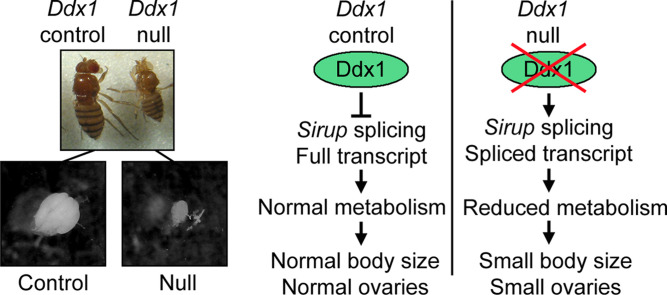
Abstract
Mammalian DDX1 has been implicated in RNA trafficking, DNA double-strand break repair and RNA processing; however, little is known about its role during animal development. Here, we report phenotypes associated with a null Ddx1 (Ddx1AX) mutation generated in Drosophila melanogaster. Ddx1 null flies are viable but significantly smaller than control and Ddx1 heterozygous flies. Female Ddx1 null flies have reduced fertility with egg chambers undergoing autophagy, whereas males are sterile due to disrupted spermatogenesis. Comparative RNA sequencing of control and Ddx1 null third instars identified several transcripts affected by Ddx1 inactivation. One of these, Sirup mRNA, was previously shown to be overexpressed under starvation conditions and implicated in mitochondrial function. We demonstrate that Sirup is a direct binding target of Ddx1 and that Sirup mRNA is differentially spliced in the presence or absence of Ddx1. Combining Ddx1 null mutation with Sirup dsRNA-mediated knock-down causes epistatic lethality not observed in either single mutant. Our data suggest a role for Drosophila Ddx1 in stress-induced regulation of splicing.
Keywords: DEAD box helicase, DDX1, Sirup, Gametogenesis, Metabolism
Graphical Abstract
Highlights
-
•
We describe a new Ddx1 null Drosophila line.
-
•
Ddx1 null flies are smaller in size and display aberrant gametogenesis.
-
•
Sirup splicing is altered in Ddx1 null flies.
-
•
We show both a physical and a genetic interaction between Ddx1 and Sirup.
1. Introduction
DEAD box proteins are a family of RNA helicases implicated in virtually every aspect of RNA metabolism (Cordin et al., 2006). These proteins are characterized by 12 conserved motifs, including the D(Asp)-E(Glu)-A(Ala)-D(Asp) motif for which they are named. DEAD box proteins function by modifying RNA secondary structure in an ATP-dependent manner (Linder and Fuller-Pace, 2013), and play a role in RNA trafficking (Linder and Stutz, 2001, Montpetit et al., 2012). There are >30 DEAD box genes in the Drosophila melanogaster genome (2014).
Several Drosophila DEAD box genes are known to play a role in early development and gametogenesis. For example, the vasa gene encodes a multifunctional DEAD box protein that localizes to the posterior pole in oocytes and is required for completion of oogenesis (Lasko, 2013). Vasa has also been implicated in chromatin condensation and generation of small non-coding RNAs (Pek and Kai, 2011, Zhang et al., 2012). Belle, which is closely related to Vasa, is essential for larval development and required for both male and female fertility (Johnstone et al., 2005). Mutation of pitchoune, encoding another DEAD box helicase, results in developmental arrest during the first instar stage. Pitchoune has been implicated in regulating cell growth and proliferation (Zaffran et al., 1998).
Only one Ddx1 mutation has been previously described in Drosophila melanogaster (Zinsmaier et al., 1994). This mutation was deemed to be recessive lethal; however, the nature of the mutation was not determined and the mutant line is no longer available. Rafti et al. described the expression of Ddx1 in Drosophila in 1996, and reported elevated levels in early embryos, and expression throughout development (Rafti et al., 1996). More recently, publicly available large scale studies using gene expression microarrays and RNA deep sequencing analysis have revealed widespread expression of Ddx1 in all tissues and cell lines tested to date (St Pierre et al., 2014). These screens show elevated Ddx1 levels in the nervous system, testes and ovaries with the highest levels observed at early embryonic stages, as previously described (Rafti et al., 1996).
Human DDX1, which is 77% similar to Drosophila Ddx1, is amplified and overexpressed in a subset of MYCN-amplified neuroblastoma and retinoblastoma cell lines and tumors (Godbout et al., 1998, Godbout and Squire, 1993, Manohar et al., 1995, Squire et al., 1995). DDX1 is also a prognostic marker in breast cancer (Balko and Arteaga, 2011, Germain et al., 2011), and plays a role in testicular tumorigenesis (Tanaka et al., 2009). Large scale screens for disease associated genes have identified DDX1 as a potential gene of interest in cervical cancer and chronic obstructive pulmonary disease (Johanneson et al., 2014, Smolonska et al., 2014). As well, DDX1 is significantly down-regulated in Down syndrome fetal brains (Kircher et al., 2002). Clues to DDX1’s role in these diseases come from in vitro analysis. For example, DDX1 was identified in a subset of RNA transport granules involved in the subcellular localization of RNA molecules in neuronal axons (Kanai et al., 2004). DDX1’s role in RNA trafficking is not limited to endogenously coded genes as HIV replication requires DDX1 for efficient export of unspliced viral genomic RNA from the nucleus to the cytoplasm (Edgcomb et al., 2012, Fang et al., 2005, Robertson-Anderson et al., 2011). This effect is mediated through an interaction between DDX1 and two virally encoded proteins, Rev and Tat, that are essential for RNA export (Lin et al., 2014). DDX1 is also required for efficient replication of coronavirus (Wu et al., 2014, Xu et al., 2010), and has been shown to transactivate hepatitis C and JC viral genes (Sunden et al., 2007a, Sunden et al., 2007b, Tingting et al., 2006).
Under normal conditions, DDX1 is mainly found in the nucleus where it forms granules that co-localize with (or reside adjacent to) nuclear organelles associated with mRNA processing such as cleavage bodies, gems and Cajal bodies (Bleoo et al., 2001, Li et al., 2006). When cells are treated with ionizing radiation, DDX1 is recruited to a subset of DNA double-strand breaks (Li et al., 2008). Biochemical analysis has shown that DDX1 can unwind RNA/RNA and RNA/DNA duplexes in vitro in an ADP-dependent manner and can efficiently digest single-stranded RNA (Li et al., 2008). Finally, DDX1 has also been implicated as a tRNA splicing factor (Popow et al., 2011, Popow et al., 2014).
While it is clear that DDX1 is a multifunctional protein, we only have a limited understanding of its biological role during development. Therefore, in order to gain insight into DDX1’s role during development, we generated a new Ddx1 null mutant Drosophila line. Here, we report that Ddx1 null flies are viable, with reduced fertility and body size. Ddx1 null flies also display aberrant gametogenesis in both testes and ovaries. We also describe a direct interaction between Sirup mRNA, previously described as up-regulated during starvation conditions and implicated in mitochondrial function (Erdi et al., 2012, Van Vranken et al., 2014), and Ddx1 protein. Finally, Sirup RNA is differentially spliced in control versus Ddx1 null flies, and an epistatic lethal effect is observed in Ddx1 AX/AX/Sirup dsRNA-mediated knock-down flies.
2. Materials and METHODS
2.1. Drosophila Stocks
All crosses were performed at 25˚C on standard Bloomington recipe media. Fly stocks were obtained from the Vienna Drosophila RNAi Center (VDRC) (Vienna, Austria) and the Bloomington Drosophila Stock Center (BDSC) (Indiana University, USA). The following fly stocks were used:
w 1118- Control line
y 1 w ⁎ ; Ly/TM3, Sb – Balancer for potential mutant allele
y 1 w 67c23 ; P{EPgy2}EY12792 – P-element upstream of Ddx1
w⁎; Dr 1 /TMS, P{ry[∆2-3]}B – Expresses P-element transposase
w⁎; Sb 1 /TM3, P{ActGFP}JMR2, Ser 1 - Mutant allele balancer
w 1118 ; Df(3L)ED230, P{3’.RS5+3.3’}ED230/TM6C, cu 1 Sb 1- Deficiency encompassing Ddx1. Referred to as Df(3L)ED230
y 1 w ⁎ ; P{Act5C-GAL4}25FO1/CyO, y + - Expresses GAL4 under the control of the actin promoter
w 1118 ; P{GD14644}v36437 – Sirup dsRNA expressing transgene
2.2. Generation of potential Ddx1 mutant alleles by P-element excision
Potential mutations of the Ddx1 locus were generated using BDSC stock #21389 (y 1 w67c23; P{EPgy2}EY12792), that contains a P-element inserted 43 bp upstream of the Ddx1 transcriptional start site. P{EPgy2}EY12792 was excised by crossing virgin P{EPgy2}EY12792 females with ∆2-3 transposase expressing males (w⁎; Dr 1 /TMS, P{ry[∆2-3]}99B, BDSC #1610). Individual white-eyed F1 single female virgin w ⁎ ; ∆P{EPgy2}EY12792/TMS, P{ry[∆2-3]}99B flies were mated to y 1 w ⁎ ; Ly/TM3, Sb males. Individual F2 w ⁎ ; ∆P{EPgy2}EY12792/TM3, Sb flies were then again mated to y 1 w ⁎ ; Ly/TM3, Sb flies, and w ⁎ ; ∆P{EPgy2}EY12792/TM3, Sb female and male progeny from each cross were further crossed to establish balanced lines.
2.3. Characterization of potential mutant lines
Individual fly lines displaying white eye color, indicating removal of P{EPgy2}EY12792 (which carries a w + transgene), were analyzed for deletion by sequential PCR reactions using staggered primers. One line (AX) was identified as containing an ~1.7 kb deletion. Subsequent genomic sequencing revealed that a 1733 bp region (3L: 22,263,326 to 22,265,059) spanning the P-element insertion site and most of the Ddx1 gene locus had been removed and replaced with the 15 bp sequence 5’-CATGATGAAATAACA-3’. This 15 bp sequence does not correspond to any part of Ddx1 or P{EPgy2}EY12792. This allele was designated Ddx1 AX. The following primers were used for PCR amplification: 5’-CCAGAAGCCGTGCATG-3’ (forward primer ~400 bp upstream of Ddx1 transcription start site), 5’-ATGAGTGTTGGCCAGCG-3’ (forward primer ~500 bp downstream of Ddx1 transcription start site), 5’-AGCTGGTGGAATTGCAC-3’ (reverse primer ~400 bp downstream of Ddx1 transcription start site), 5’-ACCATCTGCAGACGG-3’ (reverse primer ~1.4 kb downstream of Ddx1 transcription start site), 5’-GAGCTCCGACTTCCTAC-3’ (reverse primer located in Ddx1 3’ UTR). The following primer was used for genomic DNA sequencing: 5’-CTCATAAAGTCAAGTAAC-3’ (forward primer ~200 bp upstream of Ddx1 transcription start site) (Integrated DNA Technologies, Coralville, Iowa).
2.4. Western blot analysis and antibodies
Cell lysates were prepared from adult flies or larvae by grinding with a pestle in lysis buffer (1% sodium deoxycholate, 1% Triton-X-100, 0.2% SDS, 150 mM NaCl, 50 mM Tris-HCl pH 7.4 and 1X Complete® protease inhibitors (Roche, Mississauga, Canada). Samples that were analyzed for phospho-S6k were prepared in lysis buffer supplemented with 1x PhosStop (Roche). Lysates were electrophoresed in 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The following antibodies were used: rabbit anti-pS6k (1:1000) (#9209, Cell Signaling Technology, New England Biolabs, Whitby Canada), rabbit anti-Ddx1 (1:500) (custom polyclonal antibody, antigen CQKNLRTGSGYEDHV, GenScript, Piscataway, USA) and mouse anti-β-tubulin (1:2000) (DSHB, E7). The signal was detected using secondary anti-rabbit-HRP and anti-mouse-HRP antibodies (Jackson ImmunoResearch, West Grove, USA). The E7 β-tubulin antibody developed by Michael Klymkowsky was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242. Proteins were detected using the Immobilon reagent (EMD Millipore, Darmstadt, Germany).
2.5. RNA purification and RT-PCR
RNA was purified using an RNeasy Plus Universal Mini Kit (Qiagen, Toronto, Canada) as per the manufacturer’s directions. Briefly, whole flies or larvae were crushed in QIAzol reagent using plastic pestles and purified RNA isolated using a mini column. Reverse transcription was carried out using SuperScript II reverse transcriptase (Life Technologies, Burlington, Canada) and oligo dT primers as per the manufacturer’s directions. The following primers were used for RT-PCR analysis: Sirup, forward primer 5’-CCTGCGAGATTGCAATTCAG-3’, reverse primer 5’-AGTGGTTCCTTCTCCTGGTACG-5’; Sirup splice specific transcript, forward primer 5’-CAAATGGGCAAACAA⁎GTGA-3’ (asterisk indicates splice junction site), reverse primer 5’-GAATTCTTTAATAGTTTCTGCCC-3’; actin, forward primer 5’-AATCCAGAGACACCAAACCG-3’, reverse primer 5’- GAACGATACCGGTGGTACGA-3’. The forward primer for Sirup splice-specific product amplification consisted of the 15 nt sequence upstream of the splice junction followed by the 4 nt sequence downstream of the splice junction.
2.6. Viability, fertility, size and developmental delay assays and larval collections
For viability and fertility assays, single virgin females were mated with two males and left to lay eggs in standard culture tubes. After ten days, the parental flies were removed and individual pupae were counted and moved to a new tube daily to ensure no cross generational contamination. For crowded conditions, twenty females were mated to twenty males in a single tube. For size analysis, newly eclosed adults or pupae were genotyped and photographed using an Olympus SZX12 fluorescence dissecting microscope coupled to a Canon Powershot G2 color camera. Whole length, for pupae, or thorax length, for adults, was measured using Photoshop CS2 v9.0 calibrated using a ruler with 1 mm divisions. For developmental delay assays, three y 1 w ⁎; Ddx1 AX /TM3, GFP, Ser virgins were crossed with two y 1 w ⁎; Ddx1 AX /TM3, GFP, Ser males and allowed to lay eggs for 24 hours. Individual pupae were removed as above and scored for Ddx1 genotype based on GFP status. For larval collection, parental flies were placed in collection cages on apple juice agar plates with yeast paste for two hours. Plates were then incubated at 25˚C for 24, 48 or 72 hours at which point larvae were manually collected. Student’s t-test was used to compare differences between genotypes.
2.7. Immunofluorescence, microscopy and measurement
Ovaries and testes were dissected from virgin male and female flies that had been collected and held in isolation for 3 or 10 days or allowed to mate and supplemented with yeast paste for 3 days. For gross morphology, gonads were imaged immediately following dissection. For immunofluorescence, testes were fixed in 4% paraformaldehyde for 20 minutes, washed in PBS, permeabilized in 0.5% Triton X-100 for 5 minutes and washed three times in PBS. Tissues were then incubated with Alexa-546 conjugated-phalloidin (1:200, Life Technologies) for 90 minutes, washed in PBS, and mounted in polyvinyl alcohol (PVA) containing 4’,6-diamidino-2-phenylindole (DAPI) to stain the DNA. Ovaries were either incubated in LysoTracker Red (1:50, Life Technologies), fixed in 4% paraformaldehyde, dissected into individual ovarioles, or fixed, permeabilized in 0.5% Triton X-100, incubated with anti-Gurken antibody (1:30, DSHB, 1D12) followed by Alexa-546 conjugated-phalloidin and anti-mouse Alexa-488 (1:200 Life Technologies). Tissues were mounted in PVA with DAPI. The anti-Gurken antibody 1D12 developed by Schupbach T. was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 5224.
Confocal images were captured on a Zeiss LSM 710 confocal laser scanning microscope with a plan-Apochromat 63x (NA 1.4) oil immersion lens, a plan-Apochromat 40x (NA 1.3) oil immersion lens, or a plan-Apochromat 10x (NA 0.45) lens, and Zen software v7.0.4.287. An Olympus SZX12 microscope was used to photograph adult flies and gonads. Exported images were saved as TIFF files and measurements made using Photoshop.
2.8. Northern blot analysis
RNA samples were isolated from wandering 3rd instars as described above. For each sample, 5 μg of total RNA were resolved in a 10% denaturing urea polyacrylamide gel. A small RNA ladder (NEB, Whitby, Canada) was used for size determination. RNA was then transferred to Hybond-N+ membranes (GE Healthcare, Mississauga, Canada) and baked for 1 hour at 80 °C. DNA probes were generated by 32P end-labeling of single-stranded oligonucleotides. The following probes were used: tRNAtyr 5’-CTACAGTCCACCGCTCTACCAACTGAGCTATCGAAGG-3’, tRNAala 5’-TGCTAAGCGAGCGCTCTACCATCTGAGCTACATCCCC-3’, 5s rRNA 5’-CACTCGGCTCATGGGTCGATGAAGAACGCAGCAAACTG-3’. Probe hybridization was carried out in 5X SSC, 50 mM Na2HPO4 pH 6.5, 0.5X Denhardt’s solution and 0.25 mg/mL salmon sperm DNA at 42°C. The blot was then washed in 2X SSC, 0.1% SDS, followed by 0.1X SSC, 0.1% SDS. The signal was visualized using X-ray film. Membranes were initially hybridized with radioactively-labeled tRNAtyr and 5s rRNA probes. The label was then removed using stripping buffer and the blot exposed to film to ensure that the signal was sufficiently reduced to allow re-probing. The membrane was then re-hybridized with radioactively-labeled tRNAala probe.
2.9. RNA deep sequencing
RNA was isolated from three independent preparations of wandering 3rd instars for both Ddx1 null and control genotypes. RNA libraries were prepared by first removing rRNAs using a Ribo-Zero™ rRNA Removal Kit (Human/Mouse/Rat) (Epicentre, Madison, USA) followed by TruSeq Stranded Total RNA Sample Prep Kit (Illumina, San Diego, USA). Paired-end 100 nt RNA sequencing was performed on an Illumina HiSeq 2000 platform and processed using Casava V1.8.2 by the UBC Biodiversity Research Centre NextGen Sequencing Facility. Alignment, splice site identification was carried out using Bowtie v2.1.0 and Tophat v2.0.13 with standard parameters. Differential gene expression was calculated using Cufflinks v2.1.1 using standard parameters with a P-value of <0.01 considered significant.
2.10. RNA co-immunoprecipitations
Whole cell lysates were prepared from S2 cells by resuspending cell pellets in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, and 1X Complete (Roche) protease inhibitors. Four hundred micrograms of lysate were first cleared with Protein A agarose beads (GE Healthcare), followed by incubation with 5 μl of rabbit anti-Ddx1 antibody or rabbit IgG for 2 hours at 4°C. Protein A agarose beads were then added and incubation continued for 1 hour at 4°C. Co-immunoprecipitates were washed three times in lysis buffer and extracted with water-saturated phenol. An aliquot taken from the co-immunoprecipitates was saved for western blot analysis to check Ddx1 immunoprecipitation efficiency. Co-immunoprecipitated RNA was resuspended in RNase-free water and stored at -80°C until use. Reverse transcription was carried out as described above, using Sirup-specific (5’-AGTGGTTCCTTCTCCTGGTACG-3’) or Ddx1-specific (5’-TCATCGGGCAGCGTCAC-3’) reverse primers with the immunoprecipitated RNA serving as template. Following reverse transcription, PCR amplification was carried out using Sirup specific (forward 5’-CCTGCGAGATTGCAATTCAG-3’, reverse 5’-AGTGGTTCCTTCTCCTGGTACG-5’) or Ddx1 specific (forward 5’-GCATGCATTTGAGGTGAAG-3’, reverse 5’-TCATCGGGCAGCGTCAC-3’) primers.
2.11. Sirup knock-down in Ddx1 modified flies
y1 w ⁎ ; P{Act5C-GAL4}25FO1/CyO; Ddx1 AX /TM3, Sb virgin females were crossed to w 1118 ; P{GD14644}v36437; Ddx1 AX /TM3, P{ActGFP}JMR2, Ser 1 males and allowed to lay eggs for 3 days. Adults were scored for Ddx1 status by the presence of Sb or Ser alleles, and Sirup knock-down was scored based on the absence of CyO. Chi square analysis was performed to determine the significance of observed outcomes compared with expected genotype distribution.
3. RESULTS
3.1. Ddx1 null flies are viable
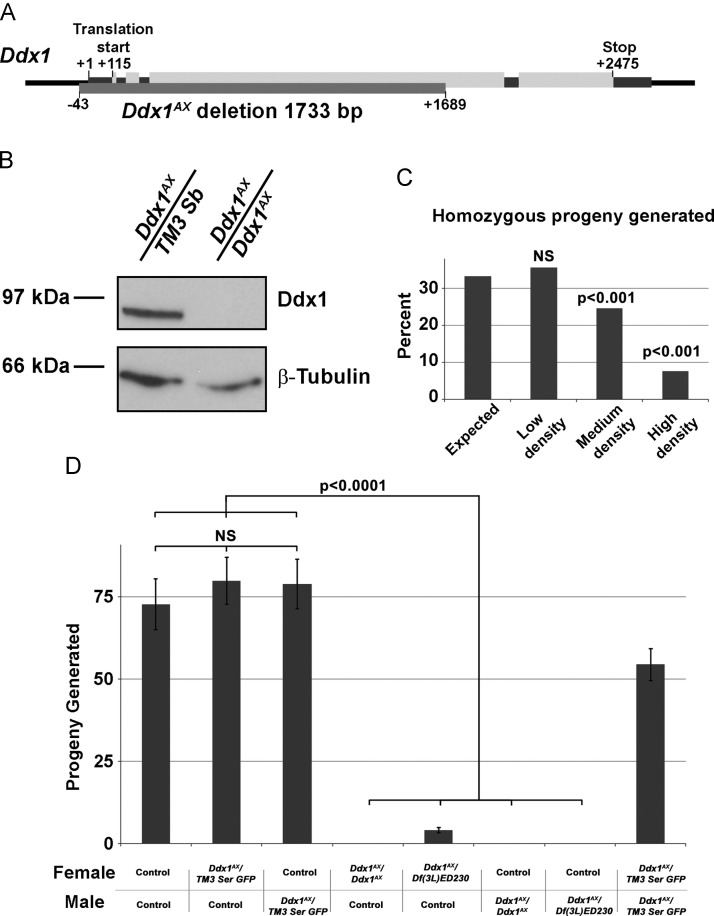
Imprecise excision of a P-element located immediately upstream of Ddx1 (y 1 w 67c23 ; P{EPgy2}EY12792) was used to screen for novel Ddx1 mutations. From this screen, we identified a new null Ddx1 allele with a 1733 bp deletion encompassing the majority of the Ddx1 open reading frame. This allele was designated Ddx1 AX ( Fig. 1A). In contrast to a previously reported Ddx1 mutation (Ddx1 9W1) that showed early embryonic lethality (Zinsmaier et al., 1994), Ddx1 AX/AX flies reached adulthood. Western blot analysis of control (w1118) and Ddx1 AX/AX adult flies showed a complete absence of Ddx1 in homozygous mutants (Fig.1B). Although a small portion of the 3’ Ddx1 open reading frame is retained in Ddx1 AX/AX flies, no novel bands were detected using an anti-Ddx1 antibody generated against the C-terminus of Ddx1. These results suggest that there is either no translation of the retained 3’-end of Ddx1 or the resulting product is unstable.
Fig. 1.
Ddx1null flies are viable but show reduced fertility. (A) The location of the Ddx1AX deletion. (B) Western blot analysis shows no detectable signal for Ddx1 protein in Ddx1AX/AXadult flies. (C) Survival of homozygous adult flies generated from Ddx1 heterozygote crosses. As the Ddx1AX mutation is carried over a recessive lethal balancer chromosome, the expected percentage of homozygous progeny is 33%. At low density, homozygous flies were generated at the expected rate. At medium density and high density, a significant reduction in the number of homozygous flies was observed; n=45 adults (low density), 1165 adults (medium density) and 499 adults (high density). (D) Progeny generated from single virgin females mated with two males (genotypes are indicated) and allowed to lay eggs for 10 days. Pupae were removed and counted daily. Homozygous mutant flies generated very low or no progeny. Heterozygous flies generated progeny at the expected rate. n≥20 crosses for all samples. Error bars indicate s.e.m.
Although Ddx1 AX/AX flies were viable, we noticed that they were consistently outcompeted under high density culturing conditions. When single eggs from heterozygote crosses were raised in isolation, we observed the expected 2:1 ratio of heterozygous to homozygous mutant progeny (heterozygous mutants were maintained over a recessive lethal balancer chromosome [TM3, P{ActGFP}JMR2, Ser 1], so no homozygous non-mutant Ddx1 progeny were generated). When a single female was allowed to lay eggs for 10 days in a standard collection tube, the ratio dropped to approximately 3:1. When 20 females were allowed to lay eggs in a standard collection tube for 10 days, the ratio of heterozygous to homozygous mutant was 13:1 (Fig. 1C).
3.2. Ddx1 null flies have reduced fertility, size and delayed development
Heterozygous y 1 w ⁎; Ddx1 AX /TM3 Ser GFP flies generated the expected number of progeny as compared to control flies (Fig. 1D). Both male and female Ddx1 AX/AX flies were sterile. To confirm that the observed infertility was due to inactivation of Ddx1, and not a line-specific effect, we out-crossed Ddx1 AX to a line containing a deficiency that encompasses the Ddx1 locus (Df[3L]ED230). Ddx1 AX /Df(3L)ED230 males were completely sterile, while females were able to generate a small number of viable progeny that survived to adulthood (Fig. 1D). Heterozygote crosses resulted in approximately 75% as many progeny as control. This was as expected, as 25% of the progeny would be homozygous for the recessive lethal balancer chromosome.
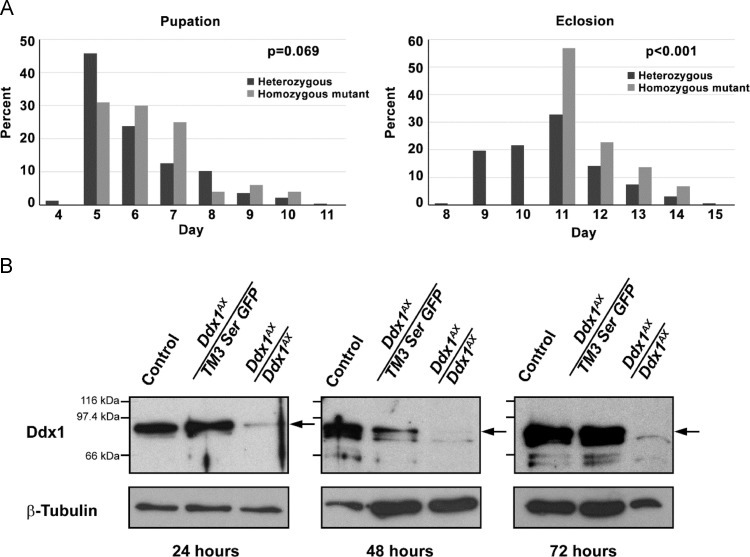
We noted that Ddx1 null progeny generally eclosed later than control and heterozygous animals. To better define the extent of the developmental delay in Ddx1 null flies, we set up heterozygote crosses, with females allowed to lay eggs for a period of 24 hours. Pupae were removed from each collection tube at 24 hour intervals and transferred to a secondary collection tube. Secondary collections were checked for eclosed adults at 24 hour intervals. Time to pupation revealed a non-significant trend (p=0.069), with heterozygous and homozygous mutant progeny having mean pupation times of 6.07 and 6.38 days, respectively ( Fig. 2A, left). A significant delay was observed for time to eclosion (p<0.001), with heterozygous and homozygous mutant progeny having mean eclosion times of 10.78 and 11.70 days, respectively (Fig. 2A, right).
Fig. 2.
Ddx1null flies show delayed development (A) and maternally contributed Ddx1 protein in larvae (B). (A) Ddx1 heterozygote crosses were allowed to lay eggs for 24 hours. Pupae were removed at 24 hour intervals and scored for Ddx1 genotype (left, n=233 for heterozygous pupae and n=104 for homozygous mutant pupae). Adults were also counted at 24 hour intervals (right, n=162 for heterozygous adults and n=44 for homozygous mutant adults). A non-significant trend was observed for pupation time, and a significant difference was observed for eclosion time. (B) Control and Ddx1AX heterozygote crosses were allowed to lay eggs on apple juice plates for a period of two hours. Protein lysates were prepared from GFP sorted (for Ddx1AX/+ and Ddx1AX/AX progeny) and control larvae collected at 24, 48 or 72 hours post-egg laying. Western blot analysis was carried out using anti-Ddx1 (GenScript) and anti-β-tubulin antibodies (E7, DSHB). A faint Ddx1 signal was observed at both 24 and 48 hours post-egg laying, indicating that maternally loaded Ddx1 is still present at these times. Ddx1 was no longer detected at 72 hours.
In order to determine if maternally loaded Ddx1 protein might be compensating for the mutant allele during early development, we examined Ddx1 protein levels in larvae at 24, 48 and 72 hours post egg laying. Significant levels of Ddx1 protein were observed in Ddx1 AX/AX larvae at 24 hours, with a weak signal detected as late as 48 hours post egg laying (Fig. 2B). These results suggest a significant amount of maternally deposited protein with a long half-life. This may explain why Ddx1 AX/AX flies only display phenotypes at later developmental stages.
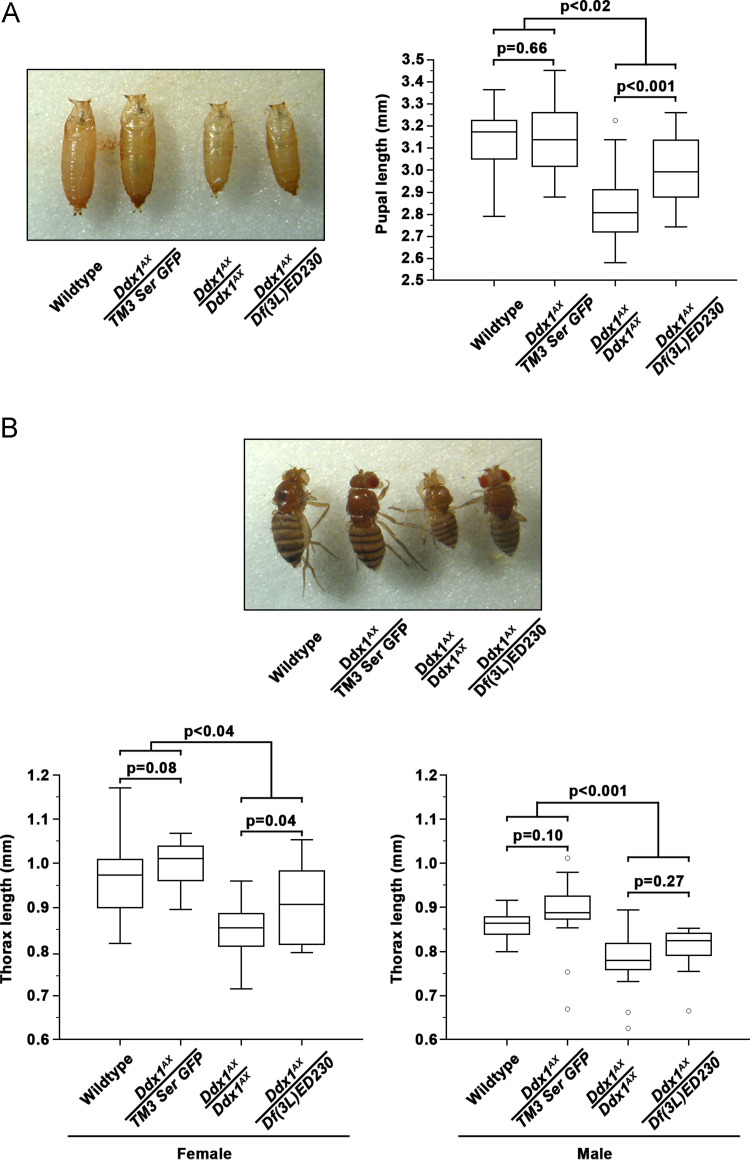
Comparison of control, heterozygous and mutant pupal lengths revealed a significant reduction in size in both Ddx1 AX/AXand Ddx1 AX /Df(3L)ED230 flies ( Fig. 3A). Of note, though both null strains were significantly smaller than control or heterozygous animals, the Ddx1 AX /Df(3L)ED230 pupae were slightly, though significantly (p<0.001), larger than Ddx1 AX/AX pupae. Similar results were observed upon measuring adult length (Fig. 3B). Adult Ddx1 null flies were smaller than control or heterozygous flies, and Ddx1 AX /Df(3L)ED230 adults were slightly, though significantly (p<0.05), larger than Ddx1 AX/AX adults.
Fig. 3.
Ddx1null flies are smaller than control. (A) Control, Ddx1 heterozygous and Ddx1 homozygous mutant pupae were collected and total pupal length was measured, n≥20 pupae for each sample. (B) Control, Ddx1 heterozygous and Ddx1 homozygous mutant one day old adults were collected and thorax length was measured, n≥19 adults for each sample. At both pupal and adult stages, Ddx1 null animals were significantly smaller than control animals, and Ddx1AX/Df(3L)ED230 animals were slightly larger than Ddx1AX/AX.
3.3. Gametogenesis is disrupted in Ddx1 null flies
To investigate the cause of reduced fertility in Ddx1 null flies, we dissected ovaries and testes from adult flies which had been held in isolation for 3 or 10 days following eclosion. At 3 days post-eclosion, heterozygous ovaries appeared essentially identical to control, containing many mature eggs. Ovaries from both Ddx1 AX/AX and Ddx1 AX /Df(3L)ED230 flies were much smaller, with Ddx1 AX /Df(3L)ED230 ovaries occasionally containing a small number of mature eggs and Ddx1 AX/AX ovaries containing no eggs ( Fig. 4A).
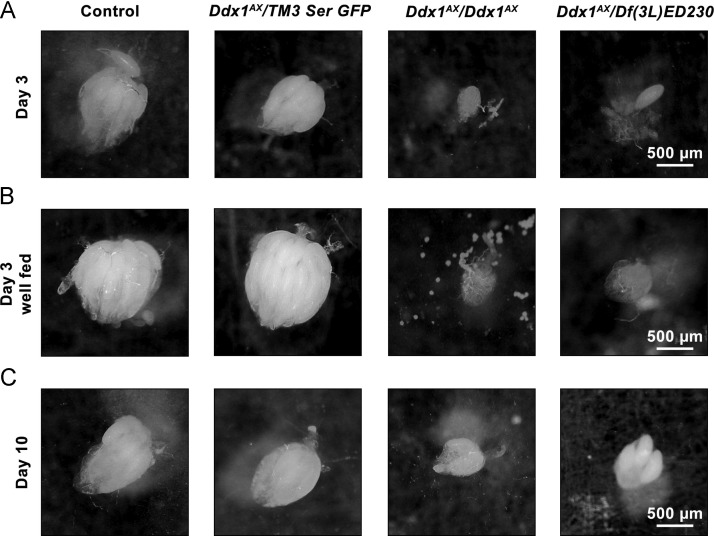
Fig. 4.
Gross ovary phenotypes inDdx1null flies. Ovaries collected from virgin females held in isolation for 3 days (A) or 10 days (C), or held with males for 3 days and supplemented with wet yeast paste (B). Under both conditions at day 3, Ddx1 null flies have much smaller ovaries, with few (Ddx1AX/Df(3L)ED230) or no (Ddx1AX/AX) mature eggs present (A and B). By day 10, a small number of full size mature eggs lacking dorsal appendages are present in Ddx1AX/AXovaries.
As holding flies in isolation on standard media can negatively impact oogenesis (Chapman and Partridge, 1996, Soller et al., 1997), ovaries were isolated from females that were supplemented with yeast paste and allowed to mate. Well-fed mated control and Ddx1 heterozygous females had notably larger ovaries than those kept in isolation, whereas Ddx1 AX/AX and Ddx1 AX /Df(3L)ED230 ovaries were similar to those from females kept in isolation (Fig. 4B). Similar results were observed with ovaries isolated from adults held in isolation for 10 days post eclosion, with Ddx1 AX /Df(3L)ED230 ovaries containing few mature eggs and some Ddx1 AX/AX ovaries containing a small number of abnormal eggs that were approximately the size of a mature egg, but lacked dorsal appendages (Fig. 4C).
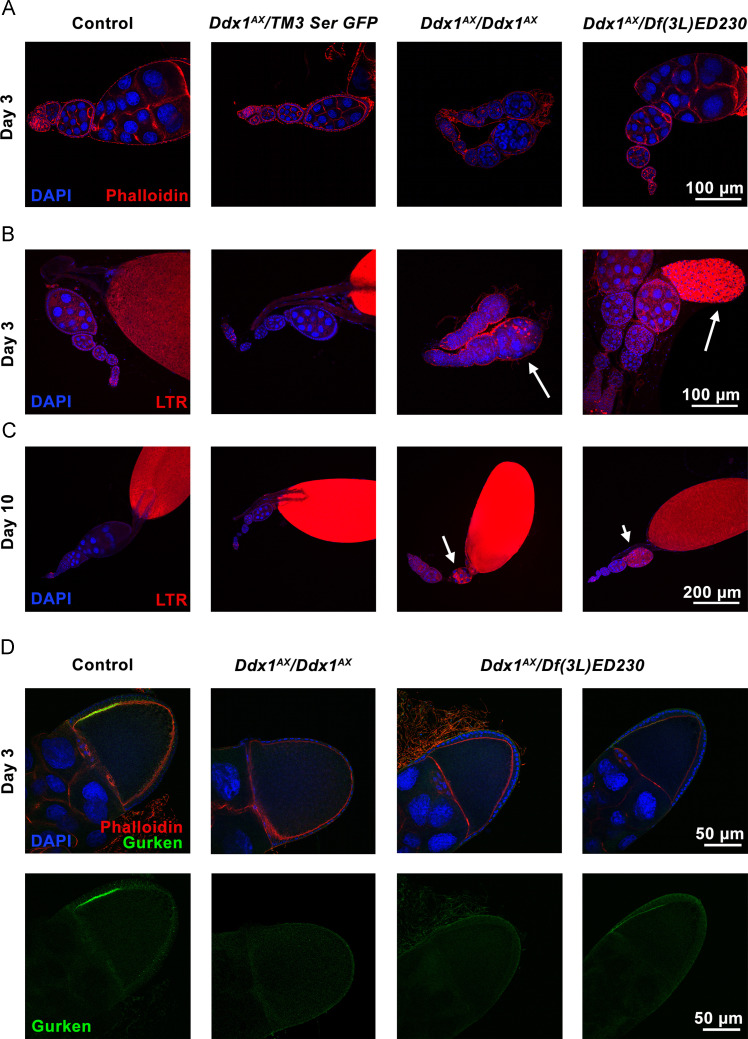
The reduced size of Ddx1 null flies is likely related to metabolism dysfunction. If this is the case, we would expect to see other phenotypes associated with reduced metabolism. When under metabolic stress or starvation conditions, developing egg chambers undergo autophagy in order to conserve energy (Barth et al., 2011, McCall, 2004). To determine if developing egg chambers in Ddx1 null flies are undergoing autophagy, we carried out immunofluorescence analysis of control and Ddx1 null ovarioles. Early stage egg chambers and germ-line stem cell niches appeared normal, indicating that the early stages of oogenesis are not disrupted in Ddx1 null flies ( Fig. 5A). Next, we stained dissected ovarioles with LysoTracker Red, a dye that is commonly used to identify autophagic cells (DeVorkin and Gorski, 2014). In Ddx1 null ovaries, at both 3 and 10 days post eclosion, we observed mid-stage autophagic egg chambers (Fig. 5B-C, arrows). Autophagic egg chambers were not observed in control and heterozygous ovaries. As mature egg chambers lacking dorsal appendages were observed in Ddx1 null ovaries, we imaged stage 8-9 egg chambers of control and Ddx1 null ovarioles for the localization of Gurken, a key factor in dorsal appendage development which localizes to the anterior dorsal corner of mid-stage oocytes (Neuman-Silberberg and Schupbach, 1996, Schupbach, 1987) (Fig. 5D). As expected, control egg chambers displayed normal Gurken localization. While a subset of egg chambers in Ddx1 AX /Df(3L)ED230 ovaries (approximately half) showed the expected Gurken localization pattern, the remaining Ddx1 AX /Df(3L)ED230 egg chambers and all egg chambers present in Ddx1 AX/AX ovaries lacked any notable Gurken expression pattern.
Fig. 5.
Aberrant development ofDdx1null egg chambers. Immunofluorescence imaging of ovaries with DAPI and Alexa 546-conjugated phalloidin (A), DAPI and LysoTracker Red (B and C), and DAPI, Alexa 546-conjugated phalloidin and anti-Gurken antibody (D). Developing egg chambers in Ddx1 null ovaries appear normal at early stages (A); however, LysoTracker Red staining reveals egg chambers undergoing autophagy (B and C, white arrows). (D) Immunostaining of stage 8-9 egg chambers with anti-Gurken antibody shows a normal Gurken pattern in control and some Ddx1AX/Df(3L)ED230 egg chambers, with absent Gurken signal in the remaining Ddx1AX/Df(3L)ED230 and all Ddx1AX/AX egg chambers.
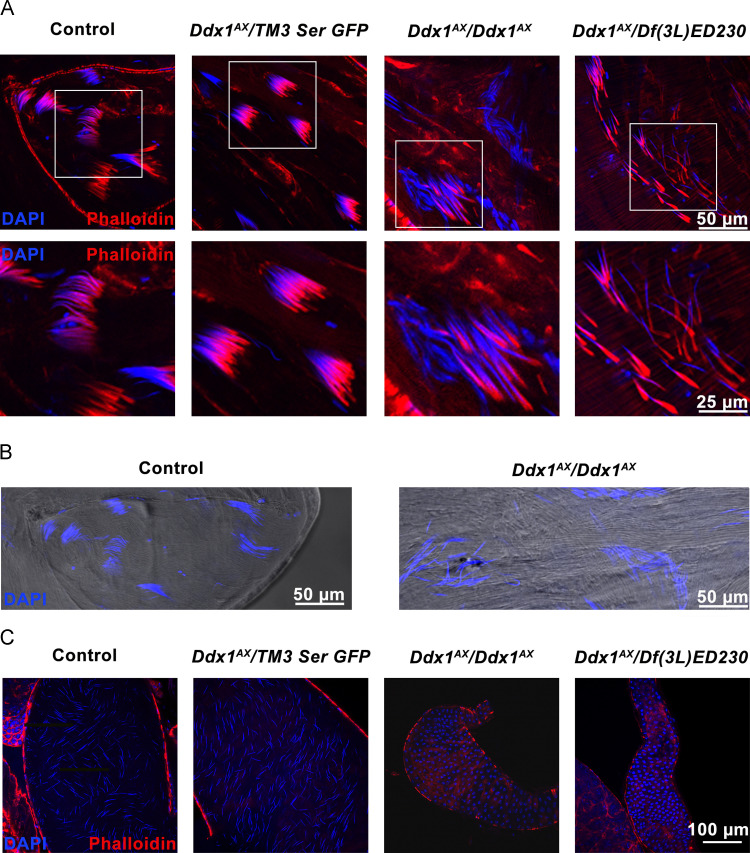
Immunofluorescence imaging of testes isolated from adult males revealed aberrant sperm development in Ddx1 null males ( Fig. 6A). Early spermatogenesis appeared unaffected in Ddx1 null males, with developing spermatids undergoing nuclear elongation (Fig. 6B). However, elongated spermatids were disrupted and scattered. To determine if mature sperm were being produced, seminal vesicles were isolated and imaged (Fig. 6C). In contrast to control and heterozygous seminal vesicles, which contained mature sperm, we did not observe any mature sperm in the seminal vesicles of homozygous mutant males.
Fig. 6.
Aberrant spermiogenesis inDdx1null testes. Analysis of testes and seminal vesicles collected from male flies held in isolation for 3 days. (A) Confocal microscopy analysis of testes from control, Ddx1 heterozygous and Ddx1 null adult male flies stained with Alexa 546-conjugated phalloidin and DAPI. The area outlined in the top diagram is magnified in the bottom diagram. (B) DIC imaging of testes from control and Ddx1 null adult male flies. (C) Confocal microscopy analysis of seminal vesicles from control, Ddx1 heterozygous and Ddx1 null adult male flies stained with Alex 546-conjugated phalloidin and DAPI. Ddx1 null testes show disordered spermatid cysts with elongated spermatid tails. No mature sperm are observed in Ddx1 null seminal vesicles.
3.4. Ddx1 null flies have reduced pS6k levels, but normal tRNA levels
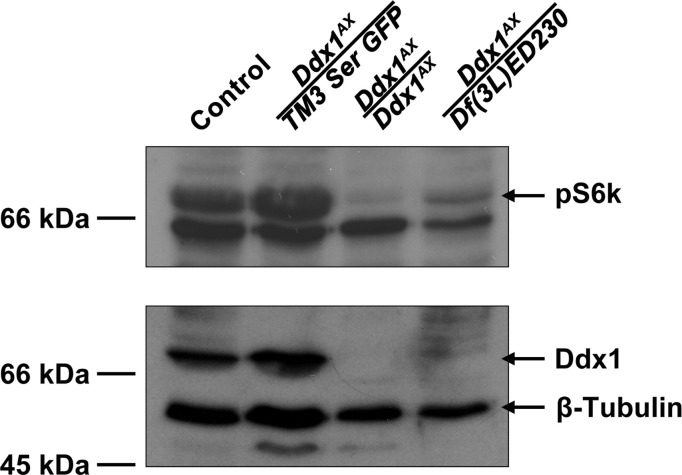
As both reduced body size and autophagy of developing oocytes are phenotypes associated with reduced metabolism (Barth et al., 2011, Edgar, 2006), we carried out western blot analysis using an antibody that recognizes the phosphorylated form of S6k, a downstream effector molecule of TOR and a common marker of active growth (Montagne et al., 1999). Phospho-S6k is down-regulated under starvation conditions (Hara et al., 1998, Zhang et al., 2000). pS6k levels were notably reduced in Ddx1 null flies at the 3rd instar stage compared to control and heterozygous animals ( Fig. 7). Notably, the pS6k signal observed in Ddx1 AX /Df(3L)ED230 larvae was stronger than that in Ddx1 AX/AX larvae. This is consistent with our previous results demonstrating that the Ddx1 AX /Df(3L)ED230 phenotype is slightly attenuated compared to Ddx1 AX/AX flies.
Fig. 7.
Reduced pS6k levels inDdx1null flies. Western blot analysis of cell lysates prepared from control, Ddx1 heterozygous and Ddx1 null 3rd instars. pS6k levels are reduced in the Ddx1 null lines, but slightly elevated in Ddx1AX/Df(3L)ED230 larvae as compared to Ddx1AX/AX.
As human DDX1 has recently been implicated in tRNA splicing, and pS6K levels are associated with RNA Pol III activity (Marshall et al., 2012), we used northern blot analysis to examine tRNAtyr (an intron-containing tRNA) and tRNAala (which contains no intron) levels in control, heterozygous and homozygous mutants. Our analysis showed no notable difference in the relative amounts of a mature tRNA that requires splicing (tRNAtyr) compared to one that does not require splicing (tRNAala). As well, there was no notable difference in relative tRNA to 5S rRNA levels (Fig. S1).
3.5. Ddx1 null animals display widespread changes in mRNA levels and splicing
DEAD box proteins have been widely implicated in the generation, maturation and degradation of RNA molecules. Therefore, we undertook RNA deep sequencing to determine the effect of knocking-out Ddx1 on the transcriptome. We utilized RNA isolated from control and Ddx1 null 3rd instars, as phenotypes are present at this point and wandering 3rd instars can be easily staged in spite of developmental delays associated with Ddx1 null mutants. We identified 72 significantly down-regulated and 261 significantly up-regulated transcripts, using a cut-off of p<0.01 (Table S1). Of note, we found that RNA is expressed from the portion of the Ddx1 AX allele retained in the genome of our Ddx1 null flies, although RNA levels were significantly reduced compared to that found in control flies. Further analysis of the region directly upstream of the Ddx1 AX allele suggests that transcription is initiated approximately 150 bp upstream of the deletion site. Immediately upstream of this putative transcriptional start site is a variant TATA box sequence (CATAAAG) that may act as a cryptic transcription initiation site in the absence of the normal start site.
We also analyzed transcripts for the presence of differentially spliced variants between Ddx1 AX/AX and control flies (Table S2). We limited our analysis to the small number of transcripts that displayed unique splice variants, with high levels in all three replicates of one genotype and absence in all three replicates of the other genotype (Tables S3-S4). It is important to note that splice site analysis must be considered within the context of total RNA levels, as a gene that is only expressed in one genotype will necessarily have splice junctions that are unique to this genotype. Using these criteria, we identified a number of splice variants uniquely found in either control or Ddx1 AX/AX flies, including Sirup. Gene ontology clustering analysis of genes with modified expression or modified splicing was uninformative.
3.6. Ddx1 interacts with Sirup mRNA
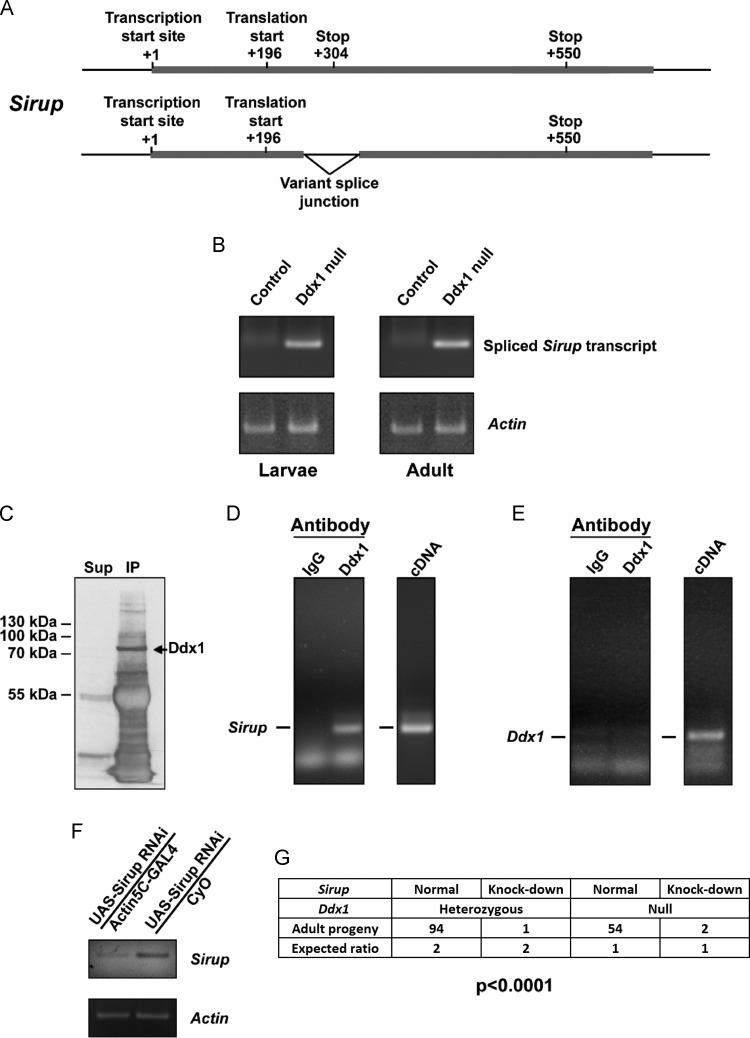
Sirup RNA has previously been shown to be up-regulated in response to starvation conditions (Erdi et al., 2012) and is required for proper mitochondrial function (Van Vranken et al., 2014). We observed up-regulation of Sirup (by 3-fold) in Ddx1 AX/AX versus control flies (Table 1). However, a much stronger effect was observed on Sirup splicing, with sequences spanning the 234-315 bp junction found uniquely and at high levels in Ddx1 null flies. Sirup is expressed as a single exon or as an alternatively spliced product characterized by the removal of a small intron located downstream of the start codon ( Fig. 8A). Using a primer that spans the junction splice site, we confirmed that Sirup displays alternative splicing in control and null flies, with Ddx1 control larvae and adult flies expressing unspliced Sirup, and Ddx1 AX/AX larvae and adults expressing spliced Sirup (Fig. 8B).
Fig. 8.
Interactions between Ddx1andSirup. (A) Pictograph showing the structure of Sirup and the variable splice junction site (top – transcript expressed in control flies; bottom – transcript expressed in Ddx1AX/AX flies). (B) RT-PCR analysis of control and null larvae and adult flies. A spliced Sirup product is only observed in Ddx1 null animals. (C) IP using anti-Ddx1 antibody demonstrating that almost all Ddx1 protein is retained in the immunoprecipitate. (D, E) Left panels – RT-PCR analysis of RNA co-immunoprecipitated with Ddx1 or IgG. Right panels – RT-PCR of cDNA generated from control flies used as a positive control for the PCR reaction. Sirup mRNA results indicate that Sirup RNA, but not Ddx1 RNA, is pulled down with Ddx1 protein. (F) RT-PCR analysis shows reduced Sirup RNA levels in Sirup knock-down adult flies. (G) Progeny generated from y1w*; Actin5C-Gal4/CyO; Ddx1AX/TM3, Sb x y1w*; UAS-Sirup-RNAi; Ddx1AX/TM3, Ser GFP crosses. Expected ratios of 2:1 for heterozygous to homozygous mutant Ddx1 and 1:1 for Sirup knock-down to CyO. A significant reduction in the number of Sirup knock-down flies was observed. Chi square analysis comparing observed distribution to expected distribution was used to determine significance.
The Sirup splice variant observed in control flies results in a longer transcript (810 nt) that retains the gene’s single intron, but a shorter open reading frame (36 aa) due to a stop codon within the intron. In Ddx1 AX/AXflies, the intron is removed, generating a shorter transcript (662 nt) but a longer open reading frame (118 aa). Intriguingly, this splice-specific regulation may be conserved in humans, as the human Sirup orthologue C6orf57 expresses two splice variants, one encoding a 108 aa protein and the other being non-coding, mirroring what we observe in Drosophila.
As Ddx1 has previously been shown to bind target RNAs, it is possible that Sirup RNA is a direct target of Ddx1. Alternatively, modified splicing of Sirup may be an indirect consequence of Ddx1 mutation. To distinguish between these possibilities, we performed RNA co-immunoprecipitations (co-IPs) using a Ddx1-specific antibody to immunoprecipitate Ddx1-binding RNAs from Schneider2 cell lysates. Using this approach, we were able to efficiently co-immunoprecipitate Ddx1 protein, as determined by western blotting, along with Sirup mRNA, but not Ddx1 mRNA, as detected by RT-PCR (Fig. 8C-E). Ddx1 mRNA was included as a negative control to ensure that the interaction between Sirup mRNA and Ddx1 protein was specific, and not the result of non-specific RNA interaction.
Next, we tested whether there is genetic interaction between Ddx1 and Sirup. We used the UAS-GAL4 system (Actin5C-GAL4 and UAS-Sirup RNAi [P{GD14644}v36437]) to generate flies that expressed a Sirup siRNA construct in Ddx1 normal, Ddx1 heterozygous and Ddx1 null flies. Sirup knock-down in a control background reduced Sirup RNA levels by approximately 75% (Fig. 8F), and generated viable, fertile flies that displayed no observable phenotypes. Sirup knock-down in both Ddx1 heterozygous and null backgrounds resulted in lethality in the majority of progeny. Although 50% of the progeny were expected to express Sirup siRNA, we found that the majority of Ddx1 mutant progeny generated from these crosses did not contain the Act5C-GAL4 transgene (with 3 mutant progeny containing both the Actin5C-GAL4 driver and the UAS-Sirup transgene versus 148 containing only the UAS-Sirup transgene) (Fig. 8G). Of note, the two Ddx1 null/Sirup knock-down progeny survived less than 1 day post eclosion, whereas the single Ddx1 heterozygous/Sirup knock-down progeny survived 14 days post eclosion.
4. Discussion
While human DDX1 has been implicated in numerous biological processes in cultured cells, we still have a poor understanding of its role in animal development. Towards this end, we generated a Ddx1 null mutant in Drosophila. In contrast to a report listing a previous Ddx1 mutation (Ddx1 9W1) as lethal (Zinsmaier et al., 1994), we found that Ddx1 null flies were viable. Although our results suggest that Ddx1 is not essential for embryonic development, it is important to note that significant levels of maternal Ddx1 protein are detected up to the 2nd instar stage. As Ddx1 levels are highest in early embryogenesis (Rafti et al., 1996), this maternal contribution is likely sufficient to fulfill an essential need for Ddx1 in early development, postponing the appearance of Ddx1 phenotypes until later stages in Ddx1 AX/AX flies.
The Ddx1 9W1 mutant described by Zinsmaier et al. (Zinsmaier et al., 1994) was generated using a P-element associated with CSP located downstream of Ddx1. The authors indirectly inferred that the described allele affected Ddx1; however, unknown at the time, there is another gene between Ddx1 and CSP, CG11523. A subsequent report showed that disrupted CG11523 is associated with a lethal phenotype (CG11523 GD7321) (Mummery-Widmer et al., 2009). It is, therefore, likely that the effect attributed to Ddx1 mutation in 1994 was actually due to disruption of CG11523. Alternatively, the lethal phenotype observed by Zinsmaier et al. may be the result of a truncated Ddx1 product, as the location of the P-element used to generate the Ddx1 9W1 allele is such that deletions would likely result in a truncation. A truncated gene product could cause a divergent phenotype from that of a null allele. We are unable to test these possibilities experimentally, as the original stock is no longer available.
Although Ddx1 null flies survive to adulthood, it is clear that Ddx1 is involved in a wide spectrum of developmental functions. Ddx1 null flies are significantly reduced in size, developmentally delayed and out-competed by heterozygous animals when raised in crowded conditions. In addition, Ddx1 null flies are infertile due to aberrant gametogenesis, with both oogenesis and spermatogenesis being affected. We observed significantly smaller ovaries containing few to no mature eggs in Ddx1 null flies. One possible explanation for reduced ovary size is disruption of stem cell maintenance and/or division; however, mutants in which the stem cell pool is affected typically display reduced numbers of early egg chambers (Li et al., 2009, Shen et al., 2009). Ddx1 null ovaries appear to have normal numbers of egg chambers, suggesting that regulation of stem cell niche is not the cause of the small ovary phenotype.
We also observed autophagic egg chambers in ovaries isolated from Ddx1 null flies. Autophagic egg chambers, in addition to small body size, are indicative of metabolic stress and starvation conditions (Barth et al., 2011, Edgar, 2006, McCall, 2004). The phenotypes observed in Ddx1 null flies therefore suggest a role for Ddx1 in metabolic function and/or regulation, with absence of Ddx1 phenocopying nutrient deficiency rather than stem cell deficiency. While our attempts to generate a transgenic Ddx1 over-expression line to rescue this phenotype were not successful, we were able to confirm comparable phenotypes in flies carrying the Ddx1 AX allele over a deficiency that encompasses the Ddx1 locus.
In addition to autophagic egg chambers, we also noted mispatterned mature eggs lacking dorsal appendages in Ddx1 null ovaries. Dorsal appendage formation is a complicated process involving more than 60 genes (Berg, 2005). We performed immunofluorescence analysis of Gurken localization, a key factor in dorsal appendage development, and observed loss of Gurken at the anterior dorsal corner in stage 8-9 egg chambers. Our RNA deep-sequencing did not identify any of the well-characterized Gurken localization factors as being differentially expressed in Ddx1 control versus null flies. Thus, in Ddx1 null flies, disruption of gurken translation, rather than gurken RNA/protein localization, may underlie the absence of Gurken protein in the anterior dorsal corner. Intriguingly, it has been shown that the DNA damage response and small RNA processing are required for proper Gurken translation (Chen et al., 2007, Klattenhoff et al., 2007). Human DDX1 has previously been implicated in both the DNA damage response and small RNA processing (Han et al., 2014, Li et al., 2008). The patterning defect observed in the few Ddx1 null eggs that do not undergo autophagy may therefore represent a secondary phenotype unrelated to metabolic disruption.
In keeping with a role for Ddx1 in metabolism, we found reduced pS6k levels in Ddx1 null flies. pS6k levels are indicative of TOR signalling, which is modified by a number of pathways including insulin signalling and nutrient sensing (Edgar, 2006). Signalling through TOR and pS6K drives protein synthesis and cell growth (Zhang et al., 2000). Our RNA deep sequencing data indicate that neither TOR nor S6k mRNA levels are significantly affected in Ddx1 AX/AX flies. RNA levels of major upstream regulators of the TOR pathway, including Rheb, Tsc1 and gigas (dTSC2) are also unchanged. The consistent levels of these key signalling factors suggest that the absence of Ddx1 does not directly affect the transcripts encoding proteins that participate in TOR signalling. Rather, absence of Ddx1 may modulate the function of RNA targets that function upstream of the TOR signalling pathway.
In contrast to other DEAD box proteins that have been directly implicated in Drosophila oogenesis (Cauchi, 2012, Johnstone et al., 2005, Styhler et al., 1998, Tomancak et al., 1998), we propose that the infertility observed in Ddx1 null females is due to disrupted metabolism. Altered metabolism also provides a possible explanation for the small number of progeny generated by Ddx1 AX /Df(3L)ED230 females, as we observed slightly higher pS6k levels and slightly larger body size in these mutants compared to the completely sterile Ddx1 AX/AX females. Thus, Ddx1AX /Df(3L)ED230 flies may have slightly higher metabolic function than Ddx1 AX/AX flies. This may be due to second-site alleles that are different between the Ddx1 AX and Df(3L)ED230 genetic background.
Spermatogenesis can also be affected by starvation conditions, with a noted reduction in the number of germinal stem cells in starved males (McLeod et al., 2010). In contrast to the phenotype observed in Ddx1 null ovaries, aberrant sperm individualization, the phenotype observed in Ddx1 null males, has not been associated with metabolic disruption. Following meiosis, spermiogenesis, the last stage of spermatogenesis, involves reshaping developing interconnected spermatids into individual mature sperm (Fabian and Brill, 2012). Spermiogenesis is characterized by gross morphological changes to the cell and mitochondria in particular. Developing spermatids in Ddx1 null males appear to undergo spermatid elongation, but fail to individualize. Instead, they fall out of the developing sperm bundle prematurely and seminal vesicles remain devoid of mature sperm.
Several DEAD box proteins have been associated with defects in sperm development. RecQ5 and Belle are both required for early spermatogenesis (Johnstone et al., 2005, Sakurai et al., 2014) and mutation of Rm62 has also been shown to cause male sterility (Buszczak et al., 2007). While the cause of sterility in Rm62 mutants has not been identified, mutation of Blanks, a binding partner for Rm62, results in spermiogenesis defects similar to those observed in Ddx1 null males (Gerbasi et al., 2011). As disrupted sperm individualization has not been linked to metabolic function, we postulate that this aspect of the Ddx1 null phenotype is the result of disruption of distinct RNA targets from those involved in metabolic function.
It has been previously established that tRNA synthesis is regulated by TOR signalling (Ciesla and Boguta, 2008) and human DDX1 has been recently identified as a tRNA splicing factor (Popow et al., 2011, Popow et al., 2014). As our analysis did not show changes in levels of spliced and unspliced tRNAs in Ddx1 null flies, it would appear that Drosophila Ddx1 is not essential for tRNA splicing. However, our northern blot analysis is only a snapshot of tRNA levels at the time of RNA isolation. Popow et al. showed that human DDX1 is necessary for efficient cycling of the RtcB-guanylate intermediate required for tRNA splicing in vitro (Popow et al., 2014). It is possible that tRNA generation efficiency is reduced in Ddx1 AX/AX flies, but a compensatory mechanism is coming into play. Alternatively, the reduction in efficiency of tRNA splicing may not be sufficient to reduce their overall steady state levels and/or our method of analysis may not be sufficiently sensitive to detect the changes in tRNA levels.
Our RNA deep sequencing analysis revealed changes in the levels of 333 RNA molecules in Ddx1 AX/AX larvae. In addition, we identified a number of transcripts that were differentially spliced in Ddx1 control versus null flies. We verified that Sirup mRNA is uniquely spliced in Ddx1 AX/AX larvae and up-regulated. We were also able to show that Ddx1 protein binds Sirup mRNA, which suggests that this splice site modification is due to a direct interaction with Ddx1, rather than being an indirect downstream consequence of the loss of Ddx1. Previous work has demonstrated that loss of Sirup results in shortened life span and neurodegeneration (Van Vranken et al., 2014). In agreement with our results, Sirup has also previously been shown to be up-regulated in response to starvation conditions (Erdi et al., 2012). We propose a role for Sirup in limiting metabolism during stress conditions. Sirup has recently been identified as the Drosophila homolog of yeast Sdh8 (Van Vranken et al., 2014). Similar to its yeast homologue, Drosophila Sirup may be required to stabilize the succinate dehydrogenase holocomplex, and enhance succinate dehydrogenase activity.
We observed spliced Sirup mRNA only in the absence of Ddx1, and propose that Ddx1 plays a role in the repression of splicing under normal conditions. This is in line with observations that human DDX1 is required for efficient trafficking of unspliced viral RNA genomes (Edgcomb et al., 2012, Fang et al., 2005, Robertson-Anderson et al., 2011). There are several possible mechanisms for splicing repression by Ddx1. A simple explanation could be that Ddx1 is remodeling RNA secondary structure into a form that is refractory to splicing. Alternatively, recent studies have shown a strong relationship between antisense non-coding RNAs (ncRNAs) and alternative splicing in humans (Morrissy et al., 2011). As DDX1 has RNA/RNA unwinding activity, Drosophila Ddx1 may act by unwinding dsRNA formed by ncRNAs bound to mRNA molecules. While it is still not clear whether ncRNAs promote or repress specific splice sites, it is possible that they may act in either fashion, depending on where they bind to target RNAs.
Although the relationship between Ddx1-dependent splice regulation of Sirup mRNA and the metabolic disruption observed in Ddx1 null flies remains a matter of speculation, Sirup’s known role in mitochondrial function provides a possible explanation for the epistatic lethality observed in Ddx1 null/Sirup knock-down flies. We propose that Sirup’s unspliced form is required for steady state metabolic function. The unspliced form of Sirup retains an early stop codon that produces a transcript that encodes a short 36 aa product missing conserved domains required for succinate dehydrogenase activity. However, there is evidence suggesting that this stop codon undergoes read-through by ribosomes, thereby generating a 145 aa length protein in spite of the stop codon (Dunn et al., 2013). Under stress conditions, suppression of Ddx1 activity may promote splicing of Sirup mRNA thereby generating a shorter 118 aa product that has reduced succinate dehydrogenase activity. The resulting slowdown in mitochondrial function would reduce resource usage by the cell, allowing it to survive periods of nutrient scarcity. Under this model, the observed epistatic lethality in Ddx1 AX/AX/Sirup knock-down flies would be the result of a reduction in the amount of Sirup mRNA expressed combined with production of the less efficient form of Sirup protein.
In conclusion, the phenotypes resulting from loss of Ddx1 expression in Drosophila are in keeping with Ddx1 being a multifunctional protein involved in a variety of biological processes. Our experiments suggest a novel role for Ddx1 in metabolism regulation mediated through interaction and modification of transcripts such as Sirup which encode products that can directly modulate metabolic activity.
Author Contributions
DG, LL and MH generated the data. DG performed the data analysis and prepared the figures. DG, SH and AS contributed to the design of the experiments. DG and RG wrote the manuscript with input from SH and AS.
Acknowledgments
We are grateful to Dr. Xuejun Sun and Gerry Barron for their technical assistance with cell imaging. We thank Dr. Michael Hendzel and Dr. Zhigang Jin for helpful discussions. This study was supported by a grant from the Alberta Cancer Foundation.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2015.09.012.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko J.M., Arteaga C.L. Dead-box or black-box: is DDX1 a potential biomarker in breast cancer? Breast Cancer Res Treat. 2011;127:65–67. doi: 10.1007/s10549-010-1105-7. [DOI] [PubMed] [Google Scholar]
- Barth J.M., Szabad J., Hafen E., Kohler K. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ. 2011;18:915–924. doi: 10.1038/cdd.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg C.A. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 2005;21:346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Bleoo S., Sun X., Hendzel M.J., Rowe J.M., Packer M., Godbout R. Association of human DEAD box protein DDX1 with a cleavage stimulation factor involved in 3’-end processing of pre-MRNA. Mol Biol Cell. 2001;12:3046–3059. doi: 10.1091/mbc.12.10.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A.D., Nystul T.G., Ohlstein B., Allen A., Wilhelm J.E., Murphy T.D., Levis R.W., Matunis E., Srivali N., Hoskins R.A., Spradling A.C. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchi R.J. Conserved requirement for DEAD-box RNA helicase Gemin3 in Drosophila oogenesis. BMC Res Notes. 2012;5:120. doi: 10.1186/1756-0500-5-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T., Partridge L. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc Biol Sci. 1996;263:755–759. doi: 10.1098/rspb.1996.0113. [DOI] [PubMed] [Google Scholar]
- Chen Y., Pane A., Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla M., Boguta M. Regulation of RNA polymerase III transcription by Maf1 protein. Acta Biochim Pol. 2008;55:215–225. [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- DeVorkin L., Gorski S.M. LysoTracker staining to aid in monitoring autophagy in Drosophila. Cold Spring Harb Protoc. 2014;2014:951–958. doi: 10.1101/pdb.prot080325. [DOI] [PubMed] [Google Scholar]
- Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Edgcomb S.P., Carmel A.B., Naji S., Ambrus-Aikelin G., Reyes J.R., Saphire A.C., Gerace L., Williamson J.R. DDX1 is an RNA-dependent ATPase involved in HIV-1 Rev function and virus replication. J Mol Biol. 2012;415:61–74. doi: 10.1016/j.jmb.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdi B., Nagy P., Zvara A., Varga A., Pircs K., Menesi D., Puskas L.G., Juhasz G. Loss of the starvation-induced gene Rack1 leads to glycogen deficiency and impaired autophagic responses in Drosophila. Autophagy. 2012;8:1124–1135. doi: 10.4161/auto.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian L., Brill J.A. Drosophila spermiogenesis: Big things come from little packages. Spermatogenesis. 2012;2:197–212. doi: 10.4161/spmg.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Acheampong E., Dave R., Wang F., Mukhtar M., Pomerantz R.J. The RNA helicase DDX1 is involved in restricted HIV-1 Rev function in human astrocytes. Virology. 2005;336:299–307. doi: 10.1016/j.virol.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Gerbasi V.R., Preall J.B., Golden D.E., Powell D.W., Cummins T.D., Sontheimer E.J. Blanks, a nuclear siRNA/dsRNA-binding complex component, is required for Drosophila spermiogenesis. Proc Natl Acad Sci U S A. 2011;108:3204–3209. doi: 10.1073/pnas.1009781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain D.R., Graham K., Glubrecht D.D., Hugh J.C., Mackey J.R., Godbout R. DEAD box 1: a novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res Treat. 2011;127:53–63. doi: 10.1007/s10549-010-0943-7. [DOI] [PubMed] [Google Scholar]
- Godbout R., Packer M., Bie W. Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J Biol Chem. 1998;273:21161–21168. doi: 10.1074/jbc.273.33.21161. [DOI] [PubMed] [Google Scholar]
- Godbout R., Squire J. Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc Natl Acad Sci U S A. 1993;90:7578–7582. doi: 10.1073/pnas.90.16.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Liu Y., Wan G., Choi H.J., Zhao L., Ivan C., He X., Sood A.K., Zhang X., Lu X. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 2014;8:1447–1460. doi: 10.1016/j.celrep.2014.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yonezawa K., Weng Q.P., Kozlowski M.T., Belham C., Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Johanneson B., Chen D., Enroth S., Cui T., Gyllensten U. Systematic validation of hypothesis-driven candidate genes for cervical cancer in a genome-wide association study. Carcinogenesis. 2014;35:2084–2088. doi: 10.1093/carcin/bgu125. [DOI] [PubMed] [Google Scholar]
- Johnstone O., Deuring R., Bock R., Linder P., Fuller M.T., Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol. 2005;277:92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kircher S.G., Kim S.H., Fountoulakis M., Lubec G. Reduced levels of DEAD-box proteins DBP-RB and p72 in fetal Down syndrome brains. Neurochem Res. 2002;27:1141–1146. doi: 10.1023/a:1020921324871. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C., Bratu D.P., McGinnis-Schultz N., Koppetsch B.S., Cook H.A., Theurkauf W.E. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Lasko P. The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim Biophys Acta. 2013;1829:810–816. doi: 10.1016/j.bbagrm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Li L., Monckton E.A., Godbout R. A role for DEAD box 1 at DNA double-strand breaks. Mol Cell Biol. 2008;28:6413–6425. doi: 10.1128/MCB.01053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Roy K., Katyal S., Sun X., Bleoo S., Godbout R. Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, Cajal bodies, and gems. Mol Biol Cell. 2006;17:1126–1140. doi: 10.1091/mbc.E05-08-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Minor N.T., Park J.K., McKearin D.M., Maines J.Z. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci U S A. 2009;106:9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.H., Sivakumaran H., Jones A., Li D., Harper C., Wei T., Jin H., Rustanti L., Meunier F.A., Spann K., Harrich D. A HIV-1 Tat mutant protein disrupts HIV-1 Rev function by targeting the DEAD-box RNA helicase DDX1. Retrovirology. 2014;11:121. doi: 10.1186/s12977-014-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Fuller-Pace F.V. Looking back on the birth of DEAD-box RNA helicases. Biochim Biophys Acta. 2013;1829:750–755. doi: 10.1016/j.bbagrm.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Linder P., Stutz F. mRNA export: travelling with DEAD box proteins. Curr Biol. 2001;11:R961–R963. doi: 10.1016/s0960-9822(01)00574-7. [DOI] [PubMed] [Google Scholar]
- Manohar C.F., Salwen H.R., Brodeur G.M., Cohn S.L. Co-amplification and concomitant high levels of expression of a DEAD box gene with MYCN in human neuroblastoma. Genes Chromosomes Cancer. 1995;14:196–203. doi: 10.1002/gcc.2870140307. [DOI] [PubMed] [Google Scholar]
- Marshall L., Rideout E.J., Grewal S.S. Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. EMBO J. 2012;31:1916–1930. doi: 10.1038/emboj.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K. Eggs over easy: cell death in the Drosophila ovary. Dev Biol. 2004;274:3–14. doi: 10.1016/j.ydbio.2004.07.017. [DOI] [PubMed] [Google Scholar]
- McLeod C.J., Wang L., Wong C., Jones D.L. Stem cell dynamics in response to nutrient availability. Curr Biol. 2010;20:2100–2105. doi: 10.1016/j.cub.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne J., Stewart M.J., Stocker H., Hafen E., Kozma S.C., Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- Montpetit B., Seeliger M.A., Weis K. Analysis of DEAD-box proteins in mRNA export. Methods Enzymol. 2012;511:239–254. doi: 10.1016/B978-0-12-396546-2.00011-5. [DOI] [PubMed] [Google Scholar]
- Morrissy A.S., Griffith M., Marra M.A. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 2011;21:1203–1212. doi: 10.1101/gr.113431.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery-Widmer J.L., Yamazaki M., Stoeger T., Novatchkova M., Bhalerao S., Chen D., Dietzl G., Dickson B.J., Knoblich J.A. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg F.S., Schupbach T. The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech Dev. 1996;59:105–113. doi: 10.1016/0925-4773(96)00567-9. [DOI] [PubMed] [Google Scholar]
- Pek J.W., Kai T. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr Biol. 2011;21:39–44. doi: 10.1016/j.cub.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C.L., Luhrmann R., Soll D., Martinez J. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- Popow J., Jurkin J., Schleiffer A., Martinez J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511:104–107. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafti F., Scarvelis D., Lasko P.F. A Drosophila melanogaster homologue of the human DEAD-box gene DDX1. Gene. 1996;171:225–229. doi: 10.1016/0378-1119(96)00034-0. [DOI] [PubMed] [Google Scholar]
- Robertson-Anderson R.M., Wang J., Edgcomb S.P., Carmel A.B., Williamson J.R., Millar D.P. Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element. J Mol Biol. 2011;410:959–971. doi: 10.1016/j.jmb.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Takai S., Kawamura K., Ogura Y., Yoshioka Y., Kawasaki K. Drosophila RecQ5 is involved in proper progression of early spermatogenesis. Biochem Biophys Res Commun. 2014;452:1071–1077. doi: 10.1016/j.bbrc.2014.09.051. [DOI] [PubMed] [Google Scholar]
- Schupbach T. Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell. 1987;49:699–707. doi: 10.1016/0092-8674(87)90546-0. [DOI] [PubMed] [Google Scholar]
- Shen R., Weng C., Yu J., Xie T. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc Natl Acad Sci U S A. 2009;106:11623–11628. doi: 10.1073/pnas.0903325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolonska J., Koppelman G.H., Wijmenga C., Vonk J.M., Zanen P., Bruinenberg M., Curjuric I., Imboden M., Thun G.A., Franke L., Probst-Hensch N.M., Nurnberg P., Riemersma R.A., van Schayck C.P., Loth D.W., Brusselle G.G., Stricker B.H., Hofman A., Uitterlinden A.G., Lahousse L., London S.J., Loehr L.R., Manichaikul A., Barr R.G., Donohue K.M., Rich S.S., Pare P., Bosse Y., Hao K., van den Berge M., Groen H.J., Lammers J.W., Mali W., Boezen H.M., Postma D.S. Common genes underlying asthma and COPD? Genome-wide analysis on the Dutch hypothesis. Eur Respir J. 2014;44:860–872. doi: 10.1183/09031936.00001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M., Bownes M., Kubli E. Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster. Eur J Biochem. 1997;243:732–738. doi: 10.1111/j.1432-1033.1997.00732.x. [DOI] [PubMed] [Google Scholar]
- Squire J.A., Thorner P.S., Weitzman S., Maggi J.D., Dirks P., Doyle J., Hale M., Godbout R. Co-amplification of MYCN and a DEAD box gene (DDX1) in primary neuroblastoma. Oncogene. 1995;10:1417–1422. [PubMed] [Google Scholar]
- Pierre S.E., Ponting L., Stefancsik R., McQuilton P. FlyBase 102--advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42:D780–D788. doi: 10.1093/nar/gkt1092. St. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S., Nakamura A., Swan A., Suter B., Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Sunden Y., Semba S., Suzuki T., Okada Y., Orba Y., Nagashima K., Umemura T., Sawa H. DDX1 promotes proliferation of the JC virus through transactivation of its promoter. Microbiol Immunol. 2007;51:339–347. doi: 10.1111/j.1348-0421.2007.tb03907.x. [DOI] [PubMed] [Google Scholar]
- Sunden Y., Semba S., Suzuki T., Okada Y., Orba Y., Nagashima K., Umemura T., Sawa H. Identification of DDX1 as a JC virus transcriptional control region-binding protein. Microbiol Immunol. 2007;51:327–337. doi: 10.1111/j.1348-0421.2007.tb03915.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Okamoto S., Ishikawa Y., Tamura H., Hara T. DDX1 is required for testicular tumorigenesis, partially through the transcriptional activation of 12p stem cell genes. Oncogene. 2009;28:2142–2151. doi: 10.1038/onc.2009.89. [DOI] [PubMed] [Google Scholar]
- Tingting P., Caiyun F., Zhigang Y., Pengyuan Y., Zhenghong Y. Subproteomic analysis of the cellular proteins associated with the 3’ untranslated region of the hepatitis C virus genome in human liver cells. Biochem Biophys Res Commun. 2006;347:683–691. doi: 10.1016/j.bbrc.2006.06.144. [DOI] [PubMed] [Google Scholar]
- Tomancak P., Guichet A., Zavorszky P., Ephrussi A. Oocyte polarity depends on regulation of gurken by Vasa. Development. 1998;125:1723–1732. doi: 10.1242/dev.125.9.1723. [DOI] [PubMed] [Google Scholar]
- Van Vranken J.G., Bricker D.K., Dephoure N., Gygi S.P., Cox J.E., Thummel C.S., Rutter J. SDHAF4 promotes mitochondrial succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab. 2014;20:241–252. doi: 10.1016/j.cmet.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Chen P.J., Yeh S.H. Nucleocapsid Phosphorylation and RNA Helicase DDX1 Recruitment Enables Coronavirus Transition from Discontinuous to Continuous Transcription. Cell Host Microbe. 2014;16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Khadijah S., Fang S., Wang L., Tay F.P., Liu D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S., Chartier A., Gallant P., Astier M., Arquier N., Doherty D., Gratecos D., Semeriva M. A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development. 1998;125:3571–3584. doi: 10.1242/dev.125.18.3571. [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang J., Xu J., Zhang Z., Koppetsch B.S., Schultz N., Vreven T., Meignin C., Davis I., Zamore P.D., Weng Z., Theurkauf W.E. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Stallock J.P., Ng J.C., Reinhard C., Neufeld T.P. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmaier K.E., Eberle K.K., Buchner E., Walter N., Benzer S. Paralysis and early death in cysteine string protein mutants of Drosophila. Science. 1994;263:977–980. doi: 10.1126/science.8310297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material