Abstract
Background
The inflammatory response in community-acquired pneumonia (CAP) depends on the host and on the challenge of the causal microorganism. Here, we analyze the patterns of inflammatory cytokines, procalcitonin (PCT), and C-reactive protein (CRP) in order to determine their diagnostic value.
Methods
This was a prospective study of 658 patients admitted with CAP. PCT and CRP were analyzed by immunoluminometric and immunoturbidimetric assays. Cytokines (tumor necrosis factor-α [TNF-α], IL-1β, IL-6, IL-8, and IL-10) were measured using enzyme immunoassay.
Results
The lowest medians of CRP, PCT, TNF-α, and IL-6 were found in CAP of unknown cause, and the highest were found in patients with positive blood cultures. Different cytokine profiles and biomarkers were found depending on cause: atypical bacteria (lower PCT and IL-6), viruses (lower PCT and higher IL-10), Enterobacteriaceae (higher IL-8), Streptococcus pneumoniae (high PCT), and Legionella pneumophila (higher CRP and TNF-α). PCT ≥ 0.36 mg/dL to predict positive blood cultures showed sensitivity of 85%, specificity of 42%, and negative predictive value (NPV) of 98%, whereas a cutoff of ≤ 0.5 mg/dL to predict viruses or atypicals vs bacteria showed sensitivity of 89%/81%, specificity of 68%/68%, positive predictive value of 12%/22%, and NPV of 99%/97%. In a multivariate Euclidean distance model, the lowest inflammatory expression was found in unknown cause and the highest was found in L pneumophila, S pneumoniae, and Enterobacteriaceae. Atypical bacteria exhibit an inflammatory pattern closer to that of viruses.
Conclusions
Different inflammatory patterns elicited by different microorganisms may provide a useful tool for diagnosis. Recognizing these patterns provides additional information that may facilitate a broader understanding of host inflammatory response to microorganisms.
Abbreviations
- CAP
community-acquired pneumonia
- CRP
C-reactive protein
- GNB
gram-negative bacilli
- GPC
gram-positive cocci
- NPV
negative predictive value
- PCT
procalcitonin
- TNF-α
tumor necrosis factor-α
The respiratory tract is constantly exposed to environmental agents and potentially pathogenic microorganisms. The ciliated epithelium, alveolar macrophages, and neutrophils are able to destroy and remove pathogenic agents and prevent the progression of tissue invasion.1, 2 When the innate response is overcome, local reactions, with activation of cytokines and inflammatory markers, promote a specific immune response against the microorganism.2 This reaction is not limited to the lungs; there is also a systemic response that has repercussions on the course of the infection and its outcome.1, 3, 4
Community-acquired pneumonia (CAP) is the leading cause of mortality due to infection in developed countries.5 The host inflammatory response is crucial to fighting the microorganism, and that interplay determines the outcome. Nevertheless, the mechanisms that trigger activation of the cytokine cascade and its different patterns (responsible for the outcome) are not sufficiently understood. An exuberant systemic activation of cytokines has been associated with a poorer outcome, although in some patients it is an adequate response, suggesting that this feature is far from understood.6 Kellum et al6 pointed out the heterogeneous cytokine pattern activation with different combinations of high, medium, and low IL-6 and IL-10 levels, although they did not evaluate the influence of causal microorganisms.
Our hypothesis is that causal microorganisms play a key role in the host response and may trigger different inflammatory responses, depending on their intrinsic properties, the presence of a capsule, lipopolysaccharides in the cell wall, virulence factors, and infection spread.1 Understanding the response of the host to the different pathogens is essential to increasing our knowledge of the course of infection in order to improve the diagnostic process and, possibly, for developing targeted therapeutic strategies.
Our objective was to investigate the cytokine systemic activation patterns (tumor necrosis factor-α [TNF-α], IL-1β, IL-6, IL-8, and IL-10) together with the biomarkers procalcitonin (PCT) and C-reactive protein (CRP) provoked by causal microorganisms in hospitalized patients with CAP. A secondary objective was to evaluate their usefulness in a causal-diagnosis approach. An abstract with some results has been published.7
Materials and Methods
We performed a prospective study of hospitalized patients with CAP in two centers from October 2004 to September 2005. The inclusion criteria were a new radiologic infiltrate and at least two compatible clinical symptoms. The exclusion criteria were admission within the previous 15 days, immunosuppressive treatments, and being HIV positive. This study was approved by the ethics committee (Comité Ético de Investigación Clínica del Hospital Universitario y Politécnico La Fe, approval number 2004/69) and patients signed informed consents. Data recorded were age, sex, toxic habits, comorbidities, and prior antibiotic treatment for the same episode prior to admission.
Cytokines, PCT, and CRP
Blood samples were taken the morning after admission, and the serum was frozen at −80°C. Determination of IL-1β, IL-6, IL-8, and IL-10 and TNF-α was made using an enzyme immunoassay (Biosource). Limits of detection were 3 pg/mL for TNF-α, 2 pg/mL for IL-6, 0.7 pg/mL for IL-8, and 1 pg/mL for IL-10. PCT was measured using an immunoluminometric technique (Liason Brahms PCT) with a detection limit of 0.3 ng/mL and CRP using an immunoturbidimetric test (Bayer Diagnostics) with a detection limit of 1.5 mg/dL.
Microbiologic Analysis
The following studies were carried out: (1) blood cultures (n = 575), (2) urinary antigens for Legionella pneumophila (n = 626) and Streptococcus pneumoniae (n = 628), (3) sputum Gram stain (n = 319) (< 10 epithelial cells and > 25 leukocytes per field × 100) and culture, (4) nasopharyngeal swab (n = 162) to detect viral nucleic acids, (5) paired serologic studies (n = 629) for Chlamydophila pneumoniae, Mycoplasma pneumoniae, Coxiella burnetii, and L pneumophila, and (6) invasive samples (n = 92) obtained by bronchoscopy and/or pleural fluid.
Microbiologic Diagnostic Criteria
Bacterial cause was established using the following criteria: (1) isolation of microorganisms in respiratory samples above the cutoffs (BAL ≥ 104 colony-forming units/mL; bronchoaspirate sample ≥ 105 colony-forming units/mL) or in pleural fluid, (2) isolation of one predominant microorganism in sputum or L pneumophila in buffered charcoal yeast extract agar, (3) microorganisms in blood culture, (4) positive urinary antigens, (5) seroconversion or fourfold antibody increase with titers of IgG ≥ 1:512 for C pneumoniae, ≥ 1:160 for M pneumoniae and C burnetii, or IgM ≥ 1:32 for C pneumoniae and ≥ 1:80 for M pneumoniae and C burnetii,8 and (6) positive detection of viral nucleic acids: ProDetect BCS RV CHIP (bcs Biotech SpA) for influenza virus A and B (gen NS), respiratory syncytial virus (gen NS2), parainfluenza virus I, II, and III (gen HN), SARS coronavirus (fragment BNI-1), and adenovirus (gen H).
Statistical Analysis
The statistical analysis was carried out using SPSS software (version 15.0; SPSS Inc). PCT, CRP, and cytokines were presented as medians and interquartile ranges, and parametric data as mean ± SD. The hypotheses were tested using the Mann-Whitney U test. Significance was established at P < .05.
The microorganisms were analyzed individually and according to the following groups: no cause, bacteria subdivided into gram-positive cocci (GPC) and gram-negative bacilli (GNB), viruses, and atypical pathogens (C pneumoniae, M pneumoniae, and C burnetii).
A multivariate Euclidean distance model was performed. The graphics were generated by means of a hierarchic cluster analysis and multidimensional scaling of the distance matrix based on the significant differences observed in the pair comparison of microorganisms using the Mann-Whitney U test.
Results
Six hundred eighty-five patients were included, and in 295 (43%) a causal diagnosis was reached: 118 S pneumoniae (17.2%), 24 L pneumophila (3.5%), 18 Pseudomonas aeruginosa (2.6%), 14 Haemophilus influenzae (2%), 13 Staphylococcus aureus (1.9%), 13 M pneumoniae (1.9%), 13 Enterobacteriaceae (1.9%), 12 viruses (1.8%) (nine influenza virus), eight C burnetii (1.2%), three C pneumoniae (0.4%), and 35 mixed infections (5%). Causal groups were 390 no cause (56.9%), 134 GPC (19.6%), 69 GNB (10.1%), 24 atypical pathogens (3.5%), and 12 viruses(1.8%) General characteristics, Fine risk classes, and mortality depending on cause are depicted in Table 1 .
Table 1.
Demographic Data, Comorbidities, Fine Risk Classes, and Mortality Depending on Causal Microorganism
| Characteristics | NE (n = 390) | GPC (n = 134) | GNB (n = 69) | ATP (n = 24) | VIR (n = 12) |
|---|---|---|---|---|---|
| Sex | |||||
| M | 252 (64.6) | 78 (58.2) | 54 (78.3) | 14 (58.3) | 9 (75) |
| F | 138 (35.4) | 56 (41.8) | 15 (21.7) | 10 (41.7) | 3 (25) |
| Mean age, y | 67.2 ± 17.2 | 66.6 ± 17.8 | 68.8 ± 13.9 | 53.9 ± 22.3 | 62.1 ± 16.9 |
| Diabetes | 77 (19.8) | 29 (21.8) | 9 (13) | 1 (4.2) | 2 (16.7) |
| Heart failure | 77 (19.8) | 21 (15.7) | 16 (23.5) | 3 (13) | 1 (8.3) |
| Chronic renal failure | 22 (5.6) | 5 (3.7) | 5 (7.2) | 1 (4.2) | 0 (0) |
| Digestive disease | 58 (14.9) | 32 (23.9) | 17 (24.6) | 5 (20.8) | 1 (8.3) |
| Cirrhosis | 11 (2.8) | 5 (3.7) | 3 (4.3) | 1 (4.2) | 1 (8.3) |
| COPD | 80 (20.5) | 20 (14.9) | 22 (31.9) | 3 (12.5) | 0 (0) |
| Neurologic disease | 87 (22.4) | 28 (21.1) | 17 (24.6) | 1 (4.2) | 1 (8.3) |
| Smoking | 75 (19.3) | 36 (27.1) | 26 (37.7) | 9 (37.5) | 5 (41.7) |
| Alcohol consumption | 27 (7) | 17 (12.8) | 9 (13) | 2 (8.3) | 1 (8.3) |
| Fine I-III | 194 (49.7) | 70 (52.2) | 27 (39.1) | 17 (70.8) | 11 (91.7) |
| Fine IV-V | 196 (50.3) | 64 (47.8) | 42 (60.9) | 7 (29.2) | 1 (8.3) |
| No sepsis | 123 (31,5) | 38 (28,4) | 16 (23,2) | 11 (45,8) | 5 (41,7) |
| Sepsis | 132 (33.8) | 30 (22.4) | 20 (29.0) | 8 (33.3) | 4 (33.3) |
| Severe sepsis | 129 (33.1) | 59 (44.0) | 27 (39.1) | 5 (20.8) | 3 (25.0) |
| Septic shock | 6 (1,5) | 7 (5,2) | 6 (8,7) | 0 (0) | 0 (0) |
| Hipoxemia | 151 (38.7) | 66 (49.3) | 35 (50.7) | 1 (4.2) | 5 (41.7) |
| Mechanical ventilation | 7 (1.8) | 8 (6.0) | 6 (8.7) | 0 (0) | 0 (0) |
| Death | 19 (4.9) | 6 (4.5) | 10 (14.5) | 0 (0) | 0 (0) |
Data are presented as No. (%) or mean ± SD. ATP = atypical pathogen (Chlamydophila pneumoniae, Mycoplasma pneumoniae, and Coxiella burnetii); F = female; GNB = gram-negative bacilli; GPC = gram-positive cocci; M = male; NE = no cause; VIR = viruses.
Bacteremia was found in 48 cases (7%): 36 S pneumoniae, seven Enterobacteriaceae, three H influenzae, four S aureus, three P aeruginosa, one Streptococcus pyogenes and one Acinetobacter baumannii. Six of the cases were mixed infections due to several bacteria, principally S pneumoniae.
CRP and PCT
Patients with a causal diagnosis showed higher CRP and PCT in comparison with those without, and the highest levels were found in those with bacteremia (Table 2 ). The diagnostic value of PCT (≥ 0.36) to predict bacteremia was as follows: 85% sensitivity, 42% specificity (E), and 98% negative predictive value (NPV).
Table 2.
Results of CRP, PCT, and Cytokines According to Causal Microorganism and Bacteremia
| Cause |
Bacteremia |
|||||
|---|---|---|---|---|---|---|
| Biomarkers | No (n = 390) | Yes (n = 295) | P Value | No (n = 527) | Yes (n = 48) | P Value |
| CRP, mg/dL | 13.7 (6.95–21.85) | 18.1 (9.7–27.3) | < .0001 | 16.1 (8.8–24.1) | 23.3 (14.9–35.1) | .002a |
| PCT, ng/mL | 0.37 (0.15–1.56) | 0.86 (0.27–4.12) | < .00001 | 0.51 (0.18–2.24) | 4.54 (0.49–11.16) | < .00001b |
| TNF-α, pg/mL | 25 (15–41) | 27 (15–48) | NS | 26 (15–45) | 30.5 (18–67) | .04c |
| IL-1, pg/mL | 15 (3–33) | 16 (4–32) | NS | 16 (4–33) | 16.5 (3–33) | NS |
| IL-6, pg/mL | 71 (25–175) | 105 (42–300) | < .0001 | 95 (34–235) | 192 (79.5–568.5) | .004d |
| IL-10, pg/mL | 5 (0–15) | 6 (0–19) | NS | 5 (0–17) | 12 (1–38.5) | NS |
| IL-8, pg/mL | 8 (2–17) | 9 (3–22) | NS | 10 (3–20) | 5 (2–30.5) | NS |
Data are presented as median (interquartile range). AUC = area under the curve; CRP = C-reactive protein; NS = not significant; PCT = procalcitonin; TNF-α = tumor necrosis factor-α.
AUC, 0.7 (0.6–0.8); P < .001.
AUC, 0.7 (0.6–0.8); P < .0001.
AUC, 0.6 (0.5–0.7); P = .03.
AUC, 0.7 (0.6–0.7); P < .001.
The medians of CRP, PCT, and cytokines depending on microorganisms are shown in Table 3 , and those depending on sepsis status and on hypoxemia or mechanical ventilation are shown in Table 4, Table 5 , respectively. The highest levels of CRP and PCT were found in CAP caused by L pneumophila, Enterobacteriaceae, and S pneumoniae. PCT was also higher in S aureus. CRP and PCT were higher in sepsis in those with GPC and GNB compared with those with unknown cause, and, after stratifying for hypoxemia or mechanical ventilation, the medians were higher in GNB. The differences among causal groups and between S pneumoniae and L pneumophila are shown in Table 6 .
Table 3.
Results of CRP, PCT, and Cytokines According to Causal Microorganisms
| Causal Microorganism | CRP, mg/dL | PCT, ng/mL | TNF-α, pg/mL | IL-1, pg/mL | IL-6, pg/mL | IL-10, pg/mL | IL-8, pg/mL |
|---|---|---|---|---|---|---|---|
| Unknown cause (n = 390) | 13.7 (6.95–21.9) | 0.37 (0.15–1.56) | 25 (15–41) | 15 (3–33) | 71 (25–175) | 5 (0–15) | 8 (2–17) |
| Gram positive | |||||||
| Streptococcus pneumoniae (n = 118) | 19.85 (10.3–28.4) | 1.71 (0.48–7.37) | 27 (16–47) | 16 (4–30) | 144 (38–305) | 7 (0–21) | 6 (2–19) |
| Staphylococcus aureus (n = 13) | 16.4 (5.6–24.8) | 1.37 (0.3–7.86) | 40.5 (22–48) | 22 (0–47.5) | 125 (63–204.5) | 7.5 (0–32.5) | 10 (3.5–16.5) |
| Gram negative | |||||||
| Legionella pneumophila (n = 24) | 24.9 (21.3–33.5) | 0.71 (0.5–3.15) | 49 (40–72) | 19 (5–35) | 202 (69–1548) | 3 (0–12) | 16 (11–35) |
| Haemophilus influenzae (n = 14) | 12.5 (2.7–17.4) | 0.36 (0.215–1.37) | 19.5 (11–33.5) | 17.5 (0–37) | 63.5 (6–155) | 7.5 (6–35.5) | 5 (1–9) |
| Pseudomonas aeruginosa (n = 18) | 10 (7.4–13.8) | 0.44 (0.14–0.62) | 23 (15–42) | 16 (4–29) | 105 (22–223) | 6 (3–12) | 12 (8–20) |
| Enterobacteriaceae (n = 13) | 20.1 (12.6–31.5) | 1.59 (0.56–8.99) | 14.5 (11.5–68) | 20 (8.5–41) | 168.5 (58–339.5) | 6 (0–25.5) | 54.5 (13.5–79.5) |
| ATP | |||||||
| Mycoplasma pneumoniae (n = 13) | 13.9 (7.6–24.6) | 0.34 (0.1–0.62) | 27.5 (15–128) | 23 (4–54) | 77 (28–98) | 4 (0–14) | 17.5 (7–24) |
| Chlamydophila pneumoniae (n = 3) | 19 (0.2–24.3) | 0.23 (0.1–0.36) | 36 (23–39) | 31 (22–35) | 26 (8–59) | 17 (10–23) | 1 (0–12) |
| Coxiella burnetii (n = 8) | 9.45 (5.5–25.3) | 0.13 (0.09–0.19) | 22 (21–38) | 17 (6.5–30.5) | 41.5 (25–122.5) | 2.5 (0.5–13) | 17 (8.5–24) |
| VIR | |||||||
| Influenza virus (n = 9) | 15.4 (12–21.6) | 0.36 (0.09–0.47) | 11.5 (8–22) | 10 (2–18) | 129 (39–405) | 24.5 (18–33) | 7 (3–16) |
Table 4.
Biomarkers and Cytokines According to Causal Microorganisms and Sepsis Status
| Biomarkers and Sepsis Status | NE | GPC | GNB | ATP | VIR | P Value |
|---|---|---|---|---|---|---|
| CRP | ||||||
| No sepsis | 13.5 (6–20) | 17.6 (5.1–30.2) | 13.8 (9.8–23.7) | 12.7 (7.6–19.1) | 4.9 (0.7–12.9) | NS |
| Sepsis | 12 (6.5–22.7) | 20.3 (12.2–28.2) | 18.5 (12.6–24.9) | 13.8 (7.5–24.3) | 12 (8.9–29.5) | < .05 |
| Severe sepsis/shock | 15.5 (8.4–23.4) | 20 (11.1–28.1) | 23.5 (10–32.8) | 11.3 (4–33.6) | 17.8 (14–21.6) | NS |
| PCT | ||||||
| No sepsis | 0.2 (0.1–0.6) | 0.7 (0.2–4.5) | 0.3 (0.1–1.1) | 0.2 (0.1–0.3) | 0.3 (0.2–0.4) | .004 |
| Sepsis | 0.3 (0.1–1.2) | 1.9 (0.7–4.1) | 0.6 (0.4–2.8) | 0.2 (0.1–0.5) | 0.1 (0.1–0.5) | < .0001 |
| Severe sepsis/shock | 0.8 (0.3–2.7) | 2.4 (0.8–9.1) | 0.7 (0.4–6) | 0.2 (0.2–0.6) | 1.2 (0.2–2.2) | .2 |
| TNF-α | ||||||
| No sepsis | 23 (13–41) | 35 (18–48) | 20 (11–58) | 23 (21–35) | 26.5 (18–35.5) | NS |
| Sepsis | 24 (14–34.5) | 29 (18–43) | 37 (13–52) | 26.5 (21–39) | 8 (8–13) | NS |
| Severe sepsis/shock | 29 (18–45) | 26 (16–48) | 37 (17–72) | 40 (36–128) | 18.5 (10–27) | NS |
| IL-1 | ||||||
| No sepsis | 10 (3–27) | 14 (3–34) | 12 (4–46) | 33 (5–50) | 14 (9–18) | NS |
| Sepsis | 17 (4–35) | 6.5 (2–19) | 22 (3–29) | 15.5 (4–26) | 2 (0–7) | NS |
| Severe sepsis/shock | 17 (2–37) | 18 (5–35) | 17.5 (6–36) | 22 (9–24) | 15.5 (13–18) | NS |
| IL- 6 | ||||||
| No sepsis | 66 (23–146) | 97 (29–225) | 71 (22–144) | 27.5 (17–77) | 54.5 (21–137.5) | NS |
| Sepsis | 65.5 (24.5–165.5) | 185 (92–380) | 79 (22–324) | 49.5 (39–79) | 39 (30–1549) | .005 |
| Severe sepsis/shock | 93 (30–273) | 150 (47–253) | 197 (69–1.288) | 166 (77–460) | 235.5 (66–405) | NS |
| IL-10 | ||||||
| No sepsis | 3 (0–9) | 2 (0–16) | 6 (0–9) | 7.5 (1–14) | 12 (1–28) | NS |
| Sepsis | 5 (0–12) | 2.5 (0–14) | 4 (0–15) | 1 (0–15) | 18 (0–27) | NS |
| Severe sepsis/shock | 8.5 (0–22) | 13 (1–31) | 6.5 (0–19) | 9 (3–14) | 29 (22–36) | NS |
| IL- 8 | ||||||
| No sepsis | 10 (4–18) | 10 (5–22) | 16 (12–35) | 16.5 (10–23) | 2 (0–4) | .006 |
| Sepsis | 7 (2–15) | 7 (3–18) | 10 (4–34) | 11 (5–19) | 5 (3–16) | NS |
| Severe sepsis/shock | 6 (2–19) | 5 (2–16) | 14 (5–61) | 13 (1–27) | 24 (9–39) | NS |
Table 5.
Biomarkers and Cytokines According to Hypoxemia and MV
| Biomarkers | NE | GPC | GNB | ATP | VIR | P Value |
|---|---|---|---|---|---|---|
| CRP | ||||||
| Hypoxemia | ||||||
| Yes | 16 (8.2–27.8) | 21.3 (10.8–28.9) | 21.3 (8.8–31.8) | 19 (19–19) | 14 (8.9–21.6) | NS |
| No | 11.7 (6.4–19.8) | 18.8 (9.2–27.3) | 17.3 (11.3–24.4) | 10.7 (7.5–24.5) | 10.5 (0.8–16.8) | .002 |
| MV | ||||||
| Yes | 15.8 (8.4–27) | 17.6 (4.9–39.9) | 26.5 (11–34.2) | … | … | NS |
| No | 13.7 (6.9–21.8) | 19.9 (10.2–28.3) | 17.8 (10.2–28) | 11.3 (7.5–24.3) | 12 (8.9–16.8) | .001 |
| PCT | ||||||
| Hypoxemia | ||||||
| Yes | 0.5 (0.2–2.7) | 1.8 (0.4–7.4) | 0.6 (0.4–3.6) | 0.4 (0.4–0.4) | 0.3 (0.2–2.2) | NS |
| No | 0.3 (0.1–0.9) | 1.6 (0.5–6.5) | 0.6 (0.3–2.8) | 0.2 (0.1–0.4) | 0.2 (0.1–0.5) | < .00001 |
| MV | ||||||
| Yes | 2.3 (0.8–7) | 1.2 (0.3–12.9) | 6.6 (0.9–38.1) | … | … | NS |
| No | 0.4 (0.1–1.4) | 1.7 (0.5–7.1) | 0.5 (0.3–2.3) | 0.2 (0.1–0.4) | 0.2 (0.2–0.5) | < .00001 |
| TNF-α | ||||||
| Hypoxemia | ||||||
| Yes | 27 (16–44) | 28.5 (15–45) | 37 (16.5–71) | 23 (23–23) | 27 (10–31) | NS |
| No | 25 (14–40) | 28 (19–48.5) | 26 (13–52) | 27.5 (21–39.5) | 13.5 (8–22) | NS |
| MV | ||||||
| Yes | 29 (16–33) | 44 (17–48) | 27 (6–166) | … | … | NS |
| No | 25 (15–41) | 28 (16–47) | 33 (15.5–56.5) | 25 (21–39) | 14 (10–27) | NS |
| IL-1 | ||||||
| Hypoxemia | ||||||
| Yes | 16 (3–36) | 16 (3–35) | 13.5 (4–35.5) | 31 (31–31) | 14 (13–18) | NS |
| No | 14 (3–29.5) | 14.5 (4–28) | 21.5 (6.5–38) | 23 (4.5–35) | 5.5 (2–14) | NS |
| MV | ||||||
| Yes | 7 (0–15) | 5 (0–16) | 36 (13–93) | … | … | NS |
| No | 15 (3–33) | 15 (4–30) | 18 (4–29) | 24 (5–35) | 13 (4–14) | NS |
| IL-6 | ||||||
| Hypoxemia | ||||||
| Yes | 95 (23–246) | 133.5 (42–284) | 197 (69.5–931.5) | 26 (26–26) | 66 (16–405) | NS |
| No | 65 (25.5–154.5) | 159.5 (59.5–302.5) | 72 (24.5–211.5) | 51 (24.5–92.5) | 61 (30–192) | .02 |
| MV | ||||||
| Yes | 34 (30–143) | 79 (71–644) | 380 (70–2.075) | … | … | NS |
| No | 72 (25–175) | 151 (47–284) | 105 (41–324) | 43 (26–87) | 66 (30–192) | .002 |
| IL-10 | ||||||
| Hypoxemia | ||||||
| Yes | 6 (0–18) | 10 (0–28) | 7 (2.5–17.5) | 10 (10–10) | 23 (22–36) | NS |
| No | 4 (0–12) | 5.5 (0–18.5) | 5 (0–9.5) | 4 (0.5–14.5) | 9.5 (1–27) | NS |
| MV | ||||||
| Yes | 25 (5–59) | 13 (1–35) | 20 (0–273) | … | … | NS |
| No | 5 (0–14) | 6 (0–21) | 6 (0–12) | 5 (1–14) | 22 (1–27) | NS |
| IL- 8 | ||||||
| Hypoxemia | ||||||
| Yes | 8 (2–18) | 6.5 (2–18) | 19.5 (11–70) | 12 (12–12) | 9 (0–39) | .005 |
| No | 9 (3–17) | 6.5 (4–19) | 11 (4.5–24) | 14 (5–23.5) | 4 (3–5) | NS |
| MV | ||||||
| Yes | 13.5 (2–49) | 10 (6–86) | 64 (0–70) | … | … | NS |
| No | 8 (2–17) | 6 (2–18) | 13 (6–34) | 13 (5–23) | 4 (3–9) | .007 |
Table 6.
Comparison of Main Groups and Microorganisms, and Diagnostic Cutoff Values
| Biomarkers | Bacteria vs ATP | Bacteria vs VIR | Gram Negative vs Gram Positive | Gram Negative vs ATP | L pneumophila vs S pneumoniae |
|---|---|---|---|---|---|
| CRP | NS | NS | NS | NS | 24.9 vs 19.9a |
| P = .01 | |||||
| PCT | 1.12 vs 0.19b | 1.12 vs 0.2c | 0.62 vs 1.67 | 0.62 vs 0.19 | NS |
| P < .0001 | P = .003 | P = .02 | P = .002 | ||
| TNF-α | NS | 29 vs 14d | NS | NS | 49 vs 27e |
| P = .03 | P = .0002 | ||||
| IL-6 | 144 vs 43f | NS | NS | 116 vs 43 | NS |
| P = .01 | P = .04 | ||||
| IL-8 | NS | NS | 13 vs 6.5 | NS | 16 vs 6g |
| P = .003 | P = .003 |
E = specificity; NPV = negative predictive value; PPV = positive predictive value; S = sensitivity. See Table 1-3 legends for expansion of other abbreviations.
CRP ≥ 22 mg/dL as cutoff to compare L pneumophila and S pneumoniae: S, 70%; E, 59%; PPV, 27%; and NPV, 90%. AUC, 0.7 (0.6–0.8); P = .01.
PCT ≤ 0.5 mg/dL as cutoff to compare bacteria and atypical: S, 81%; E, 68%; PPV, 22%; and NPV, 97%. AUC, 0.8 (0.6–0.9); P < .0001.
PCT ≤ 0.5 mg/dL as cutoff to compare bacteria and virus: S, 89%; E, 68%; PPV, 12%; and NPV, 99%. AUC, 0.8 (0.7–0.9); P < .01.
TNF-α ≤ 27 pg/mL as cutoff to compare virus and bacterial: S, 78%; E, 51%; PPV, 7%; and NPV, 98%. AUC, 0.7 (0.6–0.9); P = .03.
TNF-α ≥ 30 pg/mL as cutoff to compare L pneumophila and S pneumoniae: S, 88%; E, 57%; PPV, 39%; and NPV, 94%. AUC, 0.7 (0.6–0.8); P = .001.
IL-6 ≤ 100 pg/mL as cutoff to compare bacteria and atypical: S, 81%; E, 56%; PPV, 17%; and NPV, 96%. AUC, 0.7 (0.6–0,8); P = .01.
IL-8 ≥ 15 pg/mL as cutoff to compare L pneumophila and S pneumoniae: S, 57%; E, 69%; PPV, 27%; and NPV, 89%. AUC, 0.7 (0.6–0.8); P = .004.
Cytokines
Patients with a causal diagnosis had a higher IL-6 than those without, whereas those with bacteremia showed the highest IL-6 and TNF-α (Table 2). Causal microorganisms exhibited different cytokine patterns (Table 3): L pneumophila and S aureus had higher TNF-α, and the former also had higher IL-6; Enterobacteriaceae had higher IL-8, whereas influenza virus infections showed higher IL-10 (compared with bacteria, P = .03) and lower TNF-α. Concerning sepsis status (Table 4), IL-6 was higher in severe sepsis in those with GPC and GNB compared with those with unknown cause. In patients stratified by hypoxemia and mechanical ventilation, those with GPC and GNB had higher IL-6 (and IL-8 in GNB) compared with those with unknown cause (Table 5).
Prior Antibiotics and Inflammatory Markers
Two hundred thirty-three patients (34%) had received antibiotics prior to admission and they had lower PCT (P < .01), IL-6 (P < .03), and IL-10 (P < .01), and higher IL-8 (P < .05) (Table 7 ). PCT was higher in those with known cause who did not take prior antibiotics compared with those who did (P = .02).
Table 7.
Effect of Antibiotic Treatment on Cytokines and Biologic Markers
| Prior Antibiotic Treatment |
||||||
|---|---|---|---|---|---|---|
| No |
Yes |
|||||
| Causal Diagnosis |
Causal Diagnosis |
|||||
| Biomarkers | No (n = 256) | Yes (n = 196) | P Value | No (n = 134) | Yes (n = 99) | P Value |
| CRP, mg/dL | 13.5 (7–21.6) | 19.1 (10–28.3) | < .0001 | 15.3 (6.4–22,7) | 17 (8.7–26) | NS |
| PCT, ng/mL | 0.41 (0.16–1.86) | 1.11 (0.3–5.28) | < .0001 | 0.32 (0.14–0.9) | 0.54 (0.2–2.28) | .017 |
| TNF-α, pg/mL | 26 (16–40) | 29 (15–49) | NS | 23 (15–44) | 24 (14–44) | NS |
| IL-1, pg/mL | 16 (3–34) | 15 (4–30) | NS | 13.5 (4–31) | 19 (3–35) | NS |
| IL-6, pg/mL | 81 (25–197) | 122 (43–352) | < .001 | 60 (24–149) | 79 (29–235) | .036 |
| IL-10, pg/mL | 6 (0–19) | 8 (0–23) | NS | 3 (0–9) | 3 (0–13) | NS |
| IL-8, pg/mL | 7 (2–16) | 6.5 (2–21) | NS | 10 (5–19) | 12 (5–24) | NS |
Data are presented as median (interquartile range). See Table 2 legend for expansion of abbreviations.
Multivariate Euclidean Distance Model
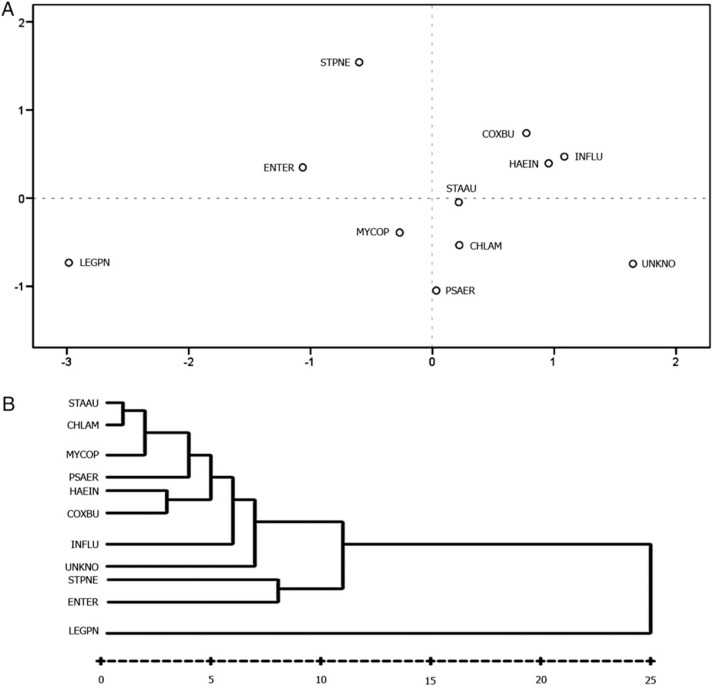
Statistical differences among cytokines and biomarkers in microorganisms converted into distances in a Euclidean two-dimensional space are depicted in Figure 1 . We found three microorganisms with higher distances: L pneumophila, S pneumoniae, and Enterobacteriaceae; a close group of bacteria (S aureus, M pneumoniae, C pneumoniae, P aeruginosa, C burnetii, and H influenzae, together with influenza virus); and a third group close to this last group, unknown-cause CAP. Hierarchic cluster analysis is also shown with microorganisms associated in similar cluster groups.
Figure 1.
A, Multidimensional scaling in two dimensions that provides a visualization of similarities or dissimilarities among cytokine and markers and cause. B, Hierarchic cluster analysis for assigning microorganisms into cluster groups that are more similar to one or another in other clusters. CHLAM = chlamydophila; COXBU = Coxiella burnetii; ENTER = Enterobacteriaceae; HAEIN = Haemophilus influenzae; INFLU = influenza virus; LEGPN = Legionella pneumophila; MYCOP = Mycoplasma pneumoniae; PSAER = Pseudomonas aeruginosa; STAAU = Staphylococcus aureus; STPNE = Streptococcus pneumoniae; UNKNO = unknown cause.
Discussion
The most outstanding findings of this study were as follows: (1) The highest levels of cytokines, CRP, and PCT were found in patients who were bacteremic and the lowest in those with unknown cause; (2) the causal microorganisms elicited different inflammatory cytokine patterns and biomarkers as corroborated in the Euclidean distances model; (3) a cutoff of PCT ≤ 0.5 to differentiate viruses or atypicals vs bacteria showed sensitivity of 89%/81%, specificity of 68%/68%, positive predictive value of 12%/22%, and NPV of 99%/97%; (4) PCT ≥ 0.36 had an excellent NPV (98%) for predicting positive blood cultures; and (5) L pneumophila CAP showed higher IL-8 and TNF-α compared with S pneumonia, with high NPV (89% and 94%, respectively).
The study of the inflammatory profile in CAP may provide better knowledge of the host-microorganism interplay and may be useful for causal diagnosis. PCT, CRP, and, to a lesser extent, cytokines were studied for their diagnostic ability in CAP.3, 8, 9, 10, 11 Interestingly, biomarkers and IL-6 were significantly lower when causal microorganisms were not found, even considering sepsis status, reflecting a lower microorganism load or virulence. On the other hand, in bacteremia, biomarkers and IL-6 were significantly higher, reflecting the greater dissemination of the infection,12 which may be useful for selecting that specific CAP population.8, 13 Müller et al13 found that PCT ≤ 0.25 ng/mL allowed a 37% reduction of blood cultures with high specificity. We chose a slightly higher cutoff (0.36 ng/mL) with very high NPV because we considered that a lower cutoff would include many atypicals and undiagnosed CAP. The role of cytokines in predicting bacteremia is less well known. We found that IL-6 ≥ 150 had an excellent NPV (96%).
Despite the interest in knowing the influence of microorganisms on triggering different inflammatory patterns, publications on the subject are few. Masiá et al14 found lower PCT and CRP in atypicals compared with bacteria, although neither of them was useful for predicting that cause. Hedlund and Hansson3 also reported lower PCT in atypical bacteria and/or viruses, with no differences in CRP. Our data confirm that PCT ≤ 0.5 provides high sensitivity and NPV for viral and/or atypical causes. Krüger et al15 used a lower PCT cutoff (≤ 0.1) to differentiate S pneumoniae from atypical or viral causes and reported a high OR of 8.3. Nevertheless, the considerable overlap in levels among microorganisms should lead us to be cautious, to avoid prescribing insufficient antibiotics.
The usefulness of CRP and PCT in distinguishing cause within the bacteria group is less clear. However, García Vázquez et al8 found that CRP (> 25 mg/dL) may be useful in diagnosing L pneumophila with high NPV (94%). We found that higher levels of TNF-α and IL-6 in L pneumophila had a high NPV compared with S pneumoniae. PCT and IL-8 showed different patterns among bacteria: higher IL-8 and lower PCT in gram-negative vs gram-positive germs. However, the clinical relevance of these findings has yet to be demonstrated, and it is possible that, for diagnostic purposes, several markers will be required.16
Prior use of antibiotics reduced the levels of PCT and cytokines, mainly IL-6 and IL-10, suggesting that the inflammatory phase was beginning to be downregulated.11, 17 We consider that PCT-guided antibiotic prescription in CAP requires extreme caution in patients with prior antibiotic treatment because it could underestimate bacterial cause,9, 10 and even in those without prior antibiotics it would appear that a low PCT has insufficient NPV to exclude pathogens. On the other hand, IL-8 was raised, as reported in in vitro experiments,18 probably because of enhanced cytokine secretion secondary to bacterial wall destruction. In fact, it was higher in those previously treated with β-lactams (12 vs 8.5, P = .06; data not shown).
Our findings highlight the differences in inflammatory cytokine activation, which were corroborated in the Euclidean distances model. The scenario with the least inflammation was found in unknown cause, whereas the greatest inflammation, though with specific expressive patterns, was found in L pneumophila, S pneumoniae, and Enterobacteriaceae. These distances reflected differences in the inflammatory profiles, probably due to variations in virulence and the recognition of different molecular patterns that activate different pathways and innate immunity, such as Toll-like receptor-9 in the case of L pneumophila, Toll-like receptor-4 in gram-negative bacteria, and Toll-like receptor-2 in gram-positive bacteria.19, 20, 21 Enterobacteriaceae showed an increase in IL-8,22 as reported in urinary infections.23 Surprisingly, P aeruginosa presented an inflammatory response closer to that in CAP of unknown cause. This colonizing microorganism, which is associated with elderly patients or severe diseases, may take on importance depending on the characteristics of the patient; furthermore, it was associated with the use of oral or inhaled corticosteroids (nine patients). Influenza virus was associated with higher IL-1024 and a lower TNF-α. This response profile plays a detrimental role in the host responses against the influenza A virus, as found in animal models.25 In fact, although IL-10 activates the natural killer lymphocytes and increases the antigen volume available to stimulate the immune system,26 it has been shown that an increase of IL-6 and TNF-α protects against influenza virus pneumonia.27
Our findings suggest that interpretation of cytokine cascade activation should take into account not only host characteristics and sepsis status, but also the causal microorganism, bacteremia, and prior antibiotic treatment. Further studies of treatments designed to modulate the cytokine response should be designed to consider microorganism-specific inflammatory patterns. Prior failures of anticytokine treatments may be partially explained by this difference in cytokine patterns.
Limitations
Not all microbiologic studies were performed on the whole population and unknown cause may correspond to underdiagnosed viruses or bacteria. Blood cultures were not obtained from patients at the same time intervals within the initial 24 h. Lower limits of PCT assay could be inadequate in milder CAP.
Conclusions
In conclusion, the main causal agents of CAP presented different inflammatory patterns, which gave each group of microorganisms a specific profile, although their usefulness in diagnosis was limited. In bacteremia, inflammation was upregulated, whereas it was lowest in CAP of unknown cause. The most notable finding is that the knowledge of specific inflammatory patterns should enable us to better understand the host response in CAP. Further studies are needed to understand the mechanisms that lead each microorganism to present its own inflammatory response and to better define this response.
Acknowledgments
Author contributions: Dr Menéndez had full access to all the data in the study and she takes responsibility for the integrity of the data and the accuracy of the data analysis and confirms that the study objectives and procedures are honestly disclosed.
Dr Menéndez: contributed to the study concept and design, data analysis, and drafting of the manuscript.
Dr Sahuquillo-Arce: contributed to the study concept and design, data analysis, and drafting of the manuscript.
Dr Reyes: contributed to the quality control of the database, statistical analysis, and critical revision of the manuscript.
Dr Martínez: contributed to the coordination of the acquisition of data and critical revision of the manuscript.
Dr Polverino: contributed to the coordination of the acquisition of data and critical revision of the manuscript.
Dr Cillóniz: contributed to the coordination of the acquisition of data, data analysis, and critical revision of the manuscript.
Dr Córdoba: contributed to the acquisition of data, data analysis, and critical revision of the manuscript.
Dr Montull: contributed to the acquisition of data and critical revision of the manuscript.
Dr Torres: contributed to the study design, analysis and interpretation of the data, and critical revision of the manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following conflicts of interest: Dr Torres is a recipient of an unrestricted research grant from Brahms. Drs Menéndez, Sahuquillo-Arce, Reyes, Martínez, Polverino, Cillóniz, Córdoba, and Montull have reported that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Role of sponsors: The funding organizations played no role in the study design, collection, or analysis of the data, or approval of the manuscript.
Other contributions: The authors gratefully acknowledge Ivan Arribas, PhD, an independent statistician, for his expertise.
Footnotes
Funding/Support: The study was funded by Centro Investigación Biomedica en Red de Enfermedades Respiratorias, an initiative of Instituto de Salud Carlos III; Sociedad Española de Neumología y Cirugía Torácica [Grant 2003]; Fondo de Investigación Sanitario [Grant PI041136]; Fondo de Investigación Sanitario [Grant PI080727]; and an IMPULSA grant.
Drs Menéndez and Sahuquillo-Arce are considered as first authors.
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians. See online for more details.
References
- 1.Deng JC, Standiford TJ. The systemic response to lung infection. Clin Chest Med. 2005;26(1):1–9. doi: 10.1016/j.ccm.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Bals R, Hiemstra PS. Innate immunity in the lung: how epithelial cells fight against respiratory pathogens. Eur Respir J. 2004;23(2):327–333. doi: 10.1183/09031936.03.00098803. [DOI] [PubMed] [Google Scholar]
- 3.Hedlund J, Hansson LO. Procalcitonin and C-reactive protein levels in community-acquired pneumonia: correlation with etiology and prognosis. Infection. 2000;28(2):68–73. doi: 10.1007/s150100050049. [DOI] [PubMed] [Google Scholar]
- 4.Menéndez R, Martínez R, Reyes S. Biomarkers improve mortality prediction by prognostic scales in community-acquired pneumonia. Thorax. 2009;64(7):587–591. doi: 10.1136/thx.2008.105312. [DOI] [PubMed] [Google Scholar]
- 5.Woodhead M. The European vision of community-acquired pneumonia. Semin Respir Crit Care Med. 2009;30(2):136–145. doi: 10.1055/s-0029-1202932. [DOI] [PubMed] [Google Scholar]
- 6.Kellum JA, Kong L, Fink MP, GenIMS Investigators Understanding the inflammatory cytokine response in pneumonia and sepsis: results of the Genetic and Inflammatory Markers of Sepsis (GenIMS) Study. Arch Intern Med. 2007;167(15):1655–1663. doi: 10.1001/archinte.167.15.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menendez R, Reyes S, Martínez R. Cytokine systemic pattern and causal microorganism in community acquired pneumonia [abstract] Am J Respir Crit Care Med. 2010;181:A5480. [Google Scholar]
- 8.García Vázquez E, Martínez JA, Mensa J. C-reactive protein levels in community-acquired pneumonia. Eur Respir J. 2003;21(4):702–705. doi: 10.1183/09031936.03.00080203. [DOI] [PubMed] [Google Scholar]
- 9.Christ-Crain M, Jaccard-Stolz D, Bingisser R. Effect of procalcitonin-guided treatment on antibiotic use and outcome in lower respiratory tract infections: cluster-randomised, single-blinded intervention trial. Lancet. 2004;363(9409):600–607. doi: 10.1016/S0140-6736(04)15591-8. [DOI] [PubMed] [Google Scholar]
- 10.Christ-Crain M, Stolz D, Bingisser R. Procalcitonin guidance of antibiotic therapy in community-acquired pneumonia: a randomized trial. Am J Respir Crit Care Med. 2006;174(1):84–93. doi: 10.1164/rccm.200512-1922OC. [DOI] [PubMed] [Google Scholar]
- 11.Bruns AH, Oosterheert JJ, Hak E, Hoepelman AI. Usefulness of consecutive C-reactive protein measurements in follow-up of severe community-acquired pneumonia. Eur Respir J. 2008;32(3):726–732. doi: 10.1183/09031936.00003608. [DOI] [PubMed] [Google Scholar]
- 12.Martínez R, Menéndez R, Reyes S. Factors associated with inflammatory cytokine patterns in community-acquired pneumonia. Eur Respir J. 2011;37(2):393–399. doi: 10.1183/09031936.00040710. [DOI] [PubMed] [Google Scholar]
- 13.Müller F, Christ-Crain M, Bregenzer T, ProHOSP Study Group Procalcitonin levels predict bacteremia in patients with community-acquired pneumonia: a prospective cohort trial. Chest. 2010;138(1):121–129. doi: 10.1378/chest.09-2920. [DOI] [PubMed] [Google Scholar]
- 14.Masiá M, Gutiérrez F, Padilla S. Clinical characterisation of pneumonia caused by atypical pathogens combining classic and novel predictors. Clin Microbiol Infect. 2007;13(2):153–161. doi: 10.1111/j.1469-0691.2006.01629.x. [DOI] [PubMed] [Google Scholar]
- 15.Krüger S, Ewig S, Papassotiriou J, CAPNETZ Study Group Inflammatory parameters predict etiologic patterns but do not allow for individual prediction of etiology in patients with CAP: results from the German competence network CAPNETZ. Respir Res. 2009;10:65. doi: 10.1186/1465-9921-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christ-Crain M, Opal SM. Clinical review: the role of biomarkers in the diagnosis and management of community-acquired pneumonia. Crit Care. 2010;14(1):203. doi: 10.1186/cc8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padrones S, Garcia-Vidal C, Fernández-Serrano S. Impact of antibiotic therapy on systemic cytokine expression in pneumococcal pneumonia. Eur J Clin Microbiol Infect Dis. 2010;29(10):1243–1251. doi: 10.1007/s10096-010-0993-0. [DOI] [PubMed] [Google Scholar]
- 18.van Langevelde P, Ravensbergen E, Grashoff P, Beekhuizen H, Groeneveld PH, van Dissel JT. Antibiotic-induced cell wall fragments of Staphylococcus aureus increase endothelial chemokine secretion and adhesiveness for granulocytes. Antimicrob Agents Chemother. 1999;43(12):2984–2989. doi: 10.1128/aac.43.12.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strieter RM, Belperio JA, Keane MP. Host innate defenses in the lung: the role of cytokines. Curr Opin Infect Dis. 2003;16(3):193–198. doi: 10.1097/00001432-200306000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Hoogerwerf JJ, de Vos AF, Bresser P. Lung inflammation induced by lipoteichoic acid or lipopolysaccharide in humans. Am J Respir Crit Care Med. 2008;178(1):34–41. doi: 10.1164/rccm.200708-1261OC. [DOI] [PubMed] [Google Scholar]
- 21.Opitz B, van Laak V, Eitel J, Suttorp N. Innate immune recognition in infectious and noninfectious diseases of the lung. Am J Respir Crit Care Med. 2010;181(12):1294–1309. doi: 10.1164/rccm.200909-1427SO. [DOI] [PubMed] [Google Scholar]
- 22.Sémiramoth N, Gleizes A, Turbica I. Escherichia coli type 1 pili trigger late IL-8 production by neutrophil-like differentiated PLB-985 cells through a Src family kinase- and MAPK-dependent mechanism. J Leukoc Biol. 2009;85(2):310–321. doi: 10.1189/jlb.0608350. [DOI] [PubMed] [Google Scholar]
- 23.Zaki Mel S. Interleukin 8 is a surrogate marker for rapid diagnosis of bacteriuria. Immunol Invest. 2008;37(7):694–703. doi: 10.1080/08820130802307278. [DOI] [PubMed] [Google Scholar]
- 24.Bermejo-Martin JF, Martin-Loeches I, Rello J. Host adaptive immunity deficiency in severe pandemic influenza. Crit Care. 2010;14(5):R167. doi: 10.1186/cc9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun K, Torres L, Metzger DW. A detrimental effect of interleukin-10 on protective pulmonary humoral immunity during primary influenza A virus infection. J Virol. 2010;84(10):5007–5014. doi: 10.1128/JVI.02408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mocellin S, Panelli MC, Wang E, Nagorsen D, Marincola FM. The dual role of IL-10. Trends Immunol. 2003;24(1):36–43. doi: 10.1016/s1471-4906(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 27.Tuvim MJ, Evans SE, Clement CG, Dickey BF, Gilbert BE. Augmented lung inflammation protects against influenza A pneumonia. PLoS ONE. 2009;4(1):e4176. doi: 10.1371/journal.pone.0004176. [DOI] [PMC free article] [PubMed] [Google Scholar]