Abstract
The pro‐apoptotic properties of severe acute respiratory syndrome coronavirus (SARS‐CoV) structural proteins were studied in vitro. By monitoring apoptosis indicators including chromatin condensation, cellular DNA fragmentation and cell membrane asymmetry, we demonstrated that the adenovirus‐mediated over‐expression of SARS‐CoV spike (S) protein and its C‐terminal domain (S2) induce apoptosis in Vero E6 cells in a time‐ and dosage‐dependent manner, whereas the expression of its N‐terminal domain (S1) and other structural proteins, including envelope (E), membrane (M) and nucleocapsid (N) protein do not. These findings suggest a possible role of S and S2 protein in SARS‐CoV induced apoptosis and the molecular pathogenesis of SARS.
Keywords: Apoptosis, Virus–cell interaction, Severe acute respiratory syndrome coronavirus, Spike protein
1. Introduction
Severe acute respiratory syndrome‐coronavirus (SARS‐CoV) was identified as the causative agent of SARS early in 2003 [1]. Fever, dyspnea [1], lymphopenia, neutropenia [2, 3] and lower tract respiratory infection [1] were commonly found in infected individuals. Comparative genomic analysis revealed that SARS‐CoV is a novel member of the viral family coronaviradae, with an RNA genome of 29.7 kbp [4, 5, 6]. At least five viral structural proteins (VSPs), namely the spike (S), envelope (E), membrane (M) and nucleocapsid (N) protein, together with the newly identified ORF3a [7, 33], were encoded from the genome [9, 10]. Among these proteins, expression of S, M and N are necessary and sufficient for pseudovirus assembly mimicking those found in SARS‐CoV infected cells [11, 12].
Accumulated evidences have demonstrated that survival of viruses depends on the successful modulation of apoptosis initiated either by the hosts or the viruses themselves [13, 14, 15, 16]. Several studies have associated apoptosis with the pathogenesis of coronaviruses [17, 18, 19, 20, 21]. Previous reports suggested that over‐expression of certain coronaviral proteins could induce apoptosis in vitro [22, 23]. For SARS‐CoV, clinical symptoms, such as depletion of hepatocytes and T lymphocytes, i.e., lymphopenia, were suggested to be related to apoptosis [24, 25, 26]. It was also demonstrated that in vitro replication of SARS‐CoV induces apoptosis [27, 28, 29, 30, 31]. Recently, the ORF3a and the accessory protein 7a, but not the N, M and E protein of SARS‐CoV, was demonstrated to induce apoptosis in Vero E6 cells [8, 32]. In contrast, it is reported that the E and N protein of SARS‐CoV induces apoptosis in Jurkat T and COS‐1 cells, respectively, under serum depletion conditions [34, 35]. It was also reported that baculovirus‐mediated expression of the N‐terminal (S1) but not the C‐terminal (S2) domain of the S protein of SARS‐CoV triggers the cell survival‐related AP‐1 signaling pathway in lung cells [36]. Nevertheless, the possible role(s) of the SARS‐CoV VSPs in the virus‐induced apoptosis is largely unknown. In this study, we demonstrated a possible role of SARS‐CoV S protein in virus‐induced apoptosis using recombinant adenovirus (rAd)‐mediated expression system. The apoptotic properties of S, S1 and S2 protein, as well as other VSPs, including E, M and N protein, were investigated in Vero E6 cells.
2. Materials and methods
2.1. Cell culture
HEK293‐derived AD‐293 cells (Stratagene) were maintained in Dulbecco's modified eagle medium (DMEM; Gibco‐BRL), supplemented with 10% heat‐inactivated fetal bovine serum (FBS; Gibco‐BRL) and 1% antibiotics–antimycotic (Gibco‐BRL) at 37 °C and 5% CO2. Vero E6 cells were maintained in minimum essential medium with eagle's salts (EMEM; Gibco‐BRL) supplemented with 10% FBS and 1% antibiotics–antimycotic.
2.2. Generation of recombinant adenoviral virus
Cloning of the SARS‐CoV VSPs from viral cDNA, including S, S1 and S2, as well as three other structural genes – E, M and N gene (Fig. 1 A), was described elsewhere [6, 37]. The cloned cDNA fragments were tagged at the carboxy‐terminal with a V5 epitope. The signal peptide of pig growth hormone (SPpGH) [38] was placed upstream of the coding sequences of S (18–1255), S1 (18–683) and also S2 (684–1255), so as to ensure a comparable post‐translational modifications for all the spike protein fragments used in the study. The transgenes were then subcloned into a modified bicistronic shuttle vector designated as pShuttle‐CMV‐GOI‐IRES‐eGFP, which is derived from the pShuttle vector of the AdEasy™ XL Adenoviral Vector System (Stratagene) and the plasmid pBMN‐I‐GFP (Dr. G.P. Nolan, Stanford University School of Medicine). The bicistronic expression cassette contains the gene of interest (GOI) and the enhanced green fluorescent protein (eGFP), which are driven by a CMV promotor and an internal ribosomal entry site (IRES), respectively (Fig. 1A). The recombinant adenovirus containing the VSPs (rAd‐VSPs) was then generated by incorporating the expression cassette into the pAdEasy‐1 vector (Stratagene) according to manufacturer's instructions (Stratagene). A control adenovirus (rAd‐Ctrl) with no transgene was also constructed. The rAds were propagated in AD‐293 cells and CsCl‐purified as described [39].
Figure 1.
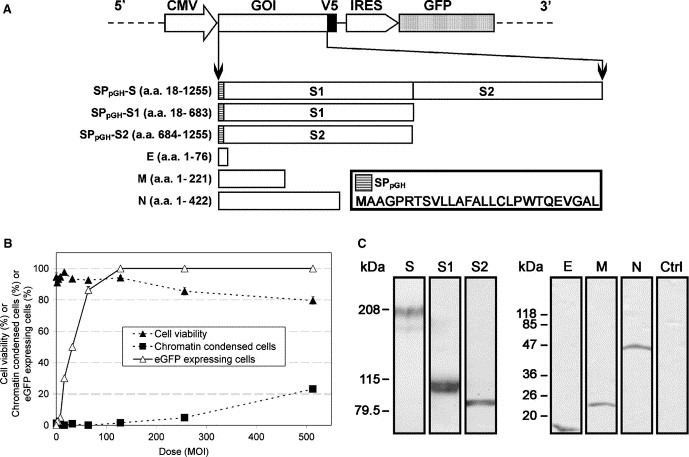
Construction of the rAd‐VSPs and the evaluation of rAd‐mediated transduction and expression. (A) A schematic diagram showing the organization of the bicistronic expression cassette of the modified shuttle vector (pShuttle‐CMV‐GOI‐IRES‐eGFP) (upper panel) used for rAd construction and the cloned coding regions of SARS‐CoV VSPs, including S, S1, S2, E, M and N (lower panel). The amino acids were numbered according to the corresponding proteins of SARS‐CoV strain HK‐39. The amino acid sequence of SPpGH is shown in the key and the detailed sequence information of the IRES‐eGFP fragment is available at “http://www.addgene.org/pgvec1?f=c&identifier=1736&cmd=findpl”. (B) Assessment of the optimal MOI for maximal transduction efficiency. The percentage of eGFP expressing cells was accessed by flow cytometer with at least 1 × 105 cells were counted for each sample. Each data point of the three assays were determined in triplicate and represents the average of three independent experiments ± standard error mean (S.E.M). (C) Expression of SARS‐CoV VSPs in Vero E6 cells. The expressed proteins were detected by using anti‐V5 antibody and the sizes of the molecular marker were shown on the left of each blot.
2.3. Immunoblotting
To access the expression of SARS‐CoV VSPs, Vero E6 cells were transduced with the corresponding rAds at multiplicity of infections (MOI) of 100. Cells were harvested 84 hours (h) post‐transduction (p.t.) and cell lysate was denatured and subjected to SDS–PAGE (S, S1 and S2 in 5% PAGE; other VSPs in 10% PAGE). To detect the expressed VSPs, Western blotting was carried out as described [37] using AP‐conjugated anti‐V5 antibody (Invitrogen).
2.4. Cell viability assay
Viability of cells transduced at indicated MOI was accessed by trypan blue exclusion assay. Cells were harvested and stained with 0.025% trypan blue dye (Invitrogen) for 10 min, and the percentage of dead cells (blue) was counted using haemocytometer.
2.5. Nuclear morphology
To detect chromatin condensation, cells transduced at indicated MOI were collected by low‐speed centrifugation and stained with Hoechst 33342 (Molecular Probes) phosphate buffered saline (PBS) solution (1:1000 v/v) at 37 °C for 5 min. At least 200 cells from three random fields of view were counted under fluorescence microscope.
2.6. DNA laddering assay
Cellular DNA fragmentation into characteristic ladders in apoptotic cells was assayed as described [40] with modifications. Briefly, cells were transduced with indicated rAds at MOI of 100. Both floating and adherent cells were collected at indicated time points p.t. and were subjected to low speed centrifugation. Cell pellets were then washed once in ice‐cold PBS and were subsequently resuspended in 80 μl of the same solution. Three hundred microliters of lysis buffer [10 mM Tris–HCl (pH 7.6), 10 mM EDTA, and 0.6% SDS] were added to the cell suspension, prior to the addition of 100 μl of 5 M NaCl. Lysates were then incubated at 4 °C overnight. Cell debris was pelleted by centrifugation and the supernatants were treated with 10 μl of 20 mg/ml proteinase K (Gibco‐BRL) at 37 °C for 1 h. Low molecular weight DNA was concentrated by ethanol precipitation overnight at −20 °C after phenol:chloroform extraction and subsequently analysed by 2% agarose gel electrophoresis.
2.7. Flow cytometry analysis of early apoptosis by 7‐AAD and Annexin V staining
The asymmetry of the plasma membrane of rAd‐S and‐S2 transduced cells at 84 h p.t. was monitored by dual staining with Annexin V‐PE and 7‐aminoactinomycin D (7‐AAD), which is a phosphatidylserine (PS)‐binding protein and an impermeable DNA‐labelling dye, respectively (Annexin V‐PE apoptosis detection Kit I, BD Pharmingen BioSciences). Data were acquired by Coulter Epics Elite Flow Cytometer and were analyzed with the WinMDI v2.81 software package (the Scripps Research Institute). Early apoptotic cells were recognized as PS‐externalized (Annexin V‐PE labeled) with intact cell membrane that resists 7‐AAD staining (lower‐right quadrant), which allows the exclusion of necrotic cells that are indistinguishable from the late apoptotic cells (upper‐right quadrant). At least 1 × 105 cells were counted for each data point.
2.8. Statistical analysis
A paired student's t‐test was used to compare the significance between specified groups, with P < 0.05, or 0.01 be defined as statistically significant.
3. Results
3.1. Adenovirus‐mediated expression of SARS‐CoV VSPs
To determine the rAd dosage needed for maximum transduction efficiency with minimal cytopathic effects, Vero E6 cells were transduced with rAd‐Ctrl at different MOIs and were examined at 84 h p.t. (Fig. 1B). At a MOI of 100, about 95% of cells were expressing eGFP, while no substantial apoptotic effect (i.e., less than 5% of non‐viable and chromatin condensed cells) was observed. Therefore, a MOI of 100 was chosen as the upper dose limit of the rAd transductions in this study. The successful and comparable transductions of all rAd‐VSPs were ensured in which at least 95% of cells showed the expression of eGFP and V5 epitope as detected by flow cytometer (data not shown), while the expression of SARS‐CoV VSPs was further confirmed by Western blots (Fig. 1C). It was noted that the well‐resolved double band pattern was observed for S and S1 at around 200 and 110 kDa, respectively, which mirrored previous reports that these two proteins are heavily glycosylated [41, 42, 43, 44, 45]. Among the bands of the S protein doublets, the one with lower molecular weight is at about 180 kDa, which is expected to be the glycosylated protein found in the endoplasmic reticulum, and the one with higher molecular weight, which is about 200 kDa, is expected to represent its more complexly glycosylated form that is found in Golgi bodies [46].
3.2. Transduction by rAd‐VSPs induces apoptosis in Vero E6 cells
We next compared the apoptotic effects induced by rAd‐VSPs transductions in terms of cell morphology, cell viability, chromatin condensation and cellular DNA fragmentation at 12 h intervals p.t. At 84 h p.t., cytopathic effects with abnormal cell morphology (i.e., shrinkage and detachment) (Fig. 2 A) and chromatin condensation (Fig. 2B) were observed in a substantial proportion of cells transduced by rAd‐S and‐S2, but neither in mock nor other rAds transduced cells. As shown in Fig. 2C, cells transduced by rAd‐S and rAd‐S2 collected at 84 h p.t. showed significantly (P < 0.01, ∗∗) stronger apoptotic effects in terms of both cell viability and chromatin condensation. Moreover, cellular DNA fragmentation into characteristic ladders was only observed in rAd‐S and ‐S2 transduced cells (Fig. 2D), in which increments of about 200 bp in size became weakly observable at 36 h p.t. Although random shearing of DNA was also observed in parallel, the intensity of the ladder was substantially increased at 84 h p.t. These observations indicate that both S and S2 protein are able to induce apoptosis in Vero E6 cells while the other VSPs do not.
Figure 2.
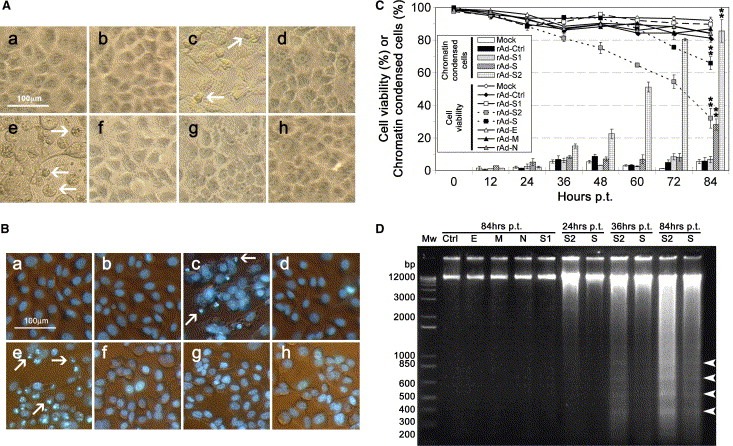
Transduction of Vero E6 cells by rAd‐S and rAd‐S2 induced cell death and apoptosis. (A) Photomicrographs showing the cell morphology of Vero E6 cells at 84 h after transduction with the indicated rAds (a–h, representing Mock, rAd‐Ctrl, ‐S, ‐S1, ‐S2, ‐E, ‐M and ‐N, respectively) at a MOI of 100. Cells undergoing cytoplasmic shrinkage are indicated in by arrows. (B) Phase contrast/Hoechst 33342 fluorescence merged photographs of the Vero E6 cells transduced as in (A). Cells undergoing chromatin condensation are indicated by arrows. (C) The apoptotic effects induced by the transduction of all rAds at a MOI of 100 were compared quantitatively in terms of cell viability (lines) and percentage of chromatin condensed cells (bars) using trypan blue exclusion assay and Hoechst 33342 staining, respectively, as in Fig 1B. (D) Cellular DNA fragmentation analysis. Characteristic DNA ladders with approximately 200 bp increments are indicated by arrowheads. Results shown in (A), (B) and (D) are representatives of three independent experiments.
3.3. Transduction of rAd‐S2 showed a stronger apoptotic effect than that of rAd‐S
To further confirm the observed apoptotic effect of the S proteins, Vero E6 cells were transduced with rAd‐Ctrl, ‐S, ‐S1 and ‐S2 at different MOIs and the percentage of apoptotic cells at 84 h p.t. were evaluated by chromatin condensation and PS‐externalization using fluorescent microscopy and flow cytometry, respectively. As shown in Fig. 3 A, the percentage of chromatin condensed cells induced by either rAd‐S or ‐S2 transduction at all indicated MOIs was significantly higher than that of the others (P < 0.05, ∗ or P < 0.01, ∗∗) in a dosage‐dependent manner. Moreover, at MOIs of 50 and 100, the percentage of chromatin condensed cells induced by rAd‐S2 transduction was significantly higher than those induced by rAd‐S transduction (P < 0.01, #). A similar phenomenon was observed when the cell membrane asymmetry of cells were examined (Fig. 3B), in which the percentage of early apoptotic cells in rAd‐S and ‐S2 transduction was at least 2 times higher than that of the controls at MOIs of 50 and 100. In summary, the above data strongly suggest that rAd‐mediated over‐expression of S and S2 protein induces apoptosis in Vero E6 cells, of which rAd‐S2 induced substantially stronger apoptosis than rAd‐S under the condition tested.
Figure 3.
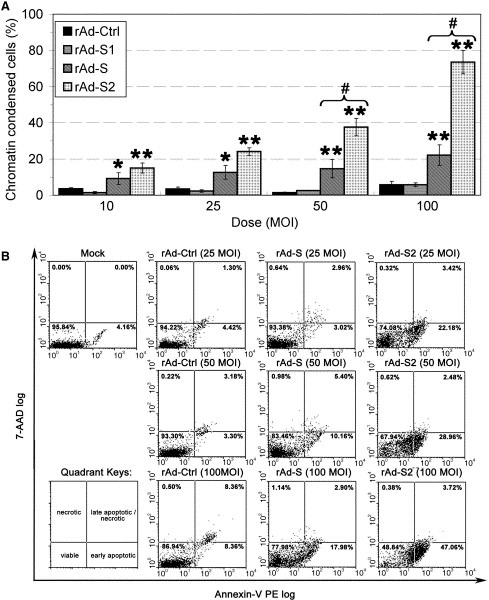
Transduction of rAd‐S2 showed a stronger apoptotic effect than that of rAd‐S in Vero E6 cells. (A) Dosage‐dependence of the apoptotic effect induced by rAd‐S and ‐S2 in terms of chromatin condensation. Cells were transduced by rAds at indicated MOIs. The percentage of apoptotic cells was determined by Hoechst 33342 staining at 84 h p.t as in Fig. 1B. (B) Early apoptosis of Vero E6 cells transduced by rAd‐S and ‐S2. The asymmetry of the plasma membrane of rAd‐S and ‐S2 transduced cells at 84 h p.t. was monitored by double staining with the Annexin V and 7‐AAD detected via flow cytometry. Quadrant keys are showed at the lower‐left corner. The percentage of cells in each quadrant is showed at the top of each dot‐plot. Results shown in (B) are representative of three independent experiments.
4. Discussion
Infection of SARS‐CoV in Vero E6 cells induces extensive apoptosis through a caspase‐3 and p38 MAPK dependent pathway [27, 28, 30, 31]. Using rAd‐mediated expression system, we assessed the apoptotic effect of the major structural proteins of SARS‐CoV, including S, S1, S2, E, M and N protein. Typical features of apoptosis such as cell rounding, shrinkage, nuclear condensation, DNA fragmentation and PE‐externalization were observed only in cells transduced with rAd carrying S and S2, but not S1, nor other structural proteins studied. These data suggest that the over‐expression of SARS‐CoV S and S2 could induce apoptosis. The present findings seem to be unique within the coronavirus family. In MHV [22] and IBV [23], overexpression of S protein did not induce observable apoptosis in vitro. On the other hand, the observed in vitro apoptotic effects of the VSPs of other coronaviruses, such as the E protein of MHV [22] and the N protein of TGEV [20], were, however, not observed when we overexpressed the SARS‐CoV homologues in Vero E6 cells. Interestingly, the E and N protein of SARS‐CoV has been reported to induce apoptosis in Jurkat T and COS‐1 cells, respectively, under serum depletion conditions [34, 35], which were not observed under the conditions tested in the current and a previously study [33]. Recently, over‐expression of two newly identified viral proteins of SARS‐CoV, ORF3a and 7a, were shown to induce apoptosis in Vero E6 cells as well, which is associated with caspase‐8 and ‐3 activity, respectively [32, 33]. Since the expression level of these viral proteins in SARS‐CoV infected cells has not been clearly demonstrated, their pro‐apoptotic properties may not be the only contributing factor in SARS‐CoV induced apoptosis. In contrast, the S protein is one of the major viral proteins in SARS‐CoV infected cells apart from N protein [47]. In SARS‐CoV infected Vero E6 cells, cleavage of the S protein into fragments was suggested in previous studies [41, 43], which includes a form that resembles the S2 protein in this study, and the inhibition of such protein processing completely abrogated the virus‐induced cytopathic effects in vitro, suggesting the potential roles of S and S2 in SARS‐CoV induced apoptosis. Although the activation of mitochondrial apoptotic pathway, caspase cascade, and the p38 MAPK‐dependent pathway are reported in several in vitro models of SARS‐CoV induced apoptosis [27, 28, 30, 31], the viral component(s) responsible for these observations remains unclear. Ren and co‐workers demonstrated that the addition of inactivated SARS‐CoV viral particles to Vero E6 cells is unable to induce apoptosis, implicating that expression of viral genes is indispensable for the viral‐induced apoptosis in vitro. Regarding these findings, the pro‐apoptotic properties of S and S2 in this report and the comparative study between the apoptotic pathways initiated by expression of individual viral genes and viral infection would certainly provide important clues in dissecting the molecular components responsible for the SARS‐CoV induced apoptosis.
The demonstrated roles of SARS‐CoV S protein in viral entry and elicitation of neutralizing immune responses make it an attractive target for antiviral therapies [10]. In this regard, investigations on the molecular basis of the S protein induced apoptosis, which are ongoing in our laboratory, together with the findings in this study, are expected to provide important insights for the rational design of anti‐viral therapies, and to the understanding of the molecular pathogenesis of SARS‐CoV infection.
Acknowledgements
This work was supported by HWF CR/3/6/3921/03 research grant funded by the Hong Kong government. We thank the colleagues in the Department of Zoology, University of Hong Kong that provided excellent technical assistances and insightful discussions; Laura Burleigh and Françoise Bachelerie (Unité d'Immunologie Virale, Institut Pasteur, Paris, France) for critical reading of this manuscript.
Chow Ken Y.C.,Yeung Yin Shan,Hon Chung Chau,Zeng Fanya,Law Ka Man and Leung Frederick C.C.(2005), Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells, FEBS Letters, 579, doi: 10.1016/j.febslet.2005.10.065
References
- 1. Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y., Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet, 361, (2003), 1319– 1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wong R.S., Wu A., To K.F., Lee N., Lam C.W., Wong C.K., Chan P.K., Ng M.H., Yu L.M., Hui D.S., Tam J.S., Cheng G., Sung J.J., Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. Bmj, 326, (2003), 1358– 1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang M., Li C.K., Li K., Hon K.L., Ng M.H., Chan P.K., Fok T.F., Hematological findings in SARS patients and possible mechanisms (review). Int. J. Mol. Med., 14, (2004), 311– 315. [PubMed] [Google Scholar]
- 4. Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L., The Genome sequence of the SARS-associated coronavirus. Science, 300, (2003), 1399– 1404. [DOI] [PubMed] [Google Scholar]
- 5. Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J., Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science, 300, (2003), 1394– 1399. [DOI] [PubMed] [Google Scholar]
- 6. Zeng F.Y., Chan C.W., Chan M.N., Chen J.D., Chow K.Y., Hon C.C., Hui K.H., Li J., Li V.Y., Wang C.Y., Wang P.Y., Guan Y., Zheng B., Poon L.L., Chan K.H., Yuen K.Y., Peiris J.S., Leung F.C., The complete genome sequence of severe acute respiratory syndrome coronavirus strain HKU-39849 (HK-39). Exp. Biol. Med. (Maywood), 228, (2003), 866– 873. [DOI] [PubMed] [Google Scholar]
- 7. Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X., Lim S.G., Hong W., Tan Y.J., The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun., 330, (2005), 286– 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan Y.J., The severe acute respiratory syndrome (SARS)-coronavirus 3a protein may function as a modulator of the trafficking properties of the spike protein. Virol. J., 2, (2005), 5– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J., Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol., 84, (2003), 2305– 2315. [DOI] [PubMed] [Google Scholar]
- 10. Chow K.Y., Hon C.C., Hui R.K., Wong R.T., Yip C.W., Zeng F., Leung F.C., Molecular advances in severe acute respiratory syndrome-associated coronavirus (SARS-CoV). Genom. Proteom. Bioinform., 1, (2003), 247– 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mortola E., Roy P., Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett., 576, (2004), 174– 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang Y., Yang Z.Y., Kong W.P., Nabel G.J., Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol., 78, (2004), 12557– 12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benedict C.A., Norris P.S., Ware C.F., To kill or be killed: viral evasion of apoptosis. Nat. Immunol., 3, (2002), 1013– 1018. [DOI] [PubMed] [Google Scholar]
- 14. Hardwick J.M., Apoptosis in viral pathogenesis. Cell Death Differ., 8, (2001), 109– 110. [DOI] [PubMed] [Google Scholar]
- 15. Koyama A.H., Adachi A., Irie H., Physiological significance of apoptosis during animal virus infection. Int. Rev. Immunol., 22, (2003), 341– 359. [DOI] [PubMed] [Google Scholar]
- 16. O'Brien V., Viruses and apoptosis. J. Gen. Virol., 79, Pt 8 (1998), 1833– 1845. [DOI] [PubMed] [Google Scholar]
- 17. Chen C.J., Makino S., Murine coronavirus-induced apoptosis in 17Cl-1 cells involves a mitochondria-mediated pathway and its downstream caspase-8 activation and bid cleavage. Virology, 302, (2002), 321– 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collins A.R., In vitro detection of apoptosis in monocytes/macrophages infected with human coronavirus. Clin. Diagn. Lab. Immunol., 9, (2002), 1392– 1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haagmans B.L., Egberink H.F., Horzinek M.C., Apoptosis and T-cell depletion during feline infectious peritonitis. J. Virol., 70, (1996), 8977– 8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eleouet J.F., Slee E.A., Saurini F., Castagne N., Poncet D., Garrido C., Solary E., Martin S.J., The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J. Virol., 74, (2000), 3975– 3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sirinarumitr T., Kluge J.P., Paul P.S., Transmissible gastroenteritis virus induced apoptosis in swine testes cell cultures. Arch. Virol., 143, (1998), 2471– 2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. An S., Chen C.J., Yu X., Leibowitz J.L., Makino S., Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol., 73, (1999), 7853– 7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C., Xu H.Y., Liu D.X., Induction of caspase-dependent apoptosis in cultured cells by the avian coronavirus infectious bronchitis virus. J. Virol., 75, (2001), 6402– 6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chau T.N., Lee K.C., Yao H., Tsang T.Y., Chow T.C., Yeung Y.C., Choi K.W., Tso Y.K., Lau T., Lai S.T., Lai C.L., SARS-associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology, 39, (2004), 302– 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Donnell R., Tasker R.C., Roe M.F., SARS: understanding the coronavirus: apoptosis may explain lymphopenia of SARS. BMJ, 327, (2003), 620– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panesar N.S., Lymphopenia in SARS. Lancet, 361, (2003), 1985– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizutani T., Fukushi S., Murakami M., Hirano T., Saijo M., Kurane I., Morikawa S., Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells. FEBS Lett., 577, (2004), 187– 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S., Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun., 319, (2004), 1228– 1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K., Zheng C., Gao G.F., SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol., 73, (2004), 323– 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bordi, L., Castilletti, C., Falasca, L., Ciccosanti, F., Calcaterra, S., Rozera, G., Di Caro, A., Zaniratti, S., Rinaldi, A., Ippolito, G., Piacentini, M., Capobianchi, M.R. in press. Bcl-2 inhibits the caspase-dependent apoptosis induced by SARS-CoV without affecting virus replication kinetics. Arch. Virol. (in press). [DOI] [PMC free article] [PubMed]
- 31. Ren L., Yang R., Guo L., Qu J., Wang J., Hung T., Apoptosis induced by the SARS-associated coronavirus in Vero cells is replication-dependent and involves caspase. DNA Cell Biol., 24, (2005), 496– 502. [DOI] [PubMed] [Google Scholar]
- 32. Law P.T., Wong C.H., Au T.C., Chuck C.P., Kong S.K., Chan P.K., To K.F., Lo A.W., Chan J.Y., Suen Y.K., Chan H.Y., Fung K.P., Waye M.M., Sung J.J., Lo Y.M., Tsui S.K., The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol., 86, (2005), 1921– 1930. [DOI] [PubMed] [Google Scholar]
- 33. Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G., Hong W., Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol., 78, (2004), 14043– 14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B., Lu H., Brendese J., Yang F., Wang H., Yang X.F., Bcl-xL inhibits T cell apotosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J., 392, Pt 1 (2005), 135– 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K., The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J., 383, (2004), 13– 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang Y.J., Liu C.Y., Chiang B.L., Chao Y.C., Chen C.C., Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J. Immunol., 173, (2004), 7602– 7614. [DOI] [PubMed] [Google Scholar]
- 37. Zeng F., Chow K.Y., Hon C.C., Law K.M., Yip C.W., Chan K.H., Peiris J.S., Leung F.C., Characterization of humoral responses in mice immunized with plasmid DNAs encoding SARS-CoV spike gene fragments. Biochem. Biophys. Res. Commun., 315, (2004), 1134– 1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kato Y., Shimokawa N., Kato T., Hirai T., Yoshihama K., Kawai H., Hattori M., Ezashi T., Shimogori Y., Wakabayashi K., Porcine growth hormone: molecular cloning of cDNA and expression in bacterial and mammalian cells. Biochim. Biophys. Acta., 1048, (1990), 290– 293. [DOI] [PubMed] [Google Scholar]
- 39. Tollefson A.E., Hermiston T.W., Wold W.S.M., Wold W.S.M. Adenovirus Methods and Protocols (1999), Humana Press ; Totowa, NJ: 1– 9. [Google Scholar]
- 40. Zhirnov O.P., Konakova T.E., Wolff T., Klenk H.D., NS1 protein of influenza A virus down-regulates apoptosis. J. Virol., 76, (2002), 1617– 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bergeron E., Vincent M.J., Wickham L., Hamelin J., Basak A., Nichol S.T., Chretien M., Seidah N.G., Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem. Biophys. Res. Commun., 326, (2005), 554– 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B., Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA, 101, (2004), 6641– 6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu X.D., Shang B., Yang R.F., Yu H., Ma Z.H., Shen X., Ji Y.Y., Lin Y., Wu Y.D., Lin G.M., Tian L., Gan X.Q., Yang S., Jiang W.H., Dai E.H., Wang X.Y., Jiang H.L., Xie Y.H., Zhu X.L., Pei G., Li L., Wu J.R., Sun B., The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res., 14, (2004), 400– 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S., The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun., 312, (2003), 1159– 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M., Murphy B.R., Subbarao K., Collins P.L., Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet, 363, (2004), 2122– 2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song H.C., Seo M.Y., Stadler K., Yoo B.J., Choo Q.L., Coates S.R., Uematsu Y., Harada T., Greer C.E., Polo J.M., Pileri P., Eickmann M., Rappuoli R., Abrignani S., Houghton M., Han J.H., Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol., 78, (2004), 10328– 10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang L.R., Chiu C.M., Yeh S.H., Huang W.H., Hsueh P.R., Yang W.Z., Yang J.Y., Su I.J., Chang S.C., Chen P.J., Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA. J. Med. Virol., 73, (2004), 338– 346. [DOI] [PMC free article] [PubMed] [Google Scholar]


