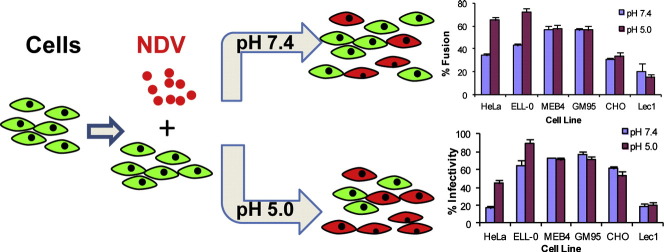
Abstract
Most paramyxoviruses enter the cell by direct fusion of the viral envelope with the plasma membrane. Our previous studies have shown the colocalization of Newcastle Disease Virus (NDV) with the early endosome marker EEA1 and the inhibition of NDV fusion by the caveolin-phosphorylating drug phorbol 12-myristate 13-acetate (PMA) prompted us to propose that NDV enters the cells via endocytosis. Here we show that the virus-cell fusion and cell-cell fusion promoted by NDV-F are increased by about 30% after brief exposure to low pH in HeLa and ELL-0 cells but not in NDV receptor- deficient cell lines such as GM95 or Lec1. After a brief low-pH exposure, the percentage of NDV fusion at 29 °C was similar to that at 37 °C without acid-pH stimulation, meaning that acid pH would decrease the energetic barrier to enhance fusion. Furthermore, preincubation of cells with the protein kinase C inhibitor bisindolylmaleimide led to the inhibition of about 30% of NDV infectivity, suggesting that a population of virus enters cells through receptor-mediated endocytosis. Moreover, the involvement of the GTPase dynamin in NDV entry is shown as its specific inhibitor, dynasore, also impaired NDV fusion and infectivity. Optimal infection of the host cells was significantly affected by drugs that inhibit endosomal acidification such as concanamycin A, monensin and chloroquine. These results support our hypothesis that entry of NDV into ELL-0 and HeLa cells occurs through the plasma membrane as well as by dynamin- low pH- and receptor- dependent endocytosis.
Abbreviations: BIM, bisindolylmaleimide; moi, multiplicity of infection; R18, octadecylrhodamine B chloride; RBCs, red blood cells
Keywords: Viral entry, Endocytosis, Low-pH, NDV, Paramyxovirus
Graphical abstract
Highlights
-
•
A pulse of low-pH enhanced NDV fusion and infectivity in a cell-dependent manner.
-
•
NDV infectivity was impaired by a protein kinase C inhibitor.
-
•
A specific inhibitor of the GTPase dynamin impaired NDV fusion and infectivity.
-
•
Inhibition of endosomal acidification inhibited NDV fusion and infectivity.
-
•
NDV may enter by dynamin-acid- and receptor-dependent endocytosis.
1. Introduction
Newcastle Disease Virus (NDV), a prototype of paramyxovirus, is an avian enveloped RNA-negative strand virus that causes respiratory disease in domestic fowl, leading to huge economic losses in the poultry industry. The envelope of NDV contains two associated glycoproteins that mediate viral entry: the hemagglutinin-neuraminidase (HN) and fusion (F) proteins. HN is the receptor-binding protein that recognizes and binds to sialoglycoconjugates at the cell surface [1] and also has receptor-cleaving (sialidase) activity [2]. Based on crystallographic studies of the HN of NDV, two sialic acid-binding sites have been described [3], [4], site II being located at the dimer interface and activated after engagement of site I to its receptor [4], [5]. Site I exhibits both receptor-binding and sialidase activities while site II only has receptor binding activity [4]. Moreover, the triggering of F protein requires the presence of its homotypic attachment protein through its fusion promotion activity.
Enveloped viruses enter the cell through two main mechanisms: by direct fusion of the viral envelope with the plasma membrane or by endocytosis. The use of the cellular endocytic machinery is commonly associated with low-pH-dependent activation of viral fusion proteins such as in influenza virus or in vesicular stomatitis virus [6].
Additionally, the fusion protein of Ebola virus is activated by acidic-pH-dependent proteases inside endosomes [7]. Moreover, recent studies suggest that viruses with pH-independent fusion proteins may use endocytosis for entry (revised in [8]). Different families of viruses may use several different endocytic routes [9], [10], [11], the major pathway being the clathrin-mediated endocytosis used by viruses such as Semliki Forest and Sindbis alpha virus [12] or Hantaan virus [13]. Recent reports indicate that HIV also uses this route productively [14]. Caveola-mediated endocytosis is the second best characterized pathway used by SV40 and Ebola viruses to enter cells [15], [16], [17], [18]. Although most viruses have been described to enter the cell through a unique mechanism, there are increasing reports showing that many viruses are able to use alternative uptake pathways to infect different target cells or even simultaneous entry mechanisms in the same cell type ([8], [19] and references therein).
With the exception of the fusion protein of the human metapneumovirus (HMPV), which depends on low-pH [20], paramyxovirus fusion proteins are activated at neutral pH, which suggests that viral entry occurs by direct fusion at the plasma membrane [2]. Nevertheless, recent reports point to the notion that the pH requirement for fusion does not necessarily imply the location of membrane fusion (revised in [21], [22]). Some members of the paramyxoviruses such as Sendai, Nipah, human metapneumovirus and human respiratory syncytial virus (hRSV) [23], [24], [25], [26] utilize the endocytic machinery for entry. Furthermore, as assessed by a R18 dequenching assay, our previous studies revealed the enhancement of NDV fusion with Cos-7 at acidic pH [27] and the colocalization of NDV with the early endosome marker EEA1 [28]. These findings prompted us to suggest that the entry of NDV would occur through caveola-mediated endocytosis as an additional mechanism to direct fusion with the plasma membrane [28]. To gain further insight into the molecular mechanisms of NDV entry, the present study focuses on the characterization of the NDV entry mechanism through the use of low-pH treatment of several cell lines that differ in the expression of sialoglycoconjugates at their surface and by analyzing the effect of different substances that either interfere with endocytosis or endosomal acidification on NDV interaction with cells. NDV entry showed to be sensitive to endosomal acid pH, and dynamin and protein kinase C inhibitors blocked NDV fusion and infectivity. Our data support the idea that NDV may penetrate HeLa and ELL-0 cells both by direct fusion at the cell surface and through receptor-mediated and dynamin-dependent endocytosis, at least in these cell lines.
2. Materials and methods
2.1. Cell lines and viruses
East Lansing Line (ELL-0) avian fibroblasts and HeLa cells were obtained from the American Type Culture Collection (ATCC); MEB4 and GM95 were obtained from Riken BRC Cell Bank (Tsukuba, Japan); ELL-0, HeLa, MEB4 and GM95 cells were maintained in Dulbecco's modified Eagle's medium (DMEM); Chinese hamster ovary (CHO) and mutant Lec1 were also purchased from ATCC and maintained in DMEM:F12 and AlphaMEM media respectively. All media were supplemented with L-GlutaMax (580 mg l− 1, Invitrogen), penicillin–streptomycin (100 U ml− 1–100 μg ml− 1) and 10% heat-inactivated fetal bovine serum (complete medium). The lentogenic “Clone 30” strain of NDV was obtained from Intervet Laboratories (Salamanca, Spain). The virus was grown and purified mainly as described previously [28]. For inectivity assays, a recombinant NDV derived from the Hitchner B1 lentogenic strain (rNDV-F3aa-mRFP [29]), which expresses a monomeric red-fluorescent protein [30] and kindly provided by Dr. Adolfo García-Sastre, was used.
2.2. Reagents and antibodies
Bisindolylmaleimide I (BIM) was from Calbiochem; dynasore, concanamycin A, monensin, chloroquine, protease inhibitor cocktail and proteinase K were from Sigma Chemical Co; octadecylrhodamine B chloride (R18), Hoechst 33258, calcein, lipofectamine and Alexa Fluor 488 donkey anti-mouse antibody were from Molecular Probes. Monoclonal anti-NP and polyclonal anti-NDV antibodies were generous gifts from Dr. García-Sastre (Emerging Pathogens Institute, Mount Sinai School of Medicine, New York, USA); monoclonal anti-HN 14f and 1b antibodies were kindly provided by Dr. M. Iorio (University of Massachusetts Medical School, Worcester, USA).
2.3. Virus titration
Virus titers were calculated in plaque-formation assays in Vero cells, as described [31]. Plaque numbers were counted under a light microscope and NDV infectivity was expressed as plaque-forming units (pfu) ml− 1.
2.4. Virus-binding assays
Monolayers of HeLa, ELL-0, MEB4, GM95, CHO and Lec1 cells on 35 mm plates were previously treated with the different inhibitors and then incubated with NDV at a moi of 1 for 1 h at 4 °C in the continuous presence of the inhibitor. Cells were washed with ice-cold PBS to remove unbound virus and then collected from the plates by incubation with EDTA (520 μM) at 37 °C. Cells were pelleted by centrifugation and fixed with 2.5% paraformaldehyde for 30 min at 4 °C. After centrifugation, cells were incubated with anti-HN 14f and 1b mAbs in blocking solution (dilution 1:100) for 1 h at 4 °C. Then, the cells were centrifuged and incubated with blocking solution containing anti-mouse Alexa Fluor 488-conjugated IgG (5 μg ml− 1) for 1 h at 4 °C. Cells were then pelleted by centrifugation and resuspended in a suitable volume of PBS for analysis in a FACScalibur flow cytometer (Becton Dickinson). At least 5 × 104 cells were analyzed for each sample. Mean fluorescence values were normalized to the control values. Data were analyzed with WinMDI 2.9 software.
2.5. NDV–cell fusion assays
Purified NDV was labeled with the fluorescent probe R18 essentially as described previously [32]. Cells plated in 24-well plates treated or untreated with the inhibitors were incubated for 30 min at 4 °C on ice with 2 μg of R18-NDV per plate. Then, cells were washed three times with cold PBS and incubated in complete medium containing Hoechst 33258 (10 μg ml− 1) for 1 h at 37 °C. They were fixed with 2% formaldehyde in PBS and the transfer of the rhodamine probe to cells was observed under an Olympus IX51 inverted fluorescence microscope. The percentage of fusion was calculated as the number of positive red-stained cells in 10 random fields with respect to the total number of cells in these areas of the well.
2.6. NDV infectivity assays
Monolayers of cells were infected for 1 h at room temperature with different dilutions of the recombinant NDV rNDV-F3aa-mRFP. After 24 h at 37 °C, the cells were observed under an Olympus IX51 inverted fluorescence microscope with a 10 × objective. The percentage of infectivity was calculated as the number of red-fluorescent cells out of the total number of cells in six random fields.
2.7. FACS infectivity assays
Monolayers of HeLa, ELL-0, MEB4, GM95, CHO and Lec1 cells on 35 mm plates were treated with the inhibitors and then incubated with 1 moi of NDV for 1 h at 37 °C in the presence of inhibitor. Cells were washed with ice-cold PBS to remove unbound virus and were collected from the plates by incubation with EDTA (520 μM) at 37 °C. Cells were pelleted by centrifugation and fixed with 2.5% paraformaldehyde for 30 min at 4 °C. Then, cells were blocked and permeabilized with PBS containing 1% BSA (blocking solution) and 0.1% TX100 for 20 min at 4 °C. After centrifugation, cells were incubated with anti-NP mAb in blocking solution (dilution 1:200) for 1 h at 4 °C. Then, they were centrifuged and incubated with blocking solution containing anti-mouse Alexa Fluor 488-conjugated IgG (5 μg ml− 1) for 1 h at 4 °C. Cells were then pelleted by centrifugation and resuspended in a suitable volume of PBS for analysis with a FACScalibur flow cytometer (Becton Dickinson). At least 5 × 104 cells were analyzed for each sample. Mean fluorescence values and the % of cells with fluorescence higher than the background were combined to quantify viral protein expression and were normalized to the control values. Mean fluorescence values were normalized to control values. Data were analyzed with WinMDI 2.9 software.
2.8. Syncytium assays
Monolayers of cells were infected with 1 moi of NDV for 1 h at 37 °C. At 7 h post-infection, viral F proteins were activated by digestion with acetyl trypsin [33]. Then, cells were incubated in complete medium overnight and stained with Giemsa or crystal violet for syncytium observation. Representative fields were captured with an inverted microscope (Olympus IX51). Quantification of syncytia was accomplished by measuring the area in pixels, referring to the total area of the field for three random fields. Areas were quantified using the analysis tool in Adobe Photoshop CS4.
2.9. Low-pH-induced fusion
Cells plated in 24-well plates were incubated for 30 min at 4 °C on ice with 3 μg of R18-NDV per plate. Then, virus–cell complexes were exposed briefly to either pH 7.4 or pH 5.0 buffer by treating them with a 3-min pulse of buffered PBS at 37 °C. After 1 h at 37 °C with regular medium at neutral pH, the percentage of fusion was calculated as the number of positive red-stained cells in 10 random fields with respect to the total number of cells in these areas of the well.
2.10. Plasmid constructs and transfection
Monolayers of HeLa cells grown in 24-well plates were cotransfected with 1 μg of pCAGGS encoding viral F protein and/or with HN or empty pCAGGS for a total amount of 2 μg of DNA per well. Transfection was performed using lipofectamine. At 24 h post-transfection, cells were washed and F protein was activated by digestion with acetyl trypsin, as described above. Following this, dye-transfer fusion or syncytium assays were performed.
2.11. Dye-transfer fusion assays
Human erythrocytes (RBCs) were double-labeled with calcein and octadecylrhodamine B chloride (R18). Briefly, RBCs at 10% hematocrit in PBS were incubated with 100 μg ml− 1 of the aqueous dye calcein for 1 h at 37 °C in the dark. After extensive washing, RBCs were collected by centrifugation and incubated with 25 μl (stock 2 mg ml− 1) of the lipid probe R18 for 30 min at room temperature in the dark, followed by several washing steps to remove unincorporated dye from labeled cells. Monolayers of HeLa cells in 24-well plates were transfected as described above. At 24 h post-transfection, F protein was activated as described above, and the cells were exposed briefly to either pH 7.4 or pH 5.0 buffer by treating them with a 5 min pulse of buffered PBS at 37 °C. Then, the media were removed and, without washing, cells were overlaid with dye-hematocrit per well in DMEM. The effector–target cell complexes were incubated at different temperatures for 1 h. Nuclei were stained by incubation with 10 μg ml− 1 of Hoechst dye. Cells then were washed several times with PBS to remove unbound RBCs, and dye transfer was visualized under an inverted fluorescence microscope at a magnification of 10 ×. The extent of fusion was calculated as the percentage of dye-labeled cells in the total number of nuclei.
2.12. Treatment of cells with chemical inhibitors
Confluent monolayers of cells in 24-well plates were pre-treated for 1.5 h for concanamycin A and monensin, 1 h for chloroquine, or 30 min for dynasore, with the inhibitor or vehicle control (control volume equal to the maximum volume of the drug) at the indicated concentrations (Table 1 ). For infectivity assays, infections were carried out such that on average 40% of the cells would be infected in the control wells. After 4 h at 37 °C in the presence of a drug or vehicle control (2 h for the dynasore experiments), the cells were washed and incubated in regular medium overnight at 37 °C. For the BIM experiments, cells were infected in the presence of the inhibitor for 1 h at room temperature and were then incubated 24 h at 37 °C in DMEM with the drug.
Table 1.
Drugs, effects and treatments of cells.
| Drug | Effect | Treatment of cells |
|---|---|---|
| Bisindolylmaleimide I (20 μM) | Protein kinase C inhibitor (receptor-mediated endocytosis) | i) Drug present during 1 h of infection ii) Drug present during1 h of infection and for 24 h post-infection iii) Addition of the drug after infection and present for 24 h post-infection |
| Dynasore (80 μM) | Dinamin-1 GTPase activity inhibitor (clathrin and caveolae mediated endocytosis) | Preincubation for 30 min; drug present during infection and for 2 h post-infection. |
| Concanamycin A (5 nM) | Vacuolar ATPase inhibitor | Preincubation for 1 h 30 min; drug present during infection and for 4 h post-infection. |
| Monensin (2.5 μM) | Ionophore | Preincubation for 1 h 30 min; drug present during infection and for 4 h post-infection. |
| Chloroquine (10 nM) | Lysosomotropic agent | i) Drug preincubated for 1 h and then removed ii) Preincubation for 1 h; drug present during infection and for 4 h post-infection. iii) Addition of the drug at 5 h post-infection and incubation for additional 6 h |
2.13. Proteinase protection assays
Proteinase K treatment of cells was carried out as previously described [34]. Virus was added to cell monolayers at a moi of 25, virus binding was performed at 4 °C for 1 h, and samples were incubated at 37 °C for different times to allow entry. Cells were treated with proteinase K (200 μg ml) in OptiMEM supplemented with 1 mM CaCl2 for 1 h at 4 °C. The protease-treated cells were pelleted and lysed in ice-cold lysis buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% NP-40) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and a cocktail of protease inhibitors. Viral proteins were detected by Western blotting using primary rabbit polyclonal anti-NDV antibodies.
2.14. Cell viability assays
Cells were detached and a small quantity of them (200 μl) were mixed with 300 μl Trypan blue 0.4% (p/v) in PBS and with 300 μl of PBS. These products were mixed for 5–10 min and then 20 μl was transferred to a counter chamber and the number of total cells and stained cells (dead cells) was counted. The percentage of viability was calculated as the number of viable cells out of total number of cells.
2.15. Statistics
The two-tailed unpaired Student's t-test was used to determine statistical significance between two groups. Probability values of p < 0.001 were considered extremely statistically significant, p < 0.01 extremely significant and p < 0.05 statistically significant. Statistical analyses were performed with the QuickCalcs program from GraphPad software.
3. Results and discussion
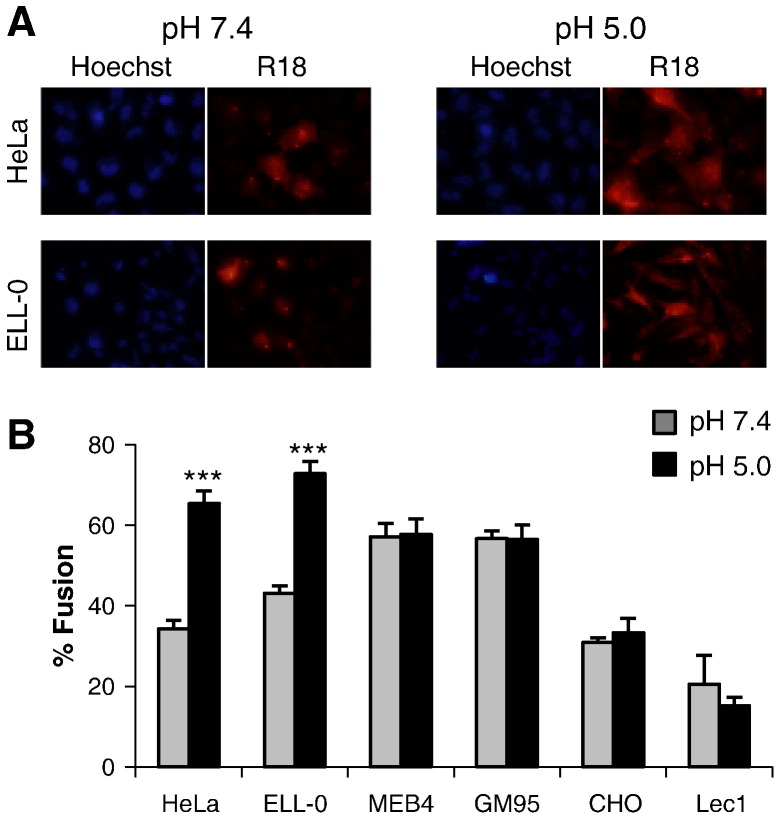
Most paramyxoviruses enter the host by fusion of their envelope with the plasma membrane at neutral pH. Nevertheless, we have previously reported the enhancement of NDV fusion with Cos-7 cells at acidic pH [27], [28]. To further investigate the role of acidic pH in NDV entry, we analyzed NDV fusion with different cell lines after a short pulse of acidic pH, as detailed in Materials and methods. The different target cell lines vary in their surface glycoconjugate expression, i.e., the NDV receptor: HeLa, which are human epithelial cells commonly used in studies of many viruses included NDV; ELL-0, a chicken fibroblast from the natural host of NDV; GM95, a glycosphingolipid-deficient cell line and its parental cell line MEB4 [35]; CHO and their mutant Lec1 cells, deficient in complex N-linked glycosylation but not in glycosphingolipids or O-glycoproteins [36]. To analyze virus–cell fusion, R18-NDV was allowed to bind to cells for 30 min at 4 °C, after which virus–cell complexes were exposed briefly to either pH 7.4 or pH 5.0 buffer by treatment with a 3 min pulse of buffered PBS. After 1 h at 37 °C with regular medium at neutral pH, the percentage of fusion was calculated as the number of positive red-stained cells from the total number of cells.
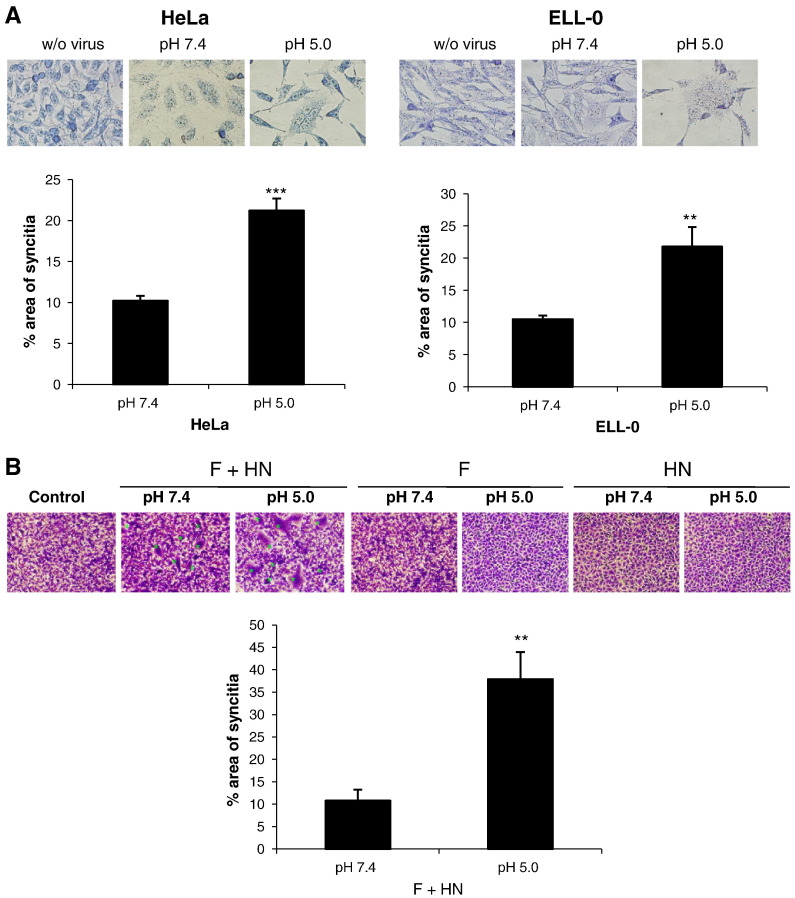
The data summarized in Fig. 1 show that low pH pulse led to an increase of about 30% of fusion in two of the cell lines, HeLa and ELL-0, suggesting that this enhancement might be cell type-specific, as we will discuss later. Next, we wished to determine whether syncytium formation was also stimulated by exposure to low pH in both virus-infected and HN/F transfected cells. HeLa and ELL-0 cells were infected with NDV at a moi of 1 for 1 h at 37 °C. Then, cells were treated with three pulses (3 min each) of pH 5.0- or pH 7.4-buffered PBS, with 1 h interval between each pulse. At 24 h post-infection syncytia were quantified (Fig. 2A). As expected, syncytium formation was enhanced about 2 times as compared with the control in both cell lines. These data correlate with the enhancement of viral infectivity observed after infection with the recombinant NDV rNDV-F3aa-mRFP (Fig. 3 ), as discussed later. Additionally, to determine whether HN protein was essential for fusion after acidic pH treatment, HeLa cells were transfected with F and/or HN vectors; at 24 h post-transfection, cells were treated with three low-pH pulses, as above. When the cells were cotransfected with both F and HN plasmids, syncytium formation was increased 3.5 times after low-pH exposure (Fig. 2B) in comparison with the controls; acidic pH treatment increased both the number and the size of the syncytia (micrograph in Fig. 2B). Nevertheless, NDV F protein did not promote the formation of syncytia in the absence of HN when treated with low pH (Fig. 2B).
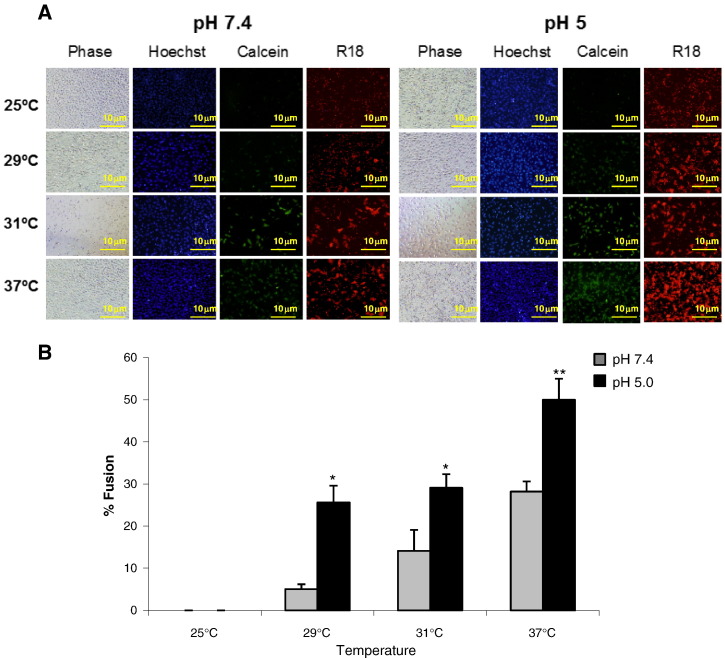
Fig. 1.
Effect of acidic pH on NDV fusion with different cell lines. R18-–cell complexes were exposed to a 3 min pulse at pH 7.4- or pH 5.0-buffered PBS at 37 °C. After 1 h at 37 °C with regular medium at neutral pH, fusion was assessed by the transfer of the R18 red dye to the cell membrane and was quantified as detailed in Materials and methods. (A) Microphotographs from a representative experiment. (B) Data are means ± SD of three independent experiments. Statistical analysis was performed using the paired t test: ***, p < 0.001, extremely significant.
Fig. 2.
Enhancement of NDV F-promoted cell–cell fusion at acidic pH. (A) Monolayers of HeLa and ELL-0 cells were infected with 1 moi of NDV for 1 h at 37 °C. Then, cells were treated with three pulses (3 min each) of pH 5.0- or pH 7.4-buffered PBS with 1 h interval between each pulse. At 7 h post-infection, viral F proteins were activated by digestion with acetyl trypsin and at 24 h post-infection, syncytia were stained with Giemsa and the occupied areas in the field were quantified with the Adobe Photoshop program, as detailed in Materials and methods. (B) HeLa cells were transfected with HN- and/or F-plasmid and the F0 precursor was activated with acetyl trypsin and immediately cells were treated with three pulses of pH 5.0- or pH 7.4-buffered PBS as in (A). At 48 h post-transfection, cells were fixed and stained with crystal violet for syncytium quantification as in (A). Control, mock-transfected cells. Data are means ± SD of three independent experiments. **, p < 0.01, highly statistically significant; ***, p < 0.001, extremely significant.
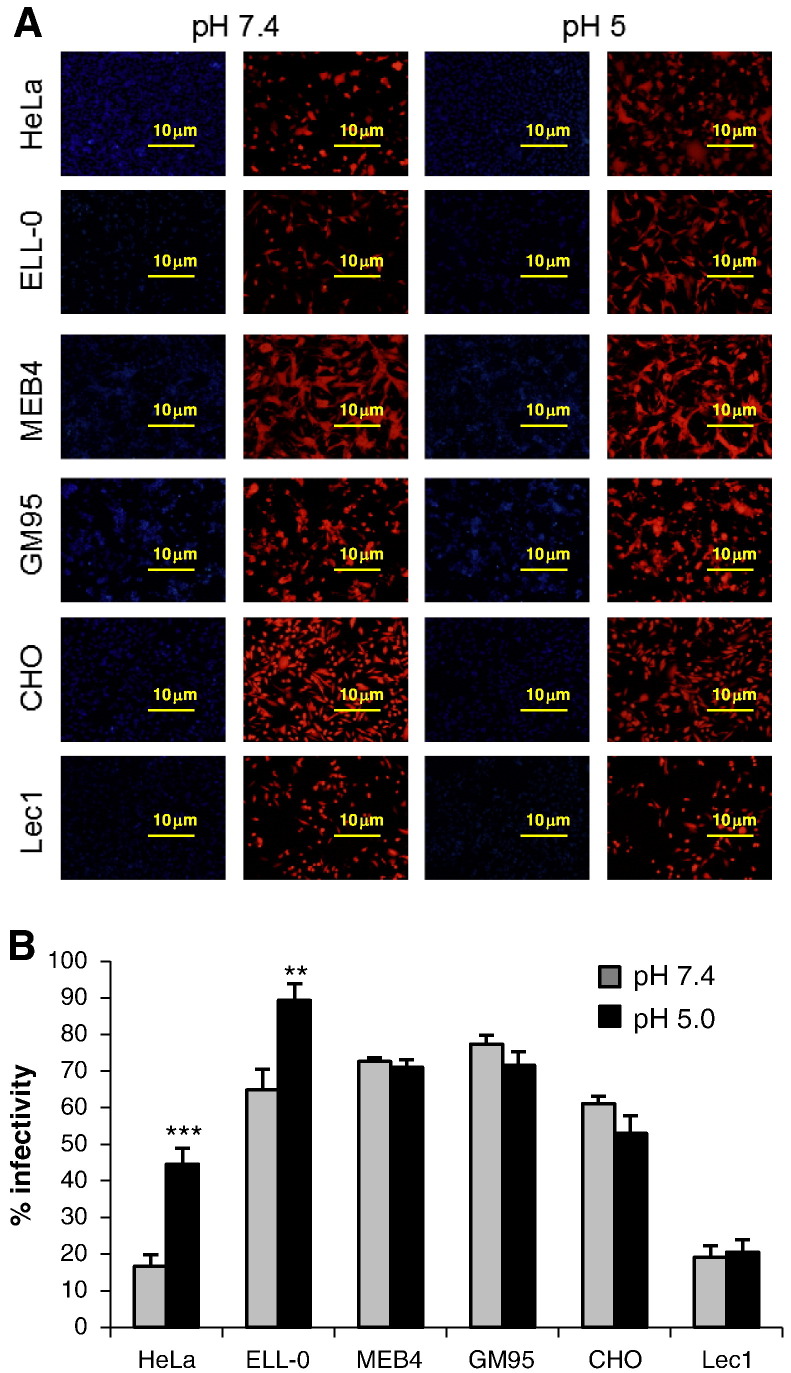
Fig. 3.
Effect of acidic pH on NDV infectivity. Monolayers of the different cell lines were infected with rNDV-F3aa-mRFP at a moi of 10 (except on HeLa cells, at a moi of 1) for 1 h at room temperature. Then, cells were treated with three pulses (3 min each) of pH 5.0- or pH 7.4-buffered PBS with 1 h interval between each pulse. Infectivity was analyzed at 24 h post-infection by calculating the percentage of red-fluorescent cells (infected cells) out of the total number of cells in three random fields. (A) Microphotographs from a representative experiment. (B) Data are means ± SD of three independent experiments. **, p < 0.01, highly statistically significant; ***, p < 0.001, extremely significant.
To analyze the effect of acidic pH exposure on NDV infectivity, ELL-0, HeLa, MEB4, GM95, CHO and Lec1 were infected with the recombinant NDV rNDV-F3aa-mRFP for 1 h at room temperature at a moi of 10, except in HeLa cells, which were infected at a moi of 1 because HeLa cells became fully infected at a moi of 10 (data not shown). Then, cells were treated with three pulses (3 min each) of pH 5.0- or pH 7.4-buffered PBS with a 1 h interval. As detailed in Materials and methods, infectivity was monitored at 24 h post-infection, calculating the percentage of the red-fluorescent infected cells from the total number of cells. The results are summarized in Fig. 3 and Supplementary Fig. S1. Similar to fusion (Fig. 1), the low-pH treatment induced an enhanced infectivity exclusively in ELL-0 and HeLa cells, again supporting the idea that there are differences in the entry mechanisms that depend on the cell line.
In the next series of experiments, we analyzed whether the acidic pH treatment was able to lower the temperature required for NDV F protein-induced fusion. In order to determine whether the possible effect was exerted differently in the fusion cascade, we used the dye-transfer-based fusion assay detailed in Materials and methods. HN- and F-transfected cells were allowed to fuse with RBCs doubly labeled with the red probe R18 in their membrane and the green probe calcein in the cytoplasm. After proteolytic activation of F protein, cells were treated with a pulse of acidic or neutral pH (control) for 5 min at 37 °C. Then, fluorescence-labeled erythrocytes were added and allowed to fuse for 1 h at 25 °C, 29 °C, 31 °C or 37 °C. As shown in Fig. 4 , fusion did not occur at 25 °C either at neutral pH or after low-pH treatment, although we did observe an increase in RBCs binding to transfected cells at acidic pH at this temperature (Fig. 4A, R18 panel, and Supplementary Fig. S2), which might contribute to the enhancement of fusion at fusion-permissive temperatures. Nevertheless, at the suboptimum temperatures of 29 °C and 31 °C, the extent of fusion after treatment at pH 5.0 was similar to that of fusion at 37 °C at neutral pH. Finally, as seen in Fig. 2, after low-pH treatment fusion was increased about 2-fold at 37 °C. In all cases, transfer of the two probes – R18 and calcein – was similar, indicating that the enhancement of fusion due to the pulse of acidic pH had a similar effect on hemifusion (transfer of R18 from the membrane of the erythrocytes to the membrane of the cell) and on complete fusion (transfer of calcein from the cytoplasm of the erythrocyte to the cytoplasm of the cell). As mentioned above, these data support the hypothesis that acid pH would decrease the energetic barrier needed for protein activation prior to the triggering of fusion, although this hypothetical decrease was not enough to allow fusion at 25 °C. Therefore, acidic pH might increase the number of F proteins that undergo the conformational changes leading to membrane fusion, even though the presence of HN protein would still be required.
Fig. 4.
Effect of acidic pH on the temperature dependence of NDV F-promoted cell–cell fusion. RBCs were double--NDV vectors. At 24 h post-transfection, F0 precursor was activated with acetyl trypsin and cells were treated with pH 5.0- or pH 7.4-buffered PBS for 5 min at 37 °C. Then, cells were incubated with labeled RBC (0.2% hematocrit) at different temperatures for 1 h. After washing unbound RBC, the extent of dye transfer was analyzed and quantified as the percentage of red- (R18) or green- (calcein) stained cells out of the total number of cells. Data concerning R18 transfer are shown and are similar to those of calcein transfer (not shown). (A) Microphotographs from a representative experiment. (B) Results are means ± SD of three independent experiments. Statistical analyses were performed using the paired t test: * p < 0.05 statistically significant: ** p < 0.01 very significant.
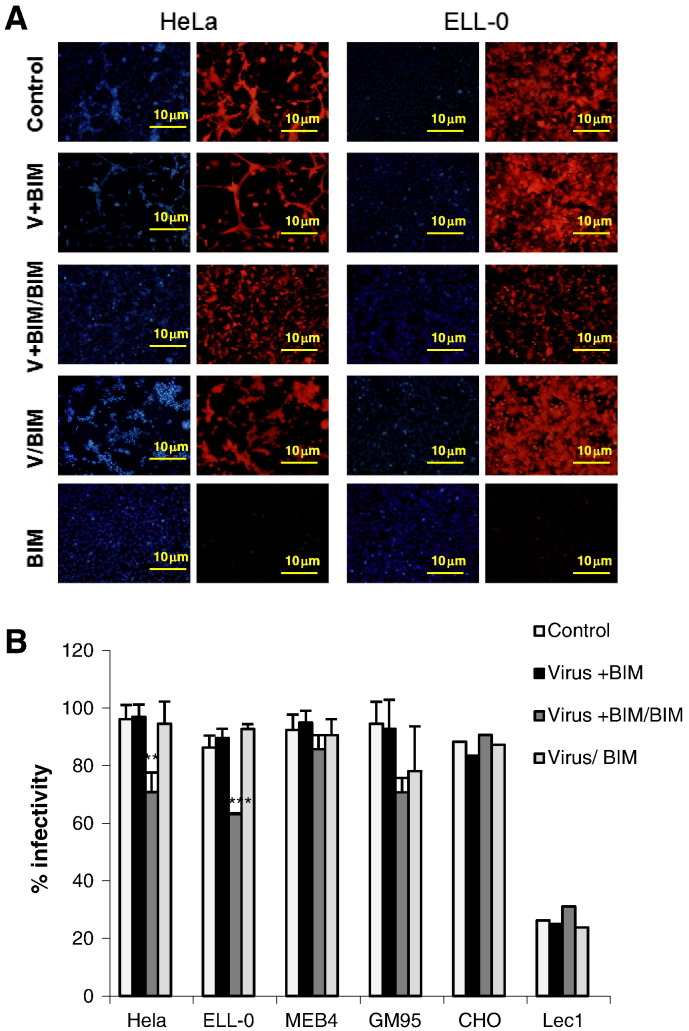
The enhancement of NDV fusion and infectivity by exposure to acidic pH reported above (Fig. 1, Fig. 2, Fig. 3) is in agreement with our previous results [27], [28] and strongly supports a role for low pH in NDV entry and the notion that a certain percentage of NDV may enter cells by endocytosis, at least in certain cell lines such as HeLa, ELL-0 and Cos-7. Since in the course of an infection a virus would encounter low pH after endocytosis and trafficking through the endocytic pathway, we wanted to further explore the possible endocytic entry of NDV studying the effect of different substances that interfere either with endocytosis or endosomal acidification (Table 1). First, we incubated the target cells in the presence of bisindolylmaleimide I (BIM), a highly selective, cell-permeable, and reversible protein kinase C (PKC) inhibitor [37], [38]. In most of the cells, cell viability as assayed by a Trypan blue exclusion method was higher than 90%, except in GM95, where a strong cytopathic effect of the drug was observed (about 60% of cell death, data not shown). We analyzed the effect of BIM preincubation on NDV infectivity under three different conditions: i) drug was present during infection and then withdrawn from the culture medium (Virus + BIM); ii) drug present for the duration of the assays, i.e., during infection and for 24 h post-infection (Virus + BIM/BIM); and iii) drug was added at 1 h post-infection (Virus/BIM). The data are summarized in Fig. 5 ; since BIM per se is a compound that exhibits red fluorescence, the background (Fig. 5A, BIM) was subtracted from the red fluorescence due to the mRFP expression occurring after rNDV-F3aa-mRFP infection. The inhibitory effect of BIM was not observed when the drug was added post-infection, meaning that it must act during an early stage of the infection. The data concerning inhibition (approximately 26% of inhibition as compared with the control) correlated well with the observed enhancement of viral fusion in both cell lines after the low-pH exposure shown in Fig. 3. The infectivity of NDV in GM95-treated cells was also reduced after BIM treatment, but the observed reduction could have been be due to the negative effect of the drug on cell viability. According to our results, NDV could penetrate in HeLa and ELL-0 cells through receptor-mediated endocytosis, since the inhibition of PKC reduced NDV infectivity by 30% in comparison with the control (Fig. 5). Similarly, the activity of PKC is essential for the entry of different non-enveloped [39] or enveloped viruses, such as influenza virus [38], alphavirus, rhabdovirus, poxvirus and herpes virus [40], which has been related to the entry through receptor-mediated endocytosis. Furthermore, we have previously suggested that caveolae/rafts could be a possible platform for NDV entry [28], [41] and caveolae seem to be the major cell surface locations for PKC [42], [43].
Fig. 5.
Effect of PKC inhibition on NDV infectivity. Monolayers of cells were treated with the PKC-inhibitor BIM at different times of the infectivity experiments. Cells were infected with rNDV-F3aa-mRFP at a moi of 10 (1 moi for HeLa cells) for 1 h at room temperature and infectivity was analyzed at 24 h post-infection by calculating the percentage of red-fluorescent cells (infected cells) out of the total number of cells in six random fields. Control, cells without the drug; Virus + BIM: cells were infected in the presence of the drug and the drug was withdrawn after infection; Virus + BIM/BIM: cells were infected in the presence of the drug that was also present for the duration of the assays; Virus/BIM: untreated cells were infected and the drug was added 1 h post-infection; BIM: non-infected cells in the presence of the drug. (A) Microphotographs from a representative experiment. (B) Data are means ± SD of three independent experiments. **, p < 0.01, highly statistically significant; ***, p < 0.001, extremely significant.
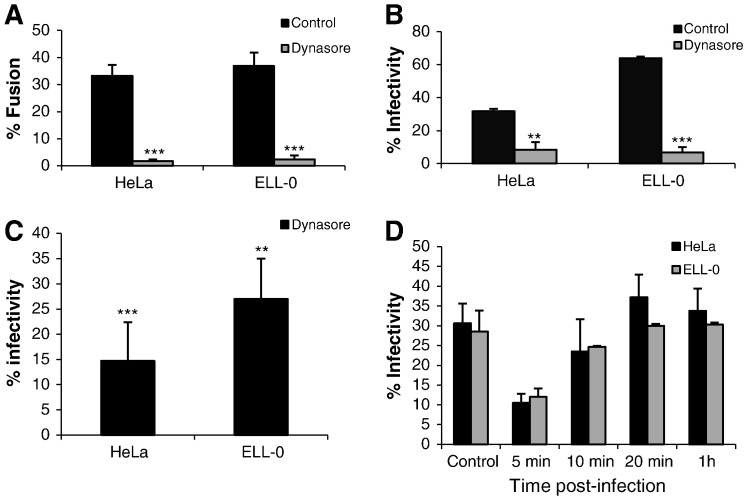
We also analyzed the effect of dynasore, a drug that inhibits the GTPase activity of dynamin 1, which has been implicated in clathrin- and caveola-mediated endocytosis [44]. Owing to the high toxicity of this drug in MEB4, GM95, CHO and Lec1 cells, these experiments were only performed in HeLa and ELL-0 cells, in which cell viability was higher than 95% after dynasore treatment (data not shown). The fusion of NDV with treated-HeLa and ELL-0 cells was almost completely abrogated (Fig. 6A), as was infectivity, when assayed both by fluorescence microscopy after rNDV-F3aa-mRFP infection (Fig. 6B) and by FACS (Fig. 6C). Nevertheless, the binding of NDV to drug-treated cells was not modified (data not shown). However, the addition of dynasore at 10 min post-infection resulted in only a minor decrease in infection, with no effect if added at 20 min post-infection (Fig. 6D), implying that the main role of dynamin was exerted during the early infection steps and again supporting a role of endocytosis in NDV entry. The rate of fusion and infectivity inhibition exerted by dynasore (more than 80%) was higher than that of BIM (Fig. 5) and than the observed enhancement of NDV fusion and infectivity after low-pH exposure (Fig. 1, Fig. 2, Fig. 3, Fig. 4). Dynamin is essential not only for the fission of endocytic vesicles in the endocytic pathways but also for membrane fusion [45], being involved in fusion pore expansion [46], [47]. Additionally, dynamin has been implicated in the regulation of the actin cytoskeleton [48], which could account for the stronger negative effect of dynasore preincubation of cells on NDV activities. The drug might negatively affect not only endocytic entry but also the direct fusion of NDV with the cell plasma membrane. In sum, the data regarding the effect of dynasore on NDV fusion and infectivity discussed above indicate that, at least in HeLa and ELL-0 cells, NDV needs dynamin activity, as described for other viruses such as HIV [14], HPV16 and BPV1 [49].
Fig. 6.
Effect of dynamin inhibition on NDV activities. (A) Effect on NDV fusion. HeLa and ELL-0 cells were previously incubated in the presence of dynasore for 30 min at 37 °C. Then, 2 μg of R18-NDV was bound to cells for 30 min at 4 °C, after which these were allowed to fuse for 1 h at 37 °C in continuous presence of the drug. Fusion was quantified as detailed in Materials and methods as the percentage of red-stained cells out of the total. (B) Effect on NDV infectivity. HeLa and ELL-0 cells were previously incubated in the presence of dynasore for 30 min at 37 °C and then infected for 1 h at 37 °C in the presence of the inhibitor with rNDV-F3aa-mRFP at a moi of 0.1 (HeLa) or 1 (ELL-0) to afford approximately 50% of cells infected in untreated control cells. Dynasore was present in the culture medium for an additional 2 h before washing and incubation over a total of 24 h. Quantification of infectivity was performed as in the legend in Fig. 3. (C) After treatment of cells with 80 μM of dynasore, cells were infected with NDV “Clone 30” at a moi of 1. At 24 h post-infection, infectivity was analyzed by a FACS-based immunoassay, as detailed in Materials and methods. Data are percentage of G mean fluorescence values from those of control untreated and infected cells taken as 100%. (D) Effect of the time of dynasore addition on NDV infectivity. Cells were infected with rNDV-F3aa-mRFP (moi of 0.1 and 1 for HeLa and ELL-0 cells respectively), and dynasore was added at different times post-infection and was present in the culture medium for additional 2 h. Infection was determined as described above for (B). Data are means ± SD of three independent experiments. **, p < 0.01, highly statistically significant; ***, p < 0.001, extremely significant.
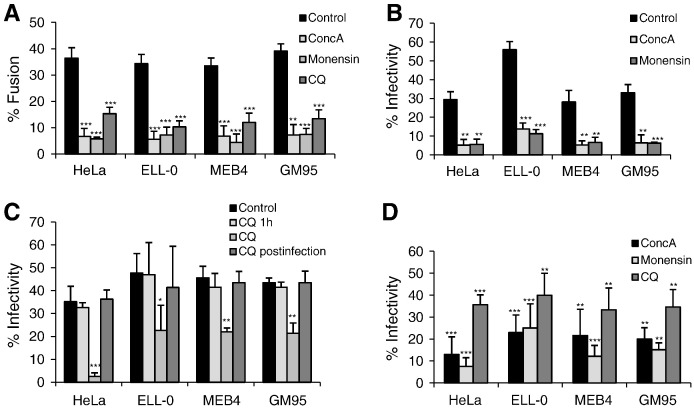
To prevent endosome acidification, cells were incubated with either concanamycin A, an inhibitor of vacuolar H+-ATPase [50], monensin, an ionophore that disrupts the proton gradient across vesicular membranes [51], or chloroquine, a lysosomotropic agent that increases pH inside endocytic vesicles [52]. The concentration and drug treatment times are specified in Table 1. Because of the high toxicity exerted by the incubation of CHO and Lec1 cells with the three drugs, these experiments were performed in HeLa, ELL-0, MEB4 and GM95 cell lines, where treatment did not interfere with cell viability (cell viability was about 95% or higher, data not shown). The data concerning the effect of the three different compounds on NDV activities are summarized in Fig. 7 and in Table 2 . The treatment of cells with one of the three compounds strongly inhibited virus–cell fusion, resulting in an approximately 80% reduction in virus–cell fusion after concanamycin A and monensin treatments, whereas chloroquine preincubation reduced viral fusion by about 60% (Fig. 7A). Similarly, the three drugs strongly reduced NDV infectivity (Fig. 7B, C, D). The negative effect of chloroquine was not observed if the drug was withdrawn when the virus was added (Fig. 7C, CQ 1 h) or was added at 5 h post-infection (Fig. 7C, CQ post-infection), meaning that the negative effect would be exerted at the early steps of the viral cycle. Viral binding to the cell surface was unaffected by any of the three compounds (Table 2). Nevertheless, in ELL-0 and HeLa cells fusion and infectivity inhibition by inhibitors of endosomal acidification (Fig. 7 and Table 2) were stronger than the enhancement observed after acid-pH treatment (Fig. 1, Fig. 3). In HeLa and ELL-0 cells, inhibition of virus infectivity resulting from monensin treatment was largely prevented by exposure to acidic pH (Supplementary Fig. S3). Moreover, concanamycin A, monensin and chloroquine would interfere with processes other than NDV fusion in endosomal compartments. In addition to raising the endosomal pH, it has been reported that chloroquine has multiple effects on mammalian cells [53]. Chloroquine inhibition of SARS-CoV infection is proposed to be exerted due to the interference with the terminal glycosylation of the viral receptor [54]. In addition, trafficking events other than NDV entry itself might be disturbed, since vesicular ATPase is involved in the regulation of trafficking events in endosomal compartments.
Fig. 7.
Effect of concanamycin A (ConcA), monensin and chloroquine (CQ) on NDV fusion and infectivity. (A) Effect on NDV fusion. Cells were incubated for 1 h and 30 min in the presence of ConcA or monensin, and for 1 h in the presence of CQ. Then, 2 μg of R18-NDV was bound to cells for 30 min at 4 °C and fusion was allowed to proceed in the continuous presence of the drug as described in the legend in Fig. 1. (B) Effect of ConcA and monensin on NDV infectivity. Cells were incubated for 1 h and 30 min in the presence of ConcA or monensin and then infected with rNDV-F3aa-mRFP at a moi of 1, except for HeLa cells that was 0.1 moi. The drugs were present in the culture medium for an additional 4 h before washing and incubation over a total of 24 h. NDV infectivity was quantified as in the legend in Fig. 3. (C) Effect of CQ on NDV infectivity. Cells were infected with rNDV-F3aa-mRFP at a moi of 1, except for HeLa cells that was 0.1 moi; the drug was added at different times: CQ 1 h: cells preincubated with the drug 1 h before infection; CQ: cells preincubated with the drug 1 h before infection and for additional 4 h post-infection; CQ post-infection: the drug was added at 5 h post-infection and then incubated for additional 6 h in the presence of the drug. At 24 h post-infection, NDV infectivity was quantified as in the legend in Fig. 3. (D) After treatment of cells with ConcA, monensin or CQ as in A, cells were infected with NDV “Clone 30” at a moi of 1. At 24 h post-infection, infectivity was analyzed by a FACS-based immunoassay, as detailed in Materials and methods. Data are percentage of G mean fluorescence values from those of control untreated and infected cells taken as 100%. Results are means ± SD of three independent experiments. * p < 0.05 statistically significant: ** p < 0.01 very significant; ***, p < 0.001, extremely significant.
Table 2.
Summary of the effect of drugs that interfere with endosome acidification on NDV interaction with different cell lines.
| Cell Line |
|||||
|---|---|---|---|---|---|
| HeLa | ELL-0 | MEB4 | GM95 | ||
| ConcA | Fusiona | 19.3 ± 3.3 | 16.3 ± 6 | 21 ± 7 | 20.8 ± 2.2 |
| Infectivitya | 18.5 ± 7 | 24.5 ± 3.7 | 25.3 ± 6.5 | 9.4 ± 2 | |
| FACSb | 13 ± 8 | 23 ± 8 | 21.6 ± 12 | 20 ± 5.2 | |
| Binding | 100 | 100 | 100 | 100 | |
| Monensin | Fusiona | 20.4 ± 5 | 20.7 ± 4.2 | 13.2 ± 6 | 31.8 ± 4.4 |
| Infectivitya | 20.1 ± 4.1 | 20.6 ± 4.3 | 17.6 ± 5 | 20.1 ± 2.4 | |
| FACSb | 7.5 ± 4 | 25.1 ± 11 | 12.1 ± 5 | 15.1 ± 3.2 | |
| Binding | 100 | 100 | 100 | 100 | |
| Chloroquine | Fusiona | 37.5 ± 5.6 | 38.3 ± 7.5 | 30 ± 5 | 34.7 ± 7 |
| Infectivitya | 7 ± 0.8 | 52.6 ± 6 | 48.3 ± 1.6 | 47.1 ± 4 | |
| FACSb | 35.7 ± 4 | 40 ± 10 | 34.6 ± 8 | ||
| Binding | 100 | 100 | 100 | 100 | |
Data are from Fig. 7, representing percentage of activity for control untreated cells.
FACS infectivity assays, data from Fig. 7D
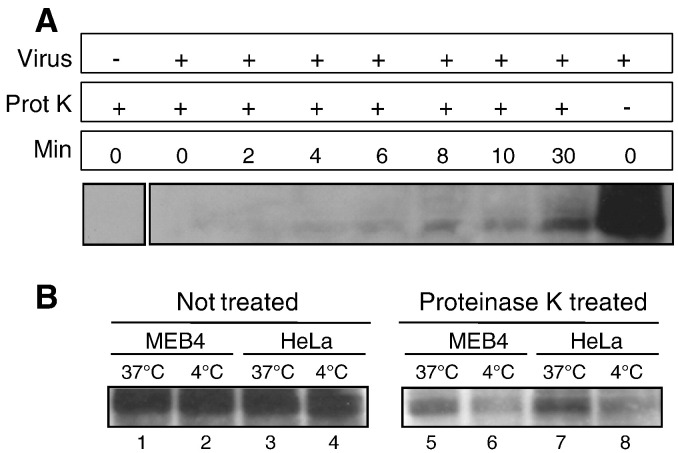
As an alternative assay of endocytic internalization, we realized proteinase K protection experiments as detailed in Materials and methods. Uninternalized virions as well as cell surface glycoproteins from virions that have fused directly with the plasma membrane would be digested by proteinase K, contrary to endocytosed virions that became protected from digestion. In ELL-0 cells a significant proportion of HN glycoprotein became protected after 6 min of incubation at 37 °C (Fig. 8A), which correlates with use of an endocytic pathway for virus entry. We also carried out the experiments with HeLa and MEB4 cells (Fig. 8B) looking for HN protection at 10 min after the initiation of entry. Although total digestion of viral glycoproteins was not observed at 4 °C (Fig. 8B, lanes 6 and 8), after incubation at 37 °C for 10 min we detected an increase in the proportion of non-digested viral HN (Fig. 8B, lanes 5 and 7), strongly supporting that a percentage of virus would also use endocytosis for entering both cell lines.
Fig. 8.
Protection of viral HN glycoprotein from proteinase K digestion. (A). Virus was attached to ELL-0 cells for 1 h at 4 °C. Entry was initiated by incubation at 37 °C for the indicated times. Cells were treated with proteinase K, lysed, and viral HN protein was detected by Western blotting with polyclonal anti-NDV antibodies. (B) Virus was attached to MEB4 (lanes 1, 2, 5, and 6) and HeLa (lanes 3, 4, 7, and 8) cells for 1 h at 4 °C. Samples were then incubated at 37 °C for 10 min (lanes 1, 3, 5, and 7) or held at 4 °C (lanes 2, 4, 6, and 8), treated with proteinase K for 1 h at 4 °C, and lysed as detailed in Materials and methods. NDV-HN protein was detected by Western blotting.
Class I viral fusion proteins can be activated by at least three different triggers: low pH, receptor binding at neutral pH, or receptor binding followed by low pH [55], [56]. Moreover, the fusion protein of the human metapneumovirus is activated by proteolysis and by acidic pH [20], [25]. The use of the endocytic pathways for paramyxovirus entry has been suggested as a common entry mechanism for the Pneumovirinae subfamily [25]. As stated above, NDV is able to fuse with cells at neutral pH; nevertheless, here we have shown in HeLa and ELL-0 cells (Fig. 1, Fig. 2, Fig. 3, Fig. 4) an enhancement of viral fusion and infectivity of approximately 30% after a brief pulse of low-pH, in agreement with our previous data [27], [28]. Although acid pH is not an absolute requirement for NDV infection, an acid pH environment would facilitate the conformational changes needed for fusion by decreasing the energetic barrier, as suggested above. This hypothetical decrease would not be enough for the activation of F in the absence of the homotypic HN (Fig. 2B), a phenotype that we have previously observed in N211A, I463A and I463F protein F mutants [57]. Another possibility is that acid pH would increase the fusion promotion activity of HN protein. As mentioned above, the second HN binding site, site II, is activated upon receptor binding to site I and exhibits higher receptor avidity than site I [5]. As speculated for the paramyxovirus HPIV3 [58], low pH may facilitate the exposure of site II and thus increase the fusion promotion activity of the protein since site II has been shown to be essential in HPIV3 and NDV F activation [59], [60].
Within the context of an in vivo infection, i.e., without lowering the pH experimentally, NDV would be able to fuse at neutral and acidic pH, depending on the entry pathways. In a cell-type-dependent mechanism, a certain percentage of viruses might be endocytosed and hence would fuse within the endosome. Similarly, some viruses utilize different cellular entry pathways to infect different target cells, such as herpes simplex virus type 1 (HSV-1), which can enter cells through the plasma membrane as well as by low-pH and neutral-pH endocytosis, depending on the cell line and receptor(s) [19], [34], [61]. Also, vaccinia virus can simultaneously use both entry mechanisms in the same cell type: fusion at the plasma membrane and endocytosis [62].
Concanamycin A, monensin and chloroquine strongly inhibited NDV fusion and infectivity in MEB4 and GM95 cells (Fig. 7), although we failed to observe any effect of the inhibitor BIM (Fig. 5) or any increase in NDV activation after low-pH exposure in these cell lines (Fig. 1, Fig. 3). This discrepancy may be explained by different possibilities: i) NDV does not enter MEB4 nor GM95 by endocytosis, and the negative effect of raising the pH in endosomes would be related to the interference with trafficking events other than NDV entry itself as discussed above; ii) the endocytic entry of NDV into MEB4 (with GM3 as the only ganglioside at the cell membrane) and GM95 (without gangliosides) cells may follow a different mechanism from that of HeLa and ELL-0 cells. Contrary to ELL-0 and HeLa cells, inhibition of virus infectivity resulting from monensin treatment was not prevented by exposure to acidic pH (Supplementary Fig. S3) supporting that the inhibition of endosomal acidification would have additional post-entry effects in these cell lines. Nevertheless, further experiments will be conducted to test these hypotheses.
The cell lines used in this study differ in the sialoglycoconjugates present at the cell surface. ELL-0 and HeLa cell membranes are abundant in α2,6 sialic acid compounds in comparison with the other cell lines ([63] and data not shown). Thus, it is tempting to speculate that this difference might be involved in diverse entry mechanisms of NDV into the host cell: only when the virus engages the proper molecule could be endocytosed, after which the viral envelope would fuse in the endocytic compartments. The binding of NDV to sialic acids in α2,6 specific linkages could allow the endocytic entry of NDV, and could be directly required either for endocytic internalization or to facilitate subsequent molecular changes enabling endocytosis. Further experiments will be needed to clarify this point.
Our data then support the notion that NDV might have evolved to use the low pH of the endocytic pathways to enhance its entry, at least in certain cell lines. In addition to direct fusion of the viral envelope with the target cell, a certain percentage of viruses might penetrate the cell through receptor-mediated and dynamin-dependent endocytosis. This entry versatility must be taken into consideration in order to design future antiviral strategies.
The following are the supplementary data related to this article.
Effect of acidic pH on NDV infection at 1 moi. Monolayers of the different cell lines were infected with rNDV-F3aa-mRFP at a moi of 1 for 1 h at room temperature. Then, cells were treated with three pulses (3 min each) of pH 5.0- or pH 7.4-buffered PBS with 1 h interval between each pulse. Infectivity was analyzed at 24 h post-infection by calculating the percentage of red-fluorescent cells (infected cells) out of the total number of cells in three random fields. For HeLa and ELL-0 cells data are means ± SD of three independent experiments; for MEB4, GM95, CHO and Lec1, data are means of two independent experiments.
Comparison of the extent of R18 dye transfer from R18-labeled RBCs to HN- and F-transfected cells at 25 °C (pH 5.0) and at 37 °C (pH 7.4). Microphotographs are from Fig. 4A. Blue arrows indicate bound but non-fused RBCs.
Cell monolayers were pretreated with monensin for 1 h and 30 min and then infected with rNDV-F3aa-mRFP at a moi of 1. After infection, cells were treated with three pulses (3 min each) of pH 5.0- or pH 7.4-buffered PBS with 1 h interval between each pulse. Data are percentage of infectivity control at 24 h post-infection. The drug was present in the culture medium for a total of 4 h post infection.
Acknowledgements
L.S.-F. received a fellowship from the Junta de Castilla y León. This work was partially supported by grants from Junta de Castilla y León (SA009A08) to I.M.B. and from the Fondo de Investigaciones Sanitarias (FIS, PI08/1813, cofinanced by FEDER funds from the EU) to E.V. We thank Dr. Adolfo García-Sastre for providing recombinant rNDV-F3aa-mRFP NDV, anti-NP and polyclonal anti-NDV antibodies; and Dr. Ronald M. Iorio for providing anti-HN monoclonal antibodies. We also thank the RIKEN cell bank for MEB4 and GM95 cell lines. Thanks are also due to N. Skinner for language corrections.
References
- 1.Villar E., Barroso I.M. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview. Glycoconj. J. 2006;23:5–17. doi: 10.1007/s10719-006-5433-0. [DOI] [PubMed] [Google Scholar]
- 2.Lamb R., Parks G. Paramyxoviridae: the viruses and their replication. In: P.M.H.E., Knipe D.M., editors. Paramyxoviridae: The Viruses and Their Replication. Wolters Kluwer/Lippincott Williams & Wilkins; 2007. pp. 1449–1496. (1[5th]) [Google Scholar]
- 3.Crennell S., Takimoto T., Portner A., Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 4.Zaitsev V., von Itzstein M., Groves D., Kiefel M., Takimoto T., Portner A., Taylor G. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porotto M., Fornabaio M., Greengard O., Murrell M.T., Kellogg G.E., Moscona A. Paramyxovirus receptor-binding molecules: engagement of one site on the hemagglutinin-neuraminidase protein modulates activity at the second site. J. Virol. 2006;80:1204–1213. doi: 10.1128/JVI.80.3.1204-1213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelkmans L., Burli T., Zerial M., Helenius A. Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell. 2004;118:767–780. doi: 10.1016/j.cell.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosset F.L., Lavillette D. Cell entry of enveloped viruses. Adv. Genet. 2011;73:121–183. doi: 10.1016/B978-0-12-380860-8.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marsh M., Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercer J., Helenius A. Virus entry by macropinocytosis. Nat. Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 11.Mercer J., Schelhaas M., Helenius A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 12.Helenius A., Kartenbeck J., Simons K., Fries E. On the entry of Semliki forest virus into BHK-21 cells. J. Cell Biol. 1980;84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin M., Park J., Lee S., Park B., Shin J., Song K.J., Ahn T.I., Hwang S.Y., Ahn B.Y., Ahn K. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology. 2002;294:60–69. doi: 10.1006/viro.2001.1303. [DOI] [PubMed] [Google Scholar]
- 14.Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G.B. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson H.A., Chen Y., Norkin L.C. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol. Biol. Cell. 1996;7:1825–1834. doi: 10.1091/mbc.7.11.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelkmans L., Puntener D., Helenius A. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science. 2002;296:535–539. doi: 10.1126/science.1069784. [DOI] [PubMed] [Google Scholar]
- 17.Empig C.J., Goldsmith M.A. Association of the caveola vesicular system with cellular entry by filoviruses. J. Virol. 2002;76:5266–5270. doi: 10.1128/JVI.76.10.5266-5270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez A. Analysis of filovirus entry into vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J. Infect. Dis. 2007;196(Suppl. 2):S251–S258. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- 19.Rahn E., Petermann P., Hsu M.J., Rixon F.J., Knebel-Morsdorf D. Entry pathways of herpes simplex virus type 1 into human keratinocytes are dynamin- and cholesterol-dependent. PLoS One. 2011;6:e25464. doi: 10.1371/journal.pone.0025464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schowalter R.M., Smith S.E., Dutch R.E. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 2006;80:10931–10941. doi: 10.1128/JVI.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith E.C., Popa A., Chang A., Masante C., Dutch R.E. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 2009;276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang A., Dutch R.E. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses. 2012;4:613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diederich S., Thiel L., Maisner A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology. 2008;375:391–400. doi: 10.1016/j.virol.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pernet O., Pohl C., Ainouze M., Kweder H., Buckland R. Nipah virus entry can occur by macropinocytosis. Virology. 2009;395:298–311. doi: 10.1016/j.virol.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 25.Schowalter R.M., Chang A., Robach J.G., Buchholz U.J., Dutch R.E. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J. Virol. 2009;83:1511–1522. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolokoltsov A.A., Deniger D., Fleming E.H., Roberts N.J., Jr., Karpilow J.M., Davey R.A. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J. Virol. 2007;81:7786–7800. doi: 10.1128/JVI.02780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.San Roman K., Villar E., Munoz-Barroso I. Acidic pH enhancement of the fusion of Newcastle disease virus with cultured cells. Virology. 1999;260:329–341. doi: 10.1006/viro.1999.9841. [DOI] [PubMed] [Google Scholar]
- 28.Cantin C., Holguera J., Ferreira L., Villar E., Munoz-Barroso I. Newcastle disease virus may enter cells by caveolae-mediated endocytosis. J. Gen. Virol. 2007;88:559–569. doi: 10.1099/vir.0.82150-0. [DOI] [PubMed] [Google Scholar]
- 29.Mibayashi M., Martinez-Sobrido L., Loo Y.M., Cardenas W.B., Gale M., Jr., García-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.San Roman K., Villar E., Munoz-Barroso I. Mode of action of two inhibitory peptides from heptad repeat domains of the fusion protein of Newcastle disease virus. Int. J. Biochem. Cell Biol. 2002;34:1207–1220. doi: 10.1016/s1357-2725(02)00045-6. [DOI] [PubMed] [Google Scholar]
- 32.Connolly S.A., Lamb R.A. Paramyxovirus fusion: real-time measurement of parainfluenza virus 5 virus–cell fusion. Virology. 2006;355:203–212. doi: 10.1016/j.virol.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Young J.K., Li D., Abramowitz M.C., Morrison T.G. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J. Virol. 1999;73:5945–5956. doi: 10.1128/jvi.73.7.5945-5956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne R.S., Nicola A.V., Whitbeck J.C., Eisenberg R.J., Cohen G.H. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 2005;79:6655–6663. doi: 10.1128/JVI.79.11.6655-6663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichikawa S., Nakajo N., Sakiyama H., Hirabayashi Y. A mouse B16 melanoma mutant deficient in glycolipids. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2703–2707. doi: 10.1073/pnas.91.7.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu V.C., Whittaker G.R. Influenza virus entry and infection require host cell N-linked glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18153–18158. doi: 10.1073/pnas.0405172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toullec D., Pianetti P., Coste H., Bellevergue P., Grand-Perret T., Ajakane M., Baudet V., Boissin P., Boursier E., Loriolle F. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 38.Root C.N., Wills E.G., McNair L.L., Whittaker G.R. Entry of influenza viruses into cells is inhibited by a highly specific protein kinase C inhibitor. J. Gen. Virol. 2000;81:2697–2705. doi: 10.1099/0022-1317-81-11-2697. [DOI] [PubMed] [Google Scholar]
- 39.Upla P., Marjomaki V., Kankaanpaa P., Ivaska J., Hyypia T., Van Der Goot F.G., Heino J. Clustering induces a lateral redistribution of alpha 2 beta 1 integrin from membrane rafts to caveolae and subsequent protein kinase C-dependent internalization. Mol. Biol. Cell. 2004;15:625–636. doi: 10.1091/mbc.E03-08-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Constantinescu S.N., Cernescu C.D., Popescu L.M. Effects of protein kinase C inhibitors on viral entry and infectivity. FEBS Lett. 1991;292:31–33. doi: 10.1016/0014-5793(91)80826-o. [DOI] [PubMed] [Google Scholar]
- 41.Martin J.J., Holguera J., Sanchez-Felipe L., Villar E., Munoz-Barroso I. Cholesterol dependence of Newcastle Disease Virus entry. Biochim. Biophys. Acta. 2012;1818:753–761. doi: 10.1016/j.bbamem.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smart E.J., Foster D.C., Ying Y.S., Kamen B.A., Anderson R.G. Protein kinase C activators inhibit receptor-mediated potocytosis by preventing internalization of caveolae. J. Cell Biol. 1994;124:307–313. doi: 10.1083/jcb.124.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mineo C., Ying Y.S., Chapline C., Jaken S., Anderson R.G. Targeting of protein kinase C alpha to caveolae. J. Cell Biol. 1998;141:601–610. doi: 10.1083/jcb.141.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Low H.H., Lowe J. Dynamin architecture—from monomer to polymer. Curr. Opin. Struct. Biol. 2010;20:791–798. doi: 10.1016/j.sbi.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Jaiswal J.K., Rivera V.M., Simon S.M. Exocytosis of post-Golgi vesicles is regulated by components of the endocytic machinery. Cell. 2009;137:1308–1319. doi: 10.1016/j.cell.2009.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anantharam A., Bittner M.A., Aikman R.L., Stuenkel E.L., Schmid S.L., Axelrod D., Holz R.W. A new role for the dynamin GTPase in the regulation of fusion pore expansion. Mol. Biol. Cell. 2011;22:1907–1918. doi: 10.1091/mbc.E11-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu C., Yaddanapudi S., Weins A., Osborn T., Reiser J., Pollak M., Hartwig J., Sever S. Direct dynamin–actin interactions regulate the actin cytoskeleton. EMBO J. 2010;29:3593–3606. doi: 10.1038/emboj.2010.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abban C.Y., Bradbury N.A., Meneses P.I. HPV16 and BPV1 infection can be blocked by the dynamin inhibitor dynasore. Am. J. Ther. 2008;15:304–311. doi: 10.1097/MJT.0b013e3181754134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drose S., Altendorf K. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 1997;200:1–8. doi: 10.1242/jeb.200.1.1. [DOI] [PubMed] [Google Scholar]
- 51.Maxfield F.R. Weak bases and ionophores rapidly and reversibly raise the pH of endocytic vesicles in cultured mouse fibroblasts. J. Cell Biol. 1982;95:676–681. doi: 10.1083/jcb.95.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Duve C., de Barsy T., Poole B., Trouet A., Tulkens P., Van Hoof F. Commentary. Lysosomotropic agents. Biochem. Pharmacol. 1974;23:2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]
- 53.Thorens B., Vassalli P. Chloroquine and ammonium chloride prevent terminal glycosylation of immunoglobulins in plasma cells without affecting secretion. Nature. 1986;321:618–620. doi: 10.1038/321618a0. [DOI] [PubMed] [Google Scholar]
- 54.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Earp L.J., Delos S.E., Park H.E., White J.M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ayllon J., Villar E., Munoz-Barroso I. Mutations in the ectodomain of Newcastle disease virus fusion protein confer a hemagglutinin-neuraminidase-independent phenotype. J. Virol. 2010;84:1066–1075. doi: 10.1128/JVI.01473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palermo L.M., Porotto M., Greengard O., Moscona A. Fusion promotion by a paramyxovirus hemagglutinin-neuraminidase protein: pH modulation of receptor avidity of binding sites I and II. J. Virol. 2007;81:9152–9161. doi: 10.1128/JVI.00888-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Porotto M., Palmer S.G., Palermo L.M., Moscona A. Mechanism of fusion triggering by human parainfluenza virus type III: communication between viral glycoproteins during entry. J. Biol. Chem. 2012;287:778–793. doi: 10.1074/jbc.M111.298059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Porotto M., Salah Z., DeVito I., Talekar A., Palmer S.G., Xu R., Wilson I.A., Moscona A. The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J. Virol. 2012;86:5730–5741. doi: 10.1128/JVI.06793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicola A.V., McEvoy A.M., Straus S.E. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 2003;77:5324–5332. doi: 10.1128/JVI.77.9.5324-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Townsley A.C., Weisberg A.S., Wagenaar T.R., Moss B. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J. Virol. 2006;80:8899–8908. doi: 10.1128/JVI.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez-Felipe L., Villar E., Munoz-Barroso I. Alpha2-3- and alpha2-6-N-linked sialic acids allow efficient interaction of Newcastle Disease Virus with target cells. Glycoconj. J. 2012;29:539–549. doi: 10.1007/s10719-012-9431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of acidic pH on NDV infection at 1 moi. Monolayers of the different cell lines were infected with rNDV-F3aa-mRFP at a moi of 1 for 1 h at room temperature. Then, cells were treated with three pulses (3 min each) of pH 5.0- or pH 7.4-buffered PBS with 1 h interval between each pulse. Infectivity was analyzed at 24 h post-infection by calculating the percentage of red-fluorescent cells (infected cells) out of the total number of cells in three random fields. For HeLa and ELL-0 cells data are means ± SD of three independent experiments; for MEB4, GM95, CHO and Lec1, data are means of two independent experiments.
Comparison of the extent of R18 dye transfer from R18-labeled RBCs to HN- and F-transfected cells at 25 °C (pH 5.0) and at 37 °C (pH 7.4). Microphotographs are from Fig. 4A. Blue arrows indicate bound but non-fused RBCs.
Cell monolayers were pretreated with monensin for 1 h and 30 min and then infected with rNDV-F3aa-mRFP at a moi of 1. After infection, cells were treated with three pulses (3 min each) of pH 5.0- or pH 7.4-buffered PBS with 1 h interval between each pulse. Data are percentage of infectivity control at 24 h post-infection. The drug was present in the culture medium for a total of 4 h post infection.