spectra are categorized as strong, medium, and weak. The sequential CαH/NH (i to i + 1) NOE cross peaks were marked in black color. The medium range CαH/NH NOE cross peaks i.e., i to i + 2/i + 3/i + 4 were marked in blue, red and green color, respectively. The primary amino acid sequence of TK9 is shown at the top. The Histogram represents the number of NOEs of TK9 in (C) DPC and (D) SDS micelles as a function of the residue number.
The average chemical shift deviation for both 13Cα and Hα is slightly larger in case of DPC micelles when compared to SDS. This might give a qualitative indication of a greater helical propensity of TK9 in DPC micellar environment.
3.2. Structural elucidation of the bound conformation of TK9 in a micellar environment
We used NOESY derived distance constraints in combination with ± 30° relaxed torsional angular restraints, derived from PREDITOR based on Hα and 13Cα chemical shift values, to elucidate the three-dimensional solution conformations of TK9 in DPC and SDS micelles. In order to calculate the structure in DPC micelles, total 81 distance restraints including 20 intra-residues, 35 sequential and 26 medium range constraints were applied while in the case of SDS micelles 20 intra-residue, 29 sequential and 8 medium range constraints (altogether 57 NOE constraints) were used (Table 1 ). From the qualitative comparison of distance restraints, it is evident that the number of medium range αN (i to i + 2/i + 3/i + 4) NOE constraints is almost 3 times less in SDS micelles compared to that in DPC micelles; this is also reflected in the trend of backbone RMSD of an ensemble of 20 structures as seen for the two micelles. In DPC micelle the ensemble of conformations of TK9 was well-organized and converged with an average backbone atom (N, Cα, and C′) and the heavy atoms root mean square deviation (RMSD) values of 0.27 ± 0.10 Å and 0.99 ± 0.21 Å, respectively (Fig. 5A). On the other hand, the ensemble conformations of TK9 in SDS was less well defined with 0.57 ± 0.15 and 1.25 ± 0.16 Å RMSD values for backbone and heavy atoms, respectively (Fig. 5D, lower panel). Close inspection suggests that the three-dimensional structure of TK9 in SDS is flexible mostly at the C-terminal end. It is also mentionworthy that the backbone dihedral angles (Φ, Ψ) were assembled in the most favorable regions and additionally allowed sections in the Ramachandran plot for both the micelle systems (Fig. S3). The overall structural backbone in either micellar environment superimposed with the corresponding free conformational structures (without angular restraints) quite reasonably, with only moderate RMSD values (Fig. S4). The superimposed RMSD was ~ 0.18 Å in case of the structures calculated in DPC micelle. This is in compliance with the significant number of αN (i to i + 2/3/4) NOEs that were accounted for the structure calculation in presence of DPC micelles. In contrary, much higher RMSD (~ 0.51 Å) was obtained in case of the structures in SDS micelle, again consistent with the comparatively lower number of medium range αN (i to i + 2/3/4) NOEs used for TK9 structure. These findings thus reinstate that the number of NOEs play a dominant role in determining the convergent 3D structure of the peptide [36]. Apart from that, we also minimized the significance of the torsional angular constraints used in the structure calculation.
Table 1.
Structural information for the 20 final NMR structures of TK9 in DPC and SDS micelles.
| DPC | SDS | |
|---|---|---|
| Distance restrains | ||
| Intra-residue (i–j = 0) | 20 | 20 |
| Sequential (| i–j | = 1) | 35 | 29 |
| Medium-range (2 ≤ | i–j | ≤ 4) | 26 | 8 |
| Long-range (| i–j | ≥ 5) | 0 | 0 |
| Total | 81 | 57 |
| Angular restraints | ||
| Ф (phi) | 8 | 8 |
| φ (psi) | 8 | 8 |
| Distance restraints from violations (≥ 0.3 Å) | 0 | 0 |
| Deviation from mean structure (Å) | ||
| Average back bone to mean structure | 0.27 ± 0.10 | 0.57 ± 0.15 |
| Average heavy atom to mean structure | 0.99 ± 0.21 | 1.25 ± 0.16 |
| Ramachandran plot analysis for ensemble | ||
| % Residues in the most favorable and additionally allowed regions | (100.0 + 0.0) | (99.3 + 0.7) |
| % Residues in the generously allowed region | 0 | 0 |
| % Residues in the disallowed region | 0 | 0 |
| PDB accession code | 5XER | 5XES |
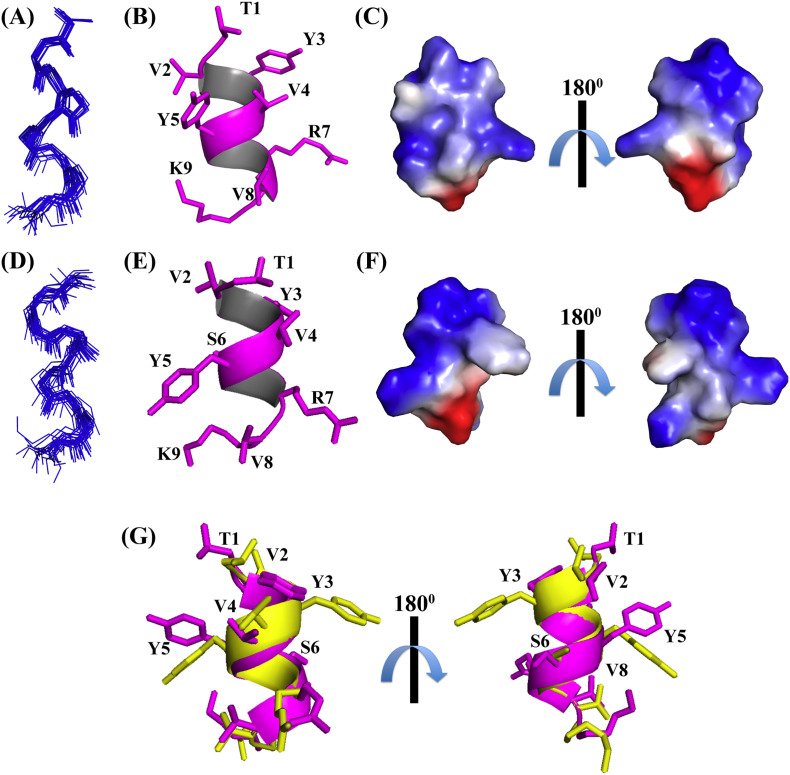
Fig. 5.
Three-dimensional solution structure of TK9 in micelles. Superposition of backbone atoms (N, Cα, C′) of the 20 lowest energy conformations of TK9 in (A) 125 mM DPC and in (D) 200 mM SDS micelles. The side chain orientation of a representative NMR structure of TK9 bound to (B) DPC or in (E) SDS is shown. The electrostatic surface potential of TK9 in (C) DPC and in (F) SDS is displayed to elaborate the segregation of the polar and the non-polar residues. The neutral, negatively charged and positively charged residues are designated in white, red and blue, respectively. (G) Overlaid structures of TK9 in DPC (pink color) and SDS (yellow color) micelles. The overall RMSD value is ~ 0.9 Å. The disagreement of the determined structures in the two micellar environments is governed mainly by the aromatic/aliphatic or aliphatic/aliphatide side chain NOEs. The images were prepared using the PyMOL software.
In both DPC (Fig. 5A and B) and SDS (Fig. 5D and E) micelles, TK9 adopts alpha-helical conformations stabilized by the cumulative hydrophobic and hydrophilic interactions, but the relative orientation of the aromatic and hydrophobic residues is different in the two micellar environments. The “VYVY” unit possibly plays a crucial role in the structural stabilization in the DPC micelle: V2 and V4 remain in close proximity to Y5 and Y3, respectively thereby facilitating CH3-aromatic (π) interaction. Other polar amino acids in the C-terminal region also might make notable electrostatic interaction with the micellar lipid head groups (which is evident from the PRE NMR experiments, as described in a later section) that further stabilizes the lipid-peptide interaction that in turn dictates the peptide orientation. However, in the case of the SDS micelle, Y5 is found to be in close proximity to K9 while Y3 remains close to V2, facilitating cumulative effect of cation-π interaction as well as CH3-aromatic (π) interaction among the residues.
The overall RMSD of 0.9 Å from the superposition of TK9 in both micellar environment is due to the deviation of the side chain orientation of the Tyrosine and Lysine residues (Fig. 5G). Nevertheless, the amphipathicity of TK9 is well conserved in its conformation in the case of both micellar systems. This is evident from the electrostatic surface potential representation of the ensemble of conformations (Fig. 5C and F) that depicts the separation of the polar (blue colored) and the neutral residues (white colored) on either side of the helix.
Taken together, the three-dimensional structures of TK9 in DPC and SDS micelle are in excellent agreement with the CD spectral results, pinpointing the propensity of the alpha helical conformation of TK9 being more pronounced in the presence of DPC micelle as against the negatively charged SDS micelle (Fig. 5G). It is also worth mentioning that the NMR derived solution structure of TK9 in SDS micelle is in good agreement with the previous structural study of the SARS-CoV E protein transmembrane domain (Fig. S4) [37].
3.3. Dynamics and orientation of TK9 in the micellar environments
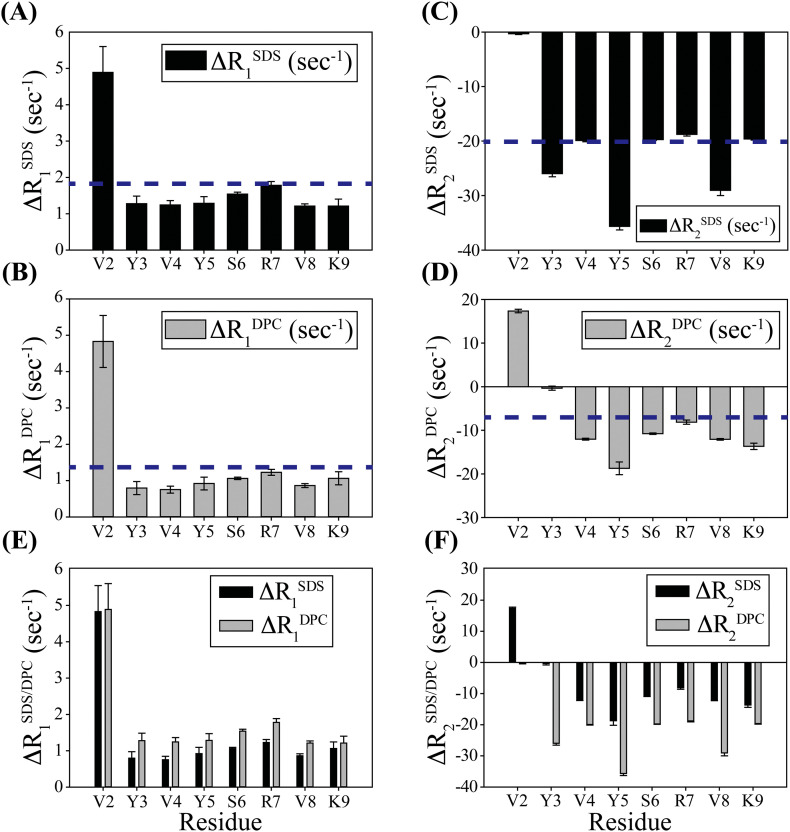
Generally, 1H NMR relaxation (transverse and longitudinal) studies are very useful to investigate the peptide-micelle binding events, which exhibit weak interactions (dissociation constant, KD ≈ μM to mM range) and polypeptide backbone dynamics falling within fast to intermediate time scale (μs–ms) [38]. The longitudinal relaxation rates (R1) of the amide protons (NH) of “micelle-bound conformations of TK9” were relatively short when compared to that of “free TK9” (Fig. 6A, B). This reduction in the relaxation rates is due to the binding effect of the peptide with the micelles. Moreover, the average ΔR1 [R1 Micelles-TK9 − R1 TK9] value was ~ 1.35 Hz and 1.85 Hz in the presence of DPC and SDS micelles, respectively, suggesting that the peptide-micelles complex formation attenuates the high-frequency motion due to the formation of larger composite systems. Similarly, the R2 value of the peptide-micelle complex is increased (negative deviation in ΔR2) in comparison to the free peptide in solution which indicates that the peptide in its bound state has an enlarged net correlation time (τc) due to slow molecular tumbling rate in its micelle-bound conformation (Fig. 6C, D, and F). Higher ΔR2 values were observed for TK9 in presence of the negatively charged SDS micelles when compared to zwitterionic DPC micelles. This trend might be due to the differential binding and relative propensities of micelle bound populations of TK9, which ultimately influence the local motional fluctuation and global tumbling pattern when exposed to the different micellar environments, where the contribution from specific structural alterations are less.
Fig. 6.
Comparison of NMR relaxation rates of amide protons of TK9 in micelle environment. Bar diagram representing the ΔR1 [R1Micelles-TK9 − R1TK9] (s− 1) values of TK9 in (A) 200 mM SDS and in (B) 125 mM DPC micelle. Similarly, bar diagram representing the ΔR2 [R2Micelles-TK9 − R2TK9] (s− 1) values of TK9 in (C) 200 mM SDS; and in (D) 125 mM DPC micelle. The blue dotted line represents the average ΔR1 or ΔR2 (s− 1) values. The comparison of ΔR1 (s− 1) and ΔR2 (s− 1) values for each micelle (SDS and DPC) is shown in (E) and (F).
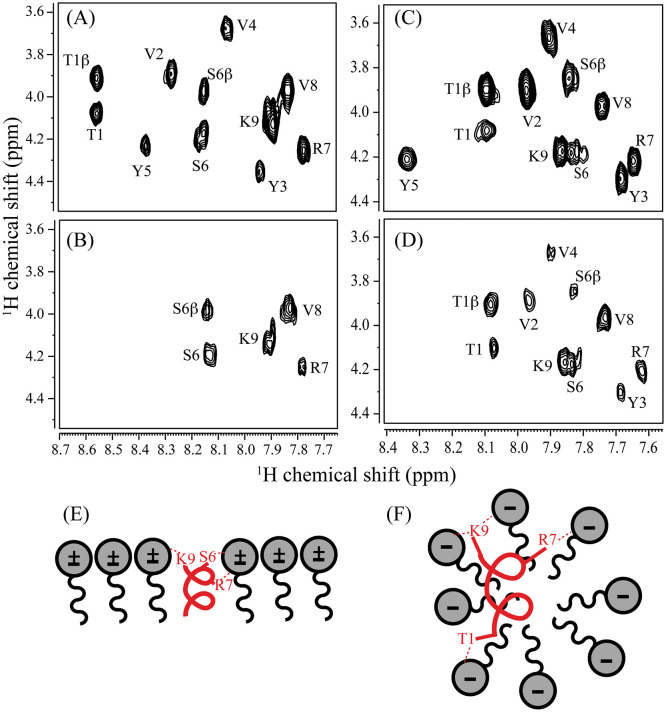
Paramagnetic Relaxation Enhancement (PRE) experiment was performed to gain insights into the specific localization of TK9 in both micellar systems. 16-Doxyl stearic acid (16-DSA), a spin-labelled probe which contains an unpaired electron, in paramagnetic nitroxide group, attached to the 16th carbon position of the chain, was used for the PRE measurements. 16-DSA usually penetrates the micelles positioning themselves near the alkyl chains of the lipid molecules and induces relaxation of the nearby residues that come in close proximity to it [39]. We observed strong PRE effect for > 60% of the residues, including T1, V2, Y3, V4 and Y5, which exhibited a characteristic peak broadening in the TOCSY spectra for DPC bound TK9. However, residues such as S6, R7, V8, and K9 did not suffer much line-broadening in presence of the quencher molecule (Fig. 7A and B). Thereby, we conclude that the hydrophobic and/or aromatic residues in the N-terminal T1-Y5 of TK9 insert deep into the DPC micelle owing to the pronounced PRE effect. However, the positively charged C-terminal residues, i.e., S6-K9 remains near the zwitterionic head groups of the micelle, and hence were less perturbed by the deep quencher probe (16-DSA) (Fig. 7E). Contrary to this, in the SDS micellar environment, Y5 is the only residue of TK9, which showed peak broadening in the presence of a similar concentration of 16-DSA (Fig. 7C–D). Based on this result, we infer that TK9 tend to bind to the surface of the SDS micelle along the lipid head group (Fig. 7F), whereas it orients itself much deeper into the hydrophobic region of the lipid layer, adopting a more or less parallel orientation in the DPC micelle.
Fig. 7.
PRE based 2D NMR of TK9 in DPC and SDS micelles. (Upper panel) Selected TOCSY spectra of TK9 (1 mM) in (A) 125 mM DPC and (B) 200 mM SDS micelles. (Middle panel) Similar TOCSY spectra of TK9 in (C) 125 mM DPC and (D) 200 mM SDS micelles after addition of 16-DSA (0.5 mM). The experiments were carried out using Bruker Avance III 500 MHz NMR spectrometer and at 310 K. The stock solution of 16-DSA was prepared in deuterated methanol. (Lower panel) Relative orientation of TK9 in (E) DPC and (F) SDS micelles.
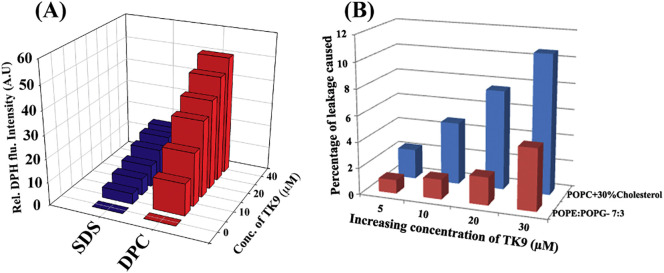
Parallel to NMR data, DPH (1,6-diphenyl-1,3,5-hexatriene) based fluorescence assay also helped us to elucidate the insertion/orientation of the molecule in the membrane environment. DPH is essentially nonfluorescent in the aqueous solution, however, its interaction with the acyl chains of the lipid molecules leads the enhancement of the fluorescence intensity. This phenomenon can be exploited to probe any biophysical interaction of the peptide that would lead to the change in the lipid arrangements of the micelles. TK9 interacts with the zwitterionic DPC micelles much readily, leading to an almost five times greater fluorescence emission of DPH (Fig. 8A) than SDS micelle. Thus, structural orientation of the peptide in the micellar environment leads to the exposure of the hydrophobic regions of the lipid molecules, in turn enabling the physical association with the DPH molecules in the solution.
Fig. 8.
Fluorescence-based assay of TK9 in the two different model membrane systems. (A) A plot showing the relative fluorescence intensity increment of DPH in the presence of different concentrations of TK9 (0 to 40 μM) either in DPC (red bars) or in SDS (blue bars) micelles. (B) Bar diagram showing the fluorescence based 6-carboxyfluorescein dye release from two different model membranes upon treatment with TK9 (0–30 μM). Dye leakage from POPE:POPG (7:3) and POPC:cholesterol membranes are represented in red and blue bars, respectively.
3.4. Membrane disruption through fluorescence based dye leakage assay in two different model membrane mimics
Fluorescence based dye-leakage assay, further helped us to understand the membrane-disrupting activity of the peptide against the zwitterionic and anionic membrane mimics. TK9 is expected to be the membrane directed motif of the viral protein that would be responsible for any membrane disrupting function of the same. In this regard, the mammalian membrane mimics (POPC with 30% cholesterol) served as the zwitterionic LUV prepared to study the dye leakage assay and the bacterial membrane mimics, composed of POPE and POPG in the ratio of 7:3 served as the anionic counterpart. The assay was performed with a membrane impermeant dye, i.e., 6-carboxyfluorescein that remains quenched while trapped inside the vesicles due to a possible dimerization of the dye molecules [40], [41]. Thus, while entrapped in the intact vesicles the dye emits only a basal level of fluorescence, being excited at 495 nm. However, any disruption caused to the lipid membranes result in the release of the dye from the vesicle interior, resulting in its dilution and subsequent dequenching, and hence an increase in fluorescence emission intensity at about 517 nm. This property of the dye allowed us to study the membrane disrupting capability of the peptide and furthermore, quantify it in terms of percentage of leakage caused. An increase in fluorescence intensity was obtained with increasing concentration of the peptide from an initial 5 μM to 30 μM concentration. A maximum of about 11% of dye leakage was obtained when studying the peptide's membrane disrupting activity against the zwitterionic mammalian membrane mimic, tested only up to a maximum of 30 μM concentration. In contrary, only 5% dye leakage was caused to anionic bacterial membrane mimicking LUVs tested with the same peptide concentrations (Fig. 8B). The interaction was directly proportional to the concentration of peptide added to the lipid micelles. Nevertheless, this selective disruption caused to the mammalian membrane mimics highlight the peptide's significance in causing host membrane disruption. The preliminary data can further lead to the in-depth analysis of the mechanism of membrane disruption that is concentration dependent.
3.5. TK9 has differential binding affinities towards the different micellar environments
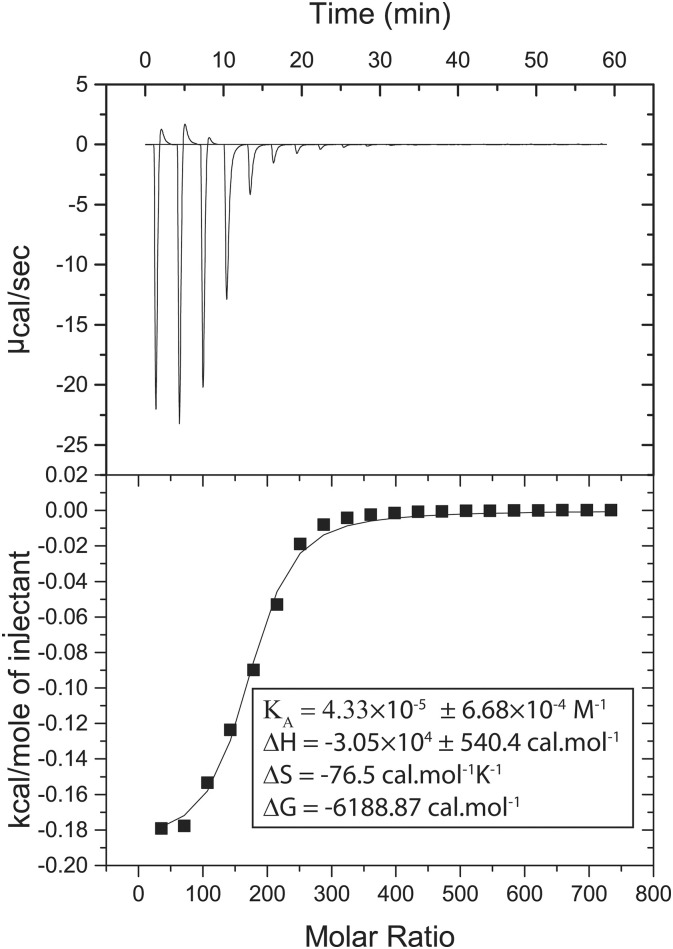
Isothermal titration calorimetry (ITC) helps provide thermodynamic parameters such as enthalpy change (ΔH), entropy change (ΔS), Gibbs free energy change (ΔG) and binding affinity (KD) for studying the receptor-ligand binding phenomenon [42], [43]. Therefore, we have performed ITC experiments to identify the crucial thermodynamic forces underlying the binding of TK9 to the DPC micelles. The binding of TK9 to DPC was exothermic because of the downward ITC thermogram (Fig. 9 ). It is also noteworthy to mention that the negative sign of ΔH (ΔH = − 3.05 × 104 ± 540.4 cal·mol− 1) and ΔS (ΔS = − 76.5 cal·mol− 1·K− 1) suggests the process to be enthalpy driven and hence, the hydrophobic, van der Waal, as well as H-bonding interactions, play a dominant role for this binding interaction [44]. The equilibrium binding affinity was in micromolar (KA = 4.33 × 10− 5 ± 6.7 × 10− 4 mol− 1) range and the corresponding change in Gibbs free energy was − 6188.87 cal·mol− 1, dictating the spontaneity of the process. This binding data is also in good accordance with our NMR and fluorescence dye leakage assay data which demonstrates significant hydrophobic interaction among the residues of TK9 and the lipid acyl chains in the micelles owing to a close physical proximity to the micellar interior (Figs. 7E and 8). Overall, these findings highlight the micromolar binding affinity of TK9 with zwitterionic DPC micelles where hydrophobic interactions play a significant role in binding. The ITC experiment with SDS, however, was not practical due to the significant detergent like frothing behaviour of the SDS micelles.
Fig. 9.
Isothermal titration calorimetry (ITC) profile showing the binding interaction between TK9 and DPC. The upper panel shows the plot of heat of reaction (μcal/s) vs time (min) upon interaction of TK9 with the DPC micelle. The lower panel shows the corresponding enthalpy change per mole of TK9 injection vs the molar ratio (TK9: DPC). All experiments were performed in 10 mM phosphate buffer (pH 7.2) and at 310 K using VP-ITC. The corresponding thermodynamic parameters are mentioned in the lower panel.
4. Conclusion
This study highlights the high-resolution structural perspective of membrane interaction of the TK9 peptide, derived from the SARS CoV E-protein in two different micellar environments. Thus, our study accentuates that the 9-residue short motif of TK9, i.e., T55-K63, integral to the membrane bound alpha helical region of the C-terminal, has a direct functional role in the host membrane identification and subsequent disruption. TK9 readily undergo conformational changes in the presence of host membrane mimicking micelles, as confirmed by CD spectroscopic analysis. The high specificity towards the zwitterionic membranes, compared to negatively charged ones, were seen from the dye-leakage assays that suggest the inherent propensity of TK9 to interact with mammalian as opposed to the bacterial cells. The binding affinity between the peptide and zwitterionic DPC was determined to be in micromolar range as suggested by the ITC experiments, which also revealed that this peptide-micelle interaction is largely governed by the van der Waal forces and is an enthalpy driven process. Further, the high-resolution solution state NMR experiments revealed the amphipathic helical conformation of TK9 in presence of the two micellar environments. Experimental evidence was put forth for the strong association of the peptide with the acyl-chains of the lipid molecules. However, the physical orientation of TK9 was shown to differ depending on the charge of the target membrane. Specifically, the “VYVY” motif of the peptide inserted deeply into the hydrophobic core of the DPC lipids as revealed by PRE experiments. It is noteworthy to mention that this same motif of TK9 had been shown earlier to self-assemble in solution to form beta-sheet rich amyloid fibrillar ordered nano-assemblies [10]. Thus, the peptide sequence that underlies the host specificity and affinity also directed helical conformation in the presence of membrane that leads to the downstream aggregation of the protein units in forming the pentameric [9] structure. Since it is well known that several amyloidogenic peptides disrupt membranes through the self-aggregation mechanism, the aggregating propensity of the “VYVY” motif in TK9 correlates well with the viroporin functionality exhibited by SARS CoV E-protein in host membranes. Moreover, in future, the mutants of “VYVY” motif subjected to kinetics studies on aggregation of TK9 in the presence of suitable host cell membrane mimicking LUVs using in vitro fluorescence based techniques (e.g. Thioflavin T emission assay), can reveal the significance of the 9-residue stretch of E-protein in mediating host membrane association and subsequent cell damage, which are imperative to viral propagation.
Author contributions
AB conceived, designed and funded the research; AG conducted all biophysical experiments, analyzed the results, calculated the three-dimensional structure using NMR spectroscopy; DB performed fluorescence experiments; AG, DB, and AB wrote the manuscript. All authors reviewed the manuscript.
Funding sources
This study was supported by Institutional Plan Project-II fund.
Dedication/notes
The paper is dedicated to Professor Dr. Thomas Peters, University of Lübeck, Germany for his 60th birthday.
Transparency document
Transparency document.
Acknowledgments
Acknowledgment
AG and DB are grateful to Bose Institute and CSIR-UGC for SRF and JRF, respectively. Central Instrument Facility (CIF) of Bose Institute is greatly acknowledged.
Footnotes
The Transparency document associated with this article can be found, in online version.
Aromatic region of NOESY spectra of TK9 in DPC and SDS micelles (Fig. S1); Comparison of the Chemical shift deviations in standard (aqueous) solution, DMSO and trisolvent for TK9 structural conformation in DPC and SDS micelle (Fig. S2); Ramachandran plot of NMR derived conformations of TK9 in DPC and SDS micelles (Fig. S3); Superimposed structures of TK9, determined in the absence (ϕ = − 30 to − 120) and presence of torsional angle constraints (Fig. S4); Superimposed conformations of TK9 in SDS micelle with the conformation of full length SARS-CoV E-protein in SDS micelle (PDB acquisition code: 2MM4) (Fig. S5). Supplementary data associated with this article can be found in the online version, at https://doi.org/10.1016/j.bbamem.2017.10.015.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Peiris J., Lai S., Poon L., Guan Y., Yam L., Lim W., Nicholls J., Yee W., Yan W., Cheung M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.-h. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Kasai T. Crisis management for infectious disease-learn from SARS experience and prepare for the influenza pandemic (summary) Jpn. Med. Assoc. J. 2007;50:117. [Google Scholar]
- 5.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Nieto-Torres J.L., DeDiego M.L., Álvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corse E., Machamer C.E. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 2002;76:1273–1284. doi: 10.1128/JVI.76.3.1273-1284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruch T.R., Machamer C.E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J. Virol. 2011;85:675–685. doi: 10.1128/JVI.01570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D., Vararattanavech A., Soong T.W., Liu D.X., Torres J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh A., Pithadia A.S., Bhat J., Bera S., Midya A., Fierke C.A., Ramamoorthy A., Bhunia A. Self-assembly of a nine-residue amyloid-forming peptide fragment of SARS corona virus E-protein: mechanism of self aggregation and amyloid-inhibition of hIAPP. Biochemistry. 2015;54:2249–2261. doi: 10.1021/acs.biochem.5b00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 12.Terzi E., Hölzemann G., Seelig J. Interaction of Alzheimer β-amyloid peptide (1–40) with lipid membranes. Biochemistry. 1997;36:14845–14852. doi: 10.1021/bi971843e. [DOI] [PubMed] [Google Scholar]
- 13.Bucciantini M., Rigacci S., Stefani M. Amyloid aggregation: role of biological membranes and the aggregate–membrane system. J. Phys. Chem. Lett. 2014;5:517–527. doi: 10.1021/jz4024354. [DOI] [PubMed] [Google Scholar]
- 14.Chan W.C., White P.D. Oxford University Press; 2000. Fmoc Solid Phase Peptide Synthesis. [Google Scholar]
- 15.Ghosh A., Datta A., Jana J., Kar R.K., Chatterjee C., Chatterjee S., Bhunia A. Sequence context induced antimicrobial activity: insight into lipopolysaccharide permeabilization. Mol. BioSyst. 2014;10:1596–1612. doi: 10.1039/c4mb00111g. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh A., Bera S., Shai Y., Mangoni M.L., Bhunia A. NMR structure and binding of esculentin-1a (1–21) NH 2 and its diastereomer to lipopolysaccharide: correlation with biological functions. Biochim. Biophys. Acta Biomembr. 2016;1858:800–812. doi: 10.1016/j.bbamem.2015.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh A., Ratha B.N., Gayen N., Mroue K.H., Kar R.K., Mandal A.K., Bhunia A. Biophysical characterization of essential phosphorylation at the flexible C-terminal region of C-Raf with 14-3-3ζ protein. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prendergast F.G., Haugland R., Callahan P. 1-[4-(Trimethylamino) phenyl]-6-phenylhexa-1, 3, 5-triene: synthesis, fluorescence properties, and use as a fluorescence probe of lipid bilayers. Biochemistry. 1981;20:7333–7338. doi: 10.1021/bi00529a002. [DOI] [PubMed] [Google Scholar]
- 19.Datta A., Bhattacharyya D., Singh S., Ghosh A., Schmidtchen A., Malmsten M., Bhunia A. Role of aromatic amino acids in lipopolysaccharide and membrane interactions of antimicrobial peptides for use in plant disease control. J. Biol. Chem. 2016;291:13301–13317. doi: 10.1074/jbc.M116.719575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saravanan R., Bhunia A., Bhattacharjya S. Micelle-bound structures and dynamics of the hinge deleted analog of melittin and its diastereomer: implications in cell selective lysis by D-amino acid containing antimicrobial peptides. Biochim. Biophys. Acta. 2010;1798:128–139. doi: 10.1016/j.bbamem.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Bhunia A., Mohanram H., Bhattacharjya S. Structural determinants of the specificity of a membrane binding domain of the scaffold protein Ste5 of budding yeast: implications in signaling by the scaffold protein in MAPK pathway. Biochim. Biophys. Acta. 2012;1818:1250–1260. doi: 10.1016/j.bbamem.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Mohanram H., Nip A., Domadia P.N., Bhunia A., Bhattacharjya S. NMR structure, localization, and vesicle fusion of Chikungunya virus fusion peptide. Biochemistry. 2012;51:7863–7872. doi: 10.1021/bi300901f. [DOI] [PubMed] [Google Scholar]
- 23.Wishart D.S., Bigam C.G., Yao J., Abildgaard F., Dyson H.J., Oldfield E., Markley J.L., Sykes B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh A., Kar R.K., Krishnamoorthy J., Chatterjee S., Bhunia A. Double GC: GC mismatch in dsDNA enhances local dynamics retaining the DNA footprint: a high-resolution NMR study. ChemMedChem. 2014;9:2059–2064. doi: 10.1002/cmdc.201402238. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh A., Kar R.K., Jana J., Saha A., Jana B., Krishnamoorthy J., Kumar D., Ghosh S., Chatterjee S., Bhunia A. Indolicidin targets duplex DNA: structural and mechanistic insight through a combination of spectroscopy and microscopy. ChemMedChem. 2014;9:2052–2058. doi: 10.1002/cmdc.201402215. [DOI] [PubMed] [Google Scholar]
- 26.Bhunia A., Ramamoorthy A., Bhattacharjya S. Helical hairpin structure of a potent antimicrobial peptide MSI-594 in lipopolysaccharide micelles by NMR spectroscopy. Chem. Eur. J. 2009;15:2036–2040. doi: 10.1002/chem.200802635. [DOI] [PubMed] [Google Scholar]
- 27.Hilty C., Wider G., Fernández C., Wüthrich K. Membrane protein–lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. ChemBioChem. 2004;5:467–473. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- 28.Güntert P. Protein NMR Techniques. 2004. Automated NMR structure calculation with CYANA; pp. 353–378. [DOI] [PubMed] [Google Scholar]
- 29.Berjanskii M.V., Neal S., Wishart D.S. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34:W63–W69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 31.Hayter J., Penfold J. Determination of micelle structure and charge by neutron small-angle scattering. Colloid Polym. Sci. 1983;261:1022–1030. [Google Scholar]
- 32.Damberg P., Jarvet J., Gräslund A. 2001. [13] Micellar Systems as Solvents in Peptide and Protein Structure Determination. [DOI] [PubMed] [Google Scholar]
- 33.Wishart D.S., Bigam C.G., Holm A., Hodges R.S., Sykes B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 34.Wishart D.S., Sykes B.D., Richards F.M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 35.Tremblay M.L., Banks A.W., Rainey J.K. The predictive accuracy of secondary chemical shifts is more affected by protein secondary structure than solvent environment. J. Biomol. NMR. 2010;46:257–270. doi: 10.1007/s10858-010-9400-5. [DOI] [PubMed] [Google Scholar]
- 36.Kwan A.H., Mobli M., Gooley P.R., King G.F., Mackay J.P. Macromolecular NMR spectroscopy for the non-spectroscopist. FEBS J. 2011;278:687–703. doi: 10.1111/j.1742-4658.2011.08004.x. [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Surya W., Claudine S., Torres J. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J. Biol. Chem. 2014;289:12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esposito V., Das R., Melacini G. Mapping polypeptide self-recognition through 1H off-resonance relaxation. J. Am. Chem. Soc. 2005;127:9358–9359. doi: 10.1021/ja051714i. [DOI] [PubMed] [Google Scholar]
- 39.Bhunia A., Domadia P.N., Bhattacharjya S. Structural and thermodynamic analyses of the interaction between melittin and lipopolysaccharide. Biochim. Biophys. Acta Biomembr. 2007;1768:3282–3291. doi: 10.1016/j.bbamem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Chen R.F., Knutson J.R. Mechanism of fluorescence concentration quenching of carboxyfluorescein in liposomes: energy transfer to nonfluorescent dimers. Anal. Biochem. 1988;172:61–77. doi: 10.1016/0003-2697(88)90412-5. [DOI] [PubMed] [Google Scholar]
- 41.Ambroggio E.E., Separovic F., Bowie J.H., Fidelio G.D., Bagatolli L.A. Direct visualization of membrane leakage induced by the antibiotic peptides: maculatin, citropin, and aurein. Biophys. J. 2005;89:1874–1881. doi: 10.1529/biophysj.105.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leavitt S., Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr. Opin. Struct. Biol. 2001;11:560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 43.Freire E., Mayorga O.L., Straume M. Isothermal titration calorimetry. Anal. Chem. 1990;62:950A–959A. [Google Scholar]
- 44.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.
Supplementary figures