Summary
Biogenesis and replication of the porcine reproductive and respiratory syndrome virus (PRRSV) include the crucial step of replicative polyprotein processing by self-encoded proteases. Whole genome bioinformatics analysis suggests that nonstructural protein 4 (nsp4) is a 3C-like serine protease (3CLSP), responsible for most of the nonstructural protein processing. The gene encoding this protease was cloned and expressed in Escherichia coli in order to confirm this prediction. The purified protein was crystallized, and the structure was solved at 1.9 Å resolution. In addition, the crystal structure of the Ser118Ala mutant was determined at 2.0 Å resolution. The monomeric enzyme folds into three domains, similar to that of the homologous protease of equine arteritis virus, which, like PRRSV, is a member of the family Arteriviridae in the order of Nidovirales. The active site of the PRRSV 3CLSP is located between domains I and II and harbors a canonical catalytic triad comprising Ser118, His39, and Asp64. The structure also shows an atypical oxyanion hole and a partially collapsed S1 specificity pocket. The proteolytic activity of the purified protein was assessed in vitro. Three sites joining nonstructural protein domains in the PRRSV replicative polyprotein are confirmed to be processed by the enzyme. Two of them, the nsp3/nsp4 and nsp11/nsp12 junctions, are shown to be cleaved in trans, while cis cleavage is demonstrated for the nsp4/nsp5 linker. Thus, we provide structural evidence as well as enzymatic proof of the nsp4 protein being a functional 3CLSP. We also show that the enzyme has a strong preference for glutamic acid at the P1 position of the substrate.
Abbreviations: nsp, nonstructural protein; 3CLSP, 3C-like serine protease; EAV, equine arteritis virus; SARS-CoV, severe acute respiratory syndrome coronavirus; 3CLpro, 3C-like proteinase; GST, glutathione S-transferase; MR, molecular replacement; SeMet, selenomethionine; PDB, Protein Data Bank; SGPE, Streptomyces griseus proteinase E; MS, mass spectrometry; MW, molecular weight; ESI, electrospray ionization
Keywords: PRRSV, 3C-like serine protease, structure, trans/cis cleavage
Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) was first discovered in North America in 19871, 2 and almost simultaneously (in 1990) in Europe.3 It manifests itself clinically with severe reproductive failure in sows and respiratory distress in neonatal pigs.2 In recent years, the virus has become endemic throughout the world, leading to significant economic losses in pig production.4 In the autumn of 2006, unparalleled outbreaks of atypical PRRS occurred in more than 10 provinces (or autonomous cities) of China, affecting more than 2 million pigs with nearly 400,000 fatal cases. Adult and growing pigs were also affected, which was different from classical PRRS.5 In 2007, an almost identical atypical PRRSV was found to be epidemic in Vietnam and China.6 Presently, only inactivated or attenuated vaccines are available to combat PRRS, but their efficacy is less than satisfactory due to adaptive evolution of PRRSV,7, 8 especially in the cases of the atypical PRRS outbreaks in China and Vietnam.5, 6 This urgent issue prompted us to exploit the possibility of drug discovery for treatment of the highly pathogenic PRRSV infection.
Like other RNA viruses in the order Nidovirales, such as equine arteritis virus (EAV), human coronavirus 229E, and severe acute respiratory syndrome coronavirus (SARS-CoV), PRRSV has a single-stranded positive-sense RNA genome that is about 15.3 kb in size, containing a large 5′-terminal replicase gene and a downstream set of structural protein genes.9 The replicase gene is translated into two large polyprotein precursors, 1a and 1ab (with molecular masses of about 269.1 and 430.3 kDa for pp1a and pp1ab, respectively), with the latter being synthesized following a ribosomal frame-shifting event.10 After the rapid autocatalytic release of nonstructural protein 1 (nsp1) and nsp2, the remainder of the polyproteins are cleaved into 10 mature nonstructural proteins (nsp3–nsp12) by the 3C-like serine protease (3CLSP), which is also often simply called 3C-like proteinase (3CLpro).9, 11 The functional importance of the 3CL proteinase in the viral life cycle makes it an attractive target for the development of drugs directed against PRRSV.
Here, we confirm that PRRSV nsp4, which we demonstrate to extend from residue G1780 to E1983 in the viral polyprotein of the virus strain JXA1, is a 3CLSP from both the structural and enzymatic points of view. The structure of the enzyme reveals two chymotrypsin-like β-barrel domains and an extra C-terminal α/β domain similar to that observed in EAV nsp4.12 A canonical catalytic triad that is composed of Ser118, His39, and Asp64 is located in the open cleft between the two β-barrel domains. An S1 subsite consisting of His133 and very likely Ser136 and an oxyanion hole without the signature helix-like turn are seen in our structure. Our results also clearly show that the recombinant enzyme displays proteolytic activity in vitro. Besides, based on the results of the cleavage assay, three nsp junction sites in the PRRSV replicase precursor are confirmed, thereby shedding light on the processing pathway of pp1a and pp1ab in the virus.
Results
Overall description of the recombinant PRRSV 3CLSP used in this study
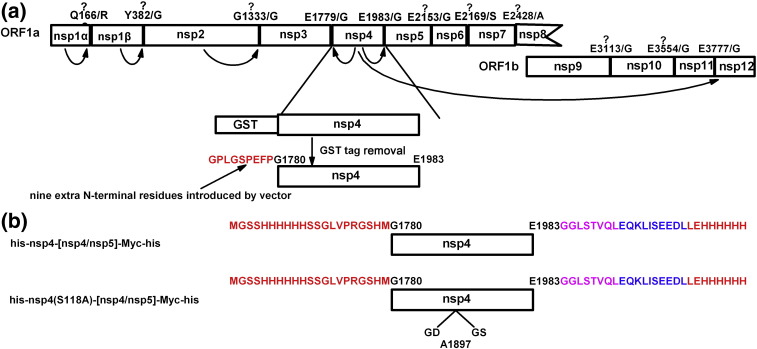
As a member of the Arteriviridae, PRRSV shows substantial similarities in genome organization to EAV—the prototype virus of this family.13, 14, 15 Accordingly, whole genome bioinformatics analysis predicts that the PRRSV nsp4 domain should be the main protease responsible for processing most of polyproteins pp1a and pp1ab.15 Previously, we reported on the successful expression of a gene construct encoding this putative serine protease in Escherichia coli as well as the crystallization of the protein.16 As shown in Fig. 1a, after removal of the glutathione S-transferase (GST) tag, nine additional amino acid residues were still left at the N-terminus of the protein. The in vitro peptide cleavage assay in this study was performed using this recombinant enzyme with the nine extra residues present, while for the cis cleavage assay, even more additional N-terminal residues were present (Fig. 1b). In both cases, the recombinant protein displayed proteolytic activity in vitro, thereby confirming the identity of nsp4 as the main protease in PRRSV replication (see results below).
Fig. 1.
Graphic representation of the recombinant 3CLSP used in this study. (a) Overview of the proteolytic processing of PRRSV replicase. The rapid autocatalytic release of nsp1α, nsp1β, and nsp2 after translation of the ORF1a or ORF1ab is indicated. The rest of the junction sites are processed by nsp4. All the predicted cleavage sites are listed above the schematic boxes representing nsp1 to nsp12. Three of the putative cleavage sites were confirmed by our work. The rest of them were not evidenced to be processed in our study and therefore are labeled with a question mark on the top. The numerals indicated in each cleavage site are based on the genomic information of PRRSV strain JXA1 (GenBank accession no. EF112445). The recombinant protein for crystallization trials and in vitro peptide cleavage assay was produced using the GST fusion expression system (GE Healthcare). Nine extra N-terminal amino acid residues remained after removal of the GST tag. (b) The translational fusion proteins used in the cis cleavage assay. The red-colored portion indicates the additional residues introduced by the pET-28b vector. The eight N-terminal amino acid residues of nsp5 are colored pink and the Myc epitope is highlighted in blue.
Structure determination and quality of the refined structure
Although the sequence identity between PRRSV and EAV nsp4s is more than 34%, we failed to solve the structure of PRRSV 3CLSP by molecular replacement (MR) using the structure of EAV nsp4 as a search model. The presence of two methionine residues in the recombinant PRRSV 3CLSP suggested that selenomethionine (SeMet)-based multiple-wavelength anomalous dispersion17 could be used to solve the phase problem. The unit-cell dimensions of the crystals (a = 112.4 Å, b = 49.2 Å, c = 42.9 Å, β = 110.3°; space group C2) indicate that there is only one molecule per asymmetric unit, and we successfully located both of the two crystallographically independent selenium sites. Based on the obtained phases, a final model with good stereochemistry was built (for details, see Materials and Methods).
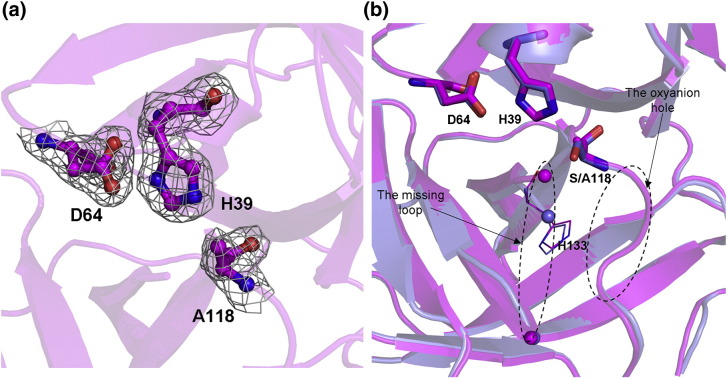
As shown in Table 1 , the structure was refined to 1.9 Å resolution, with R cryst = 19.6% and R free = 24.6%; 89.5% of the residues lie in the most favored regions of the Ramachandran plot and 10.5% lie in the additionally allowed regions.18 The quality of the electron density map of the recombinant protease at 1.9 Å is excellent, with an overall temperature factor of 22.2 Å2, and allowed us to model the positions of the main chain, most side-chain atoms, and 208 solvent molecules unambiguously. At the polypeptide chain termini, electron density is visible starting from residue 3 and ending at residue 199, but loop 136–140 is not seen in the maps due to disorder.
Table 1.
Data collection and refinement statistics
| Native protein | S118A mutant | |||
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2 | C2 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 112.4, 49.2, 42.9 | 113.3, 48.6, 42.9 | ||
| α, β, γ (°) | 90.0, 110.3, 90.0 | 90.0, 110.4, 90.0 | ||
| SeMet multiple-wavelength anomalous dispersion | ||||
| Native | Peak | Remote | ||
| Wavelength (Å) | 1.5418 | 0.9793 | 0.9000 | 1.5418 |
| Resolution range (Å) | 32.3–1.9 | 53.0–1.9 | 50.0–2.0 | 44.19–2.01 |
| Rmergea,b | 0.045 (0.167) | 0.213 (0.459) | 0.123 (0.216) | 0.049 (0.169) |
| Average I/σ(I)a | 22.1 (4.3) | 12.5 (1.4) | 10.1 (2.4) | 47.1 (11.8) |
| Completeness (%)a | 95.2 (91.6) | 95.7 (77.6) | 89.8 (75.7) | 98.9 (96.3) |
| Redundancy | 4.5 (4.3) | 4.5 (3.4) | 2.4 (2.0) | 5.2 (4.8) |
| Refinement | ||||
| Rcryst/Rfree (%)c | 19.6/24.6 | 18.2/21.5 | ||
| Ramachandran plot (%)d | ||||
| Most favored regions | 89.5 | 90.2 | ||
| Additionally allowed regions | 10.5 | 9.8 | ||
| Generously allowed regions | 0 | 0 | ||
| Disallowed regions | 0 | 0 | ||
| r.m.s.d. from ideality | ||||
| Bonds (Å) | 0.017 | 0.018 | ||
| Angles (°) | 1.572 | 1.529 | ||
| Average B-factor (Å2) | ||||
| Protein | 19.933 | 28.120 | ||
| Water | 36.572 | 37.980 | ||
Values for the outermost resolution shell are given in parentheses.
Rmerge = ∑i∑hkl|Ii − 〈I〉|/∑i∑hklIi, where Ii is the observed intensity and 〈I〉 is the average intensity from multiple measurements.
Rcryst = ∑||Fo| − |Fc||/∑|Fo|, where Fo and Fc are the structure-factor amplitudes from the data and the model, respectively. Rfree is the R-factor for a subset (5%) of reflections that was selected prior to refinement calculations and was not included in the refinement.
Ramachandran plots were generated by using the program PROCHECK.
The overall structure of PRRSV 3CLSP and comparison with EAV nsp4
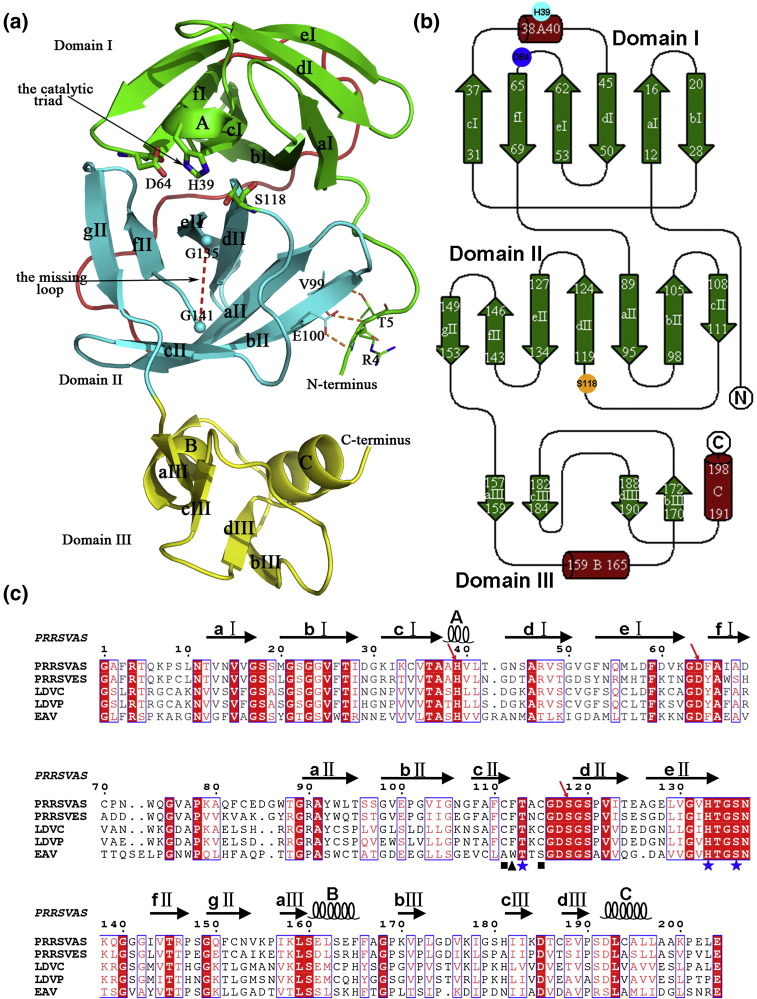
The PRRSV 3CLSP is folded into three domains, including two antiparallel β-barrels and an extra C-terminal α/β domain (Fig. 2a and b). The N-terminal β-barrel (domain I) consists of six β-strands (aI to fI) and a short α-helix (helix A) that closes the barrel like a lid, whereas the middle β-barrel (domain II) comprises seven β-strands (aII to gII) and is connected to domain I via a very long loop (amino acids 69–89) (Fig. 2a). The catalytic site is located at the opening of the cleft between domains I and II and consists of residues Ser118, His39, and Asp64 (Fig. 2a), which are totally conserved among members of the Arteriviridae (Fig. 2c). The extra C-terminal domain (domain III) comprises residues 157–199 and consists of two pairs of short antiparallel β-sheets (strands aIII–cIII and bIII–dIII) and two α-helices. The crossing angle is ∼ 90° between helix B and helix C (Fig. 2a). A long N-terminal loop extends over domain II to the vicinity of domain III. The orientation of this loop is stabilized by the three hydrogen bonds Arg4 N…Glu100 OE1, Thr5 N…Val99 O, and Thr5 OG1…Val99 N (Fig. 2a). The phenyl ring of Phe3 is directed into the hydrophobic interface between domains II and III and, therefore, also helps to stabilize the orientation of the N-terminal loop (Fig. 5a, left image).
Fig. 2.
Crystal structure of PRRSV nsp4. (a) Cartoon representation of the overall structure of PRRSV nsp4. The N-terminal (aI–fI) and middle (aII–gII) β-barrels are colored green and cyan, respectively, with the red broken line indicating the loop 136–140, which is totally devoid of visible electron density. The C-terminal extra domain is shown in yellow. Members of the catalytic triad and the three hydrogen bonds that are believed to stabilize the orientation of the long N-terminus are featured. The long loop connecting domains I and II is highlighted in red. (b) Topological presentation of the same structure. (c) Multiple sequence alignment of the 3CLSPs of the members in the family Arteriviridae. The catalytic triad is indicated by red arrows, and the three residues that are believed to constitute the S1 specificity pocket in EAV nsp4 are marked with blue stars. The Phe residue, which is thought to lead to the atypical oxyanion hole in PRRSV 3CLSP, is labeled by a dark triangle. Cys111 and Cys115, which have been referred to in the text, are marked with black squares. LDVC and LDVP, lactate dehydrogenase-elevating virus neurovirulent type C and strain Plagemann, respectively; PRRSVAS and PRRSVES, PRRSV American strain and European strain, respectively.
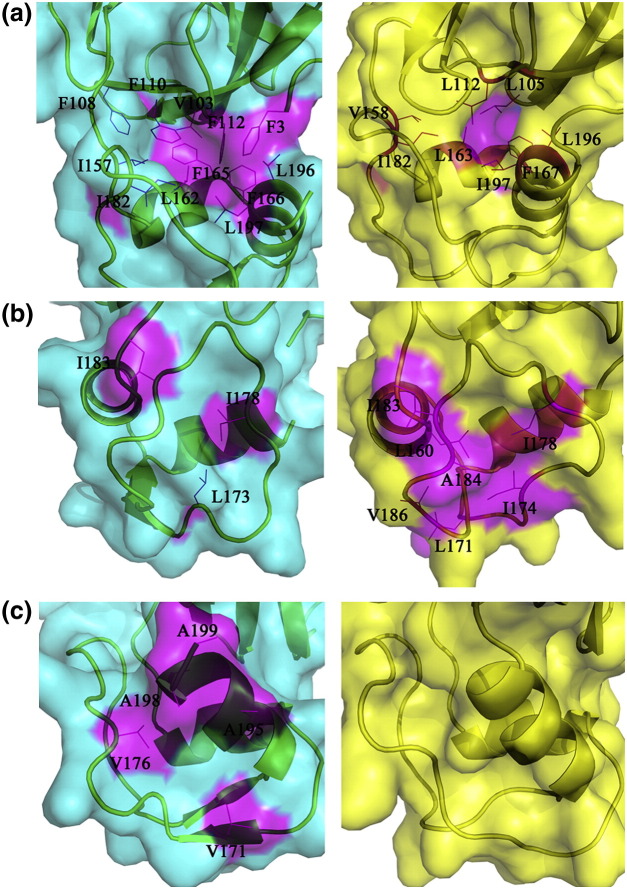
Fig. 5.
Surface representation of (a) the hydrophobic interface between domains II and III; (b) the exposed hydrophobic patch on one face of the C-terminal extra domain and (c) another patch of exposed hydrophobic residues located on a different face of the same extra domain of both PRRSV (left) and EAV (right) nsp4s. Residues involved are labeled and colored magenta. The surface is shown in cyan for PRRSV nsp4 and in yellow for EAV nsp4.
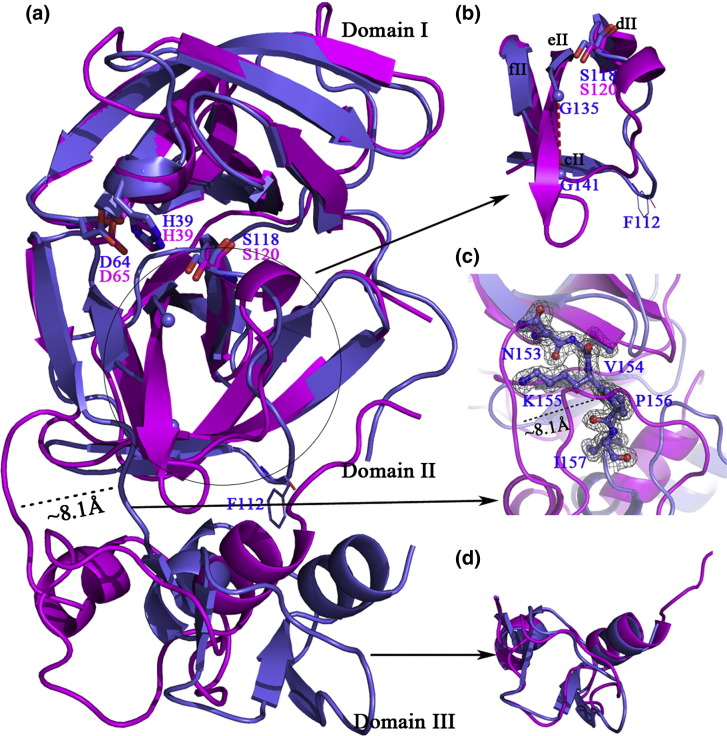
A Dali search19 of the Protein Data Bank (PDB) database gave a variety of proteins with significant structural similarities to our structure, among which the EAV nsp4 expectedly had the highest Z score (21.7). As the prototype structure of nsp4s in Arteriviridae, the EAV 3CLSP also folds into three domains.12 Superposing domains I and II of PRRSV 3CLSP onto those of the EAV nsp4 (copy A of the four copies in the asymmetric unit) yielded an r.m.s.d. of 2.04 Å for 122 equivalent (out of 134 compared) Cα pairs (Fig. 3a), while domain III alone exhibited an r.m.s.d. of 0.52 Å for 30 (out of 38) Cα pairs (Fig. 3d), indicating that the two enzymes share great structural similarity at the tertiary level. However, the steric arrangement of the domains in the two proteases is quite different. An ∼ 8 Å shift of domain III relative to domains I and II was observed in our structure, when compared to EAV nsp4. This shift causes the whole domain III to move away from the cleft where the catalytic triad is located (Fig. 3a). Clear electron density for the loop Asn153-Ile157 that connects domains II and III, as shown in Fig. 3c, leaves no doubt about the chain tracing of the loop and verifies the 8-Å shift of domain III, compared to EAV nsp4. The shift may also account for our failure to solve the structure by MR using EAV nsp4 as the search model.
Fig. 3.
Structural alignments between EAV (magenta) and PRRSV (light blue) nsp4s. (a) Superposition of the two β-barrel domains of PRRSV 3CLSP with those of EAV nsp4. A shift of about 8 Å observed in the C-terminal extra domain is highlighted. The well-aligned catalytic triad of the two proteases is shown in stick representation. The two loop regions that display great discrepancies between the two enzymes are also featured. Residue Phe112, which makes hydrophobic interactions with domain III in PRRSV nsp4, is highlighted by line representation. (b) Magnified view of the two loop regions referred to in the text. (c) Stereo view of the electron density map of the loop (N153-I157) connecting domains II and III in PRRSV nsp4. This 2|Fo| − |Fc| map is contoured at 1.0 σ above the mean. (d) Alignment of domain III alone of PRRSV and EAV nsp4 to show their well superposition.
Further discrepancies were also observed in two loop regions. One is the loop connecting strands cII and dII (Cys111-Pro121 in PRRSV nsp4). In EAV nsp4, this loop is much like a tadpole with its head lying next to the catalytic nucleophile Ser120 (Fig. 3b). However, the corresponding loop in our structure is approximately “⨼” shaped with a protuberance pointing to domain III (Fig. 3a and b) on which the hydrophobic side chain of Phe112 resides. This residue is conserved in PRRSV and lactate dehydrogenase-elevating virus 3CLSP but not in EAV nsp4, where it is substituted by Trp (Fig. 2c). The phenyl ring of Phe112 protrudes into the hydrophobic interface between domains II and III (Fig. 5a, left image) and thereby causes the “⨼”-shaped conformation of the cII–dII loop.
Another loop of great variance is the one connecting strands eII and fII. In PRRSV 3CLSP, this loop is eight residues in length, from Gly135 to Gly142, and is highly flexible as proved by the total lack of electron density for residues 136–140. However, in EAV nsp4, the corresponding region is mainly a β-strand with a much shorter loop, indicating that the conformation of this part is relatively rigid (Fig. 3b).
Interestingly these two regions showing significant variance are actually believed to cover, respectively, the S1 specificity pocket and the oxyanion hole in EAV nsp4,12 two highly important features involved in substrate recognition and catalysis by 3CL proteases.
The S1 specificity pocket
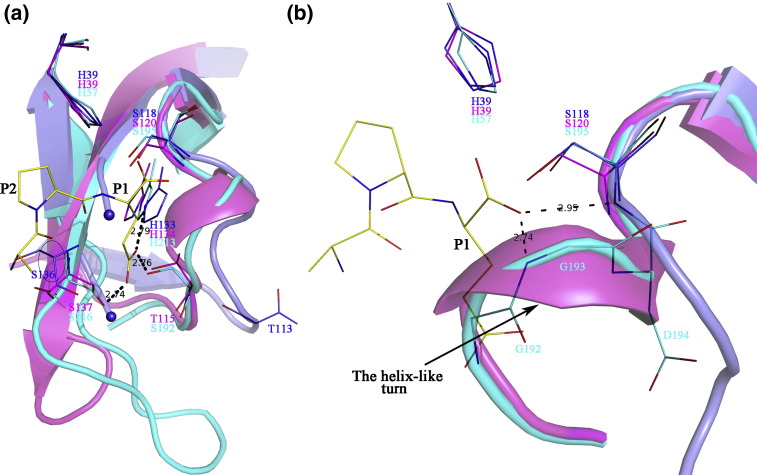
In 3CL proteases, the S1 specificity pocket accommodates the side chain of the substrate P1 residue and therefore determines the enzyme's preference for specific amino acid residues at this position. To identify the possible residues that may be involved in the formation of the S1 pocket in PRRSV 3CLSP, we used the structure of Streptomyces griseus proteinase E (SGPE) in complex with a tetrapeptide as the reference structure for superposition. SGPE is another chymotrypsin-like proteinase that has a substrate preference for Glu at the P1 position.20 Both the EAV nsp4 (copy A) and our structure were aligned. As shown in Fig. 4a, the S1 specificity pocket of both the SGPE and EAV nsp4 is composed of three residues, with a conserved His (His134 in EAV nsp4 and His213 in SGPE) lying at the bottom of the pocket and one Ser (EAV nsp4: Ser137; SGPE: Ser216) lining one side and another Thr/Ser (EAV nsp4: Thr115; SGPE: Ser192) located on the opposite side. However, only the histidine (His133) could be found in our structure. The first serine involved in the S1 subsite is conserved in PRRSV 3CLSP (Ser136) (Fig. 2c), but we cannot define its position due to the absence of electron density for the loop 136–140 as mentioned before. Nevertheless, there is no residue in PRRSV nsp4 to match the Thr/Ser (Thr115 in EAV nsp4, Ser192 in SGPE) on the other side of the pocket. Thr113 of PRRSV nsp4, which is also highly conserved among members of Arteriviridae (Fig. 2c), is too far away to contact the substrate P1 residue (Fig. 4a).
Fig. 4.
The S1 specificity pocket and the oxyanion hole of PRRSV nsp4. Black broken lines indicate hydrogen bonds. Residues referred to in the text are labeled. The position of Ser136 in PRRSV 3CLSP is extrapolated from the position of the residue directly preceding it (Gly135) and is therefore encircled in black. (a) Superposition of the S1 subsites of PRRSV nsp4 (light blue), EAV nsp4 copy A (magenta) (PDB code: 1MBM), and SGPE (cyan) (PDB code: 1HPG). (b) Comparison of the oxyanion holes of the three serine proteases. Gly192-Ser195 of SGPE is also shown with line representation. The signature helix-like turn is highlighted.
An atypical oxyanion hole without the signature helix-like turn
Both EAV nsp4 and SGPE form a canonical oxyanion hole that is characterized by a helix-like turn preceding the nucleophilic serine. As shown in Fig. 4b, the backbone amides of Ser195 and Gly193 in SGPE can form hydrogen bonds with and therefore stabilize the carbonyl oxyanion in the tetrahedral intermediate during catalysis. Although, in EAV nsp4, only copy B has the normal active oxyanion hole geometry while the other three copies only form collapsed ones, this difference is mainly caused by flipping of the peptide bond between Ser117 and Gly118 rather than a big change of the orientation of the signature turn.12 However, in PRRSV nsp4, this oxyanion-specific turn is missing. Instead, a much more stretched loop leaves only the amide group of the catalytic nucleophile (Ser118) in contact with the oxyanion. We term this rare property an atypical oxyanion hole.
A strong hydrophobic interaction between domains II and III
Compared to that of the EAV nsp4, an even stronger hydrophobic interaction between domains II and III is observed in our structure. With Phe3, Val103, Phe108, Phe110, Phe112, Ile157, Leu162, Phe165, Phe166, Ile182, L196, and L197, the interface harbors 12 hydrophobic residues, whereas in EAV nsp4, this interface only includes eight such residues (Fig. 5a). This hydrophobic core holds domains II and III together and likely forces the loop connecting the two domains to adopt its observed orientation, leading to the 8-Å shift as described above.
Another unexpected feature of the extra domain is the presence of two patches of solvent-exposed hydrophobic residues, implying possible interactions of PRRSV nsp4 with other proteins. One patch, which is also present in EAV nsp4 (Fig. 5b, right image),12 consists of Leu173, Ile178, and Ile183 (Fig. 5b, left image). Another patch, comprising Val171, Val176, Ala195, Ala198, and Ala199, is even more extended and locates on a surface that is almost perpendicular to the former one (Fig. 5c, left image, and 5b, left image). It is noteworthy that this second hydrophobic patch is totally absent in EAV nsp4 (Fig. 5c, right image).
Trans cleavage of domain junctions in pp1a/pp1ab
Peptides corresponding to eight predicted junctions in PRRSV pp1a or pp1ab (Fig. 1a; Table 2 ) were synthesized, tested for trans cleavage, and further analyzed by high-performance liquid chromatography (HPLC) in order to see whether the recombinant nsp4 is proteolytically active in vitro. In our assay, we showed that peptides corresponding to two junction sites can be cleaved by the enzyme. For each predicted cleavage site, the scissile peptide bond was between Glu and Gly.
Table 2.
Comparative cleavage of peptides corresponding to polyprotein junctions in PRRSV JXA1
| Peptide | Polyprotein junction | Sequence(P–P′) |
Cleavage | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P10 | P9 | P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | P5′ | P6′ | P7′ | P8′ | |||
| SP1 | nsp3/nsp4 | S | Q | L | G | S | L | L | E | G | A | F | R | T | Q | K | P | + | ||
| SP2 | nsp4/nsp5 | L | A | A | K | P | E | L | E | G | G | L | S | T | V | Q | L | − | ||
| SP3 | nsp5/nsp6 | F | F | L | R | Y | F | A | E | G | K | L | R | E | G | V | S | − | ||
| SP4 | nsp6/nsp7 | Q | S | C | G | M | S | H | E | S | L | T | G | A | L | A | M | − | ||
| SP5 | nsp7/nsp8 | E | N | R | R | A | Q | W | E | A | A | K | L | S | V | E | Q | − | ||
| SP6 | nsp9/nsp10 | E | K | L | R | S | N | H | E | G | K | K | S | R | M | C | G | − | ||
| SP7 | nsp10/nsp11 | R | A | I | C | A | D | L | E | G | S | S | S | P | L | P | K | − | ||
| SP8 | nsp11/nsp12 | K | T | A | Y | F | Q | L | E | G | R | H | F | T | W | Y | Q | +++ | ||
| SP8-1 | nsp11/nsp12 | K | D | K | T | A | Y | F | Q | L | E | G | R | H | F | T | W | +++ | ||
| SP8-M | K | D | K | T | A | Y | F | Q | L | Q | G | R | H | F | T | W | +/− | |||
Synthetic 16-mer peptides that represent the eight predicted junction sequences in the replicative polyprotein were incubated with 25 μM recombinant PRRSV nsp4 as described in Materials and Methods and analyzed by HPLC for the extent of cleavage. After incubation at 37 °C for 15 h, if more than 50% of the peptide was found to be cleaved as is determined by the decrease in area of the peak representing the parental peptide, “+++” is given; if 10–20% of the parental peptide is cleaved, we perceive it as “+”; if less than 10% is cleaved but still detectable amounts of products are observed as marked by the appearance of new peaks in the HPLC profile, we mark it “+/−”. “−” means no visible product peaks in the HPLC profile could be observed at all. All the cleaved products were verified by MS and/or comparison with HPLC traces of peptide standards. SP8-1 and SP8 correspond to the same junction site in pp1b. SP8-1 was synthesized and analyzed for the purpose of separating the two product peaks from each other.
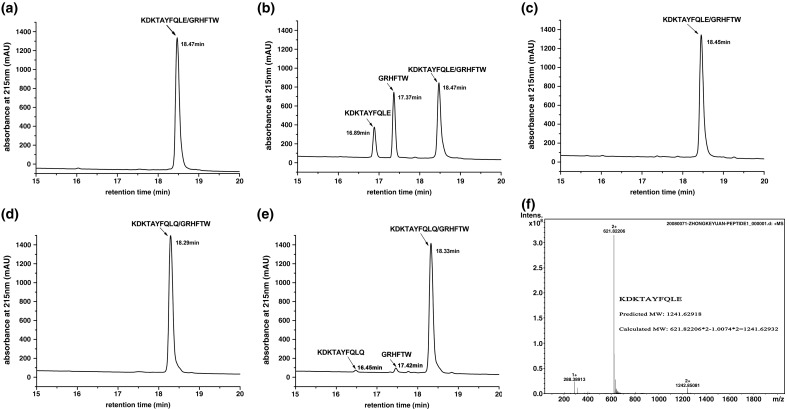
Just like other reports on peptide-based cleavage assays,21, 22 our enzyme also shows a remarkable difference in efficiency towards different peptides. Of the nine potential substrates investigated, only three can be cleaved to the extent of being detectable on an HPLC system (see Table 2). Peptide SP8-1, derived from the junction sequence between nsp11 and 12, is most susceptible to this enzyme. After incubation with the recombinant enzyme, two new peaks with retention times of 16.9 and 17.4 min, respectively, were observed after analyzing a reaction volume of 40 μl on a C18 column (Fig. 6b). The uncleaved peptide gave a peak at 18.5 min, consistent with that for the peptide-alone control (Fig. 6a). The two product peaks were collected and analyzed by mass spectrometry (MS). The results showed that the calculated molecular masses corresponding to the peaks were 1241.6293 and 802.3875, respectively, matching their theoretical monoisotopic molecular weight (MW) perfectly (the theoretical MW for fragment KDKTAYFQLE is 1241.6291, and for GRHFTW, it is 802.3874). A typical representation of the MS result is shown in Fig. 6f. When the catalytic serine residue was replaced by alanine (Ser118Ala mutant), no detectable peaks of cleaved peptides were observed in the HPLC profile (Fig. 6c). This confirmed the crucial function of Ser118 as the nucleophile in catalysis.
Fig. 6.
Proteolytic activity of recombinant PRRSV nsp4 in an in vitro peptide cleavage assay using either SP8-1 or SP8-M as substrate. Two hundred fifty micromolar of either peptide was incubated either with 25 μM nsp4 or 25 μM S118A mutant protein or without any enzyme added at 37 °C for 15 h and analyzed by HPLC (for details, see Materials and Methods). Two additional peaks were observed for the peptides incubated with nsp4. The retention time and the identity of each peptide are given. (a) SP8-1 alone. (b) SP8-1 plus nsp4. (c) SP8-1 plus nsp4 (S118A). (d) SP8-M alone. (e) SP8-M plus nsp4. (f) A typical MS result of the product peptide with a retention time of 16.89 min in the HPLC profile for SP8-1 incubated with nsp4. Its calculated MW is determined using the formula shown in the figure. 1.0074 represents the accurate MW of the hydrogen atom.
Another peptide that was shown to be processed is SP1, which corresponds to the nsp3/nsp4 junction. However, the proteolytic efficiency for SP1 was much lower than that for SP8-1. Only a small portion (< 20%) of the peptide was cleaved after a 15-h incubation (data not shown).
The kinetic parameters of the protein were determined in vitro using a fluorescent assay system to quantitatively test the proteolytic activity of the recombinant enzyme. The results showed that the enzyme exhibits a K m of about 2.24 mM and a k cat of about 0.435 min− 1 for the substrate Dabsyl-KTAYFQLE↓GRHFE-Edans.
The glutamic acid at the P1 position of the peptide substrate is crucial for effective cleavage and cannot be replaced by glutamine
3C-like proteases of RNA viruses are specific for either Gln or Glu at the P1 position of the substrate. Among the nidoviruses, coronavirus 3CLpro cleavage sites always have a Gln at the P1 position.9, 23, 24 However, in the same order Nidovirales, the 3CLSPs of Arteriviridae, which are represented by the prototype EAV nsp4, show a clear preference for glutamic acid over glutamine at the P1 position of the substrate.9 In EAV, of the eight cleavage sites of the replicative polyprotein processed by the main protease, only one junction contains a glutamine at the P1 position (Q2837/S joining nsp10/11), while all other cleavage sites have glutamic acid in this position.11, 25, 26 Based on the similarity in genome organization between PRRSV and EAV, the putative cleavage sites processed by 3CLSP were predicted in PRRSV, all of which harbor a Glu at the P1 position.15 Therefore, it is a reasonable assumption that the 3CLSP of PRRSV also shows preference for Glu over Gln at the P1 position of the substrate. The peptide SP8-M was synthesized and tested for cleavage by 3CLSP to confirm this prediction. SP8-M is identical in sequence to SP8-1 except that the Glu in P1 was replaced by Gln. As shown in Fig. 6d, the retention time for this peptide is 18.3 min. As expected, after incubation of the peptide with 3CLSP for 15 h at 37 °C, only tiny amounts of cleavage products can be detected (Fig. 6e), indicating that the glutamic acid at the P1 position in the substrate is crucial for effective cleavage by 3CLSP and cannot be replaced by glutamine.
The nsp4/nsp5 junction site is processed by 3CLSP via cis cleavage
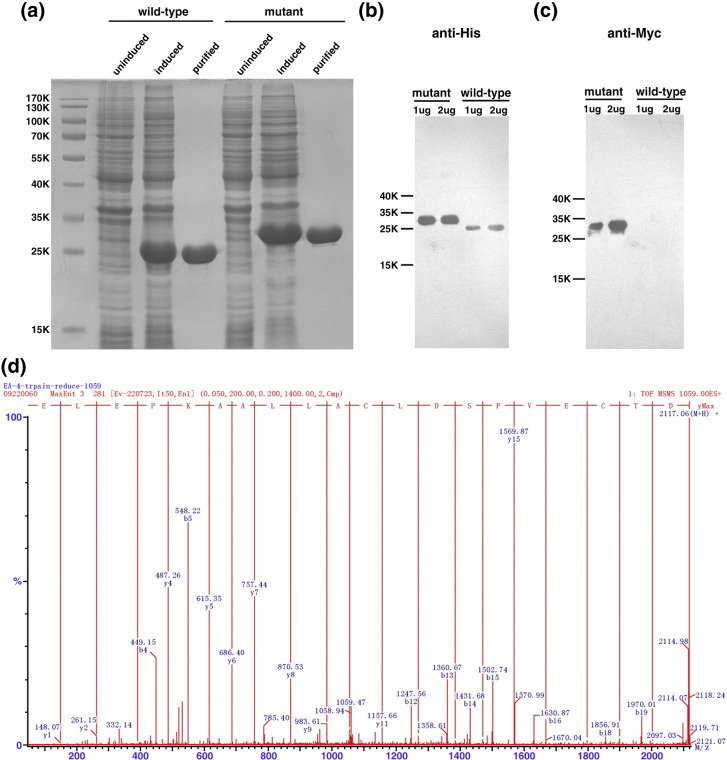
Whereas the linker between nsp3 and nsp4 was shown to be processed in vitro, the SP2 peptide that harbors the junction of nsp4 and nsp5 was not observed to be cleaved in trans in our in vitro assay (Table 2). However, processing of the nsp4/nsp5 junction is crucial for the release of the enzyme to free itself from the polyprotein precursor. Therefore, a cis cleavage assay was designed in order to investigate processing of this site. As shown in Fig. 1b, two constructs bearing the nsp4/nsp5 junction fused to a Myc epitope at the C-terminus of wild-type 3CLSP or its S118A mutant were made. Extra N-terminal and C-terminal hexa-histidine tags were introduced into the fusion proteins by the pET-28b vector to facilitate detection, resulting in the wild-type protein his-nsp4-[nsp4/nsp5]-Myc-his and the mutant product his-nsp4(S118A)-[nsp4/nsp5]-Myc-his. After expression of the two gene constructs in E. coli, an apparent molecular mass difference between the two protein products was observed in SDS-PAGE, where the mass of the wild-type protein is smaller than that of the mutant by about 3 kDa, indicating the occurrence of cis cleavage at the C-terminal junction site of his-nsp4-[nsp4/nsp5]-Myc-his (Fig. 7a). Both the wild-type and the mutant proteins were purified and subjected to Western blot analysis for detection of the Myc epitope as well as the hexa-histidine tag to confirm this conclusion. Two different amounts (2 and 1 μg) of both proteins were loaded. At both amounts, the wild-type protein showed no signal for the Myc epitope, in contrast to a strong Myc signal given by the mutant protein (Fig. 7c). Both proteins displayed an unambiguous His-tag signal, which was much stronger in the case of the mutant product, consistent with the fact that it contains a hexa-His tag at both termini while the wild-type protein only has the N-terminal tag left (Fig. 7b). These results clearly showed that his-nsp4-[nsp4/nsp5]-Myc-his removed its C-terminal tail via cis cleavage after expression.
Fig. 7.
The cis cleavage activity of PRRSV nsp4. Both his-nsp4-[nsp4/nsp5]-Myc-his and his-nsp4(S118A)-[nsp4/nsp5]-Myc-his proteins bear the nsp4/nsp5 junction sequence fused to a Myc epitope at the C-terminus. The N- and C-terminal hexa-His tags were introduced by the pET-28b vector. After induction by IPTG at 37 °C for 5 h, the cells were harvested, lysed, and then purified by affinity chromatography on a Ni-NTA column followed by gel filtration. Coomassie staining as well as Western blot analysis results are shown. (a) The 12% SDS-PAGE separation profile of both the wild-type and the mutant fusion proteins. The uninduced cell sample, cell lysate after induction, and the purified protein are indicated. (b) Detection of the wild-type and the mutant fusion proteins by the anti-His antibody. (c) Detection of both proteins using an anti-Myc antibody. (d) C-terminal sequencing result of the purified his-nsp4-[nsp4/nsp5]-Myc-his protein. After digestion of the protein by trypsin, a peptide fragment of 2117.06 Da was chosen to sequence by nano-ESI-MS/MS. The deduced sequence (read from right to left) is identical with that of the C-terminal portion of the his-nsp4-[nsp4/nsp5]-Myc-his protein after cleavage at the E1983/G junction, confirming the cleavage.
The purified wild-type protein was sent to the National Center of Biochemical Analysis, Academy of Military Medical Sciences, for C-terminal sequencing to further ascertain the exact peptide bond cut by the enzyme. After digestion of the protein using trypsin, a peptide fragment with observed mass of 2117.06 was found and selected for further sequencing. As shown in Fig. 7d, the amino acid sequence determined for this fragment is 100% identical with that of the C-terminal portion of the wild-type protein after cleavage at the E1983/G junction, which, in turn, confirms this site to be the nsp4/nsp5 junction in pp1a or pp1ab.
The structure of the S118A mutant of 3CLSP
The 2. 0-Å structure of the S118A mutant protein was solved by MR using the wild-type structure as the search model and refined to R cryst = 18.2% and R free = 21.3 (see Table 1). The overall structure of the mutant protein is almost identical with that of the native 3CLSP, with an r.m.s.d. of only 0.19 Å. The structure contains residues 3–199 but, like the wild-type structure, lacks electron density for the loop 136–140. As shown in Fig. 8a, there is clear electron density for the catalytic center, demonstrating that substitution of Ser118 by Ala was successful. At the same time, the substitution did not change the microenvironment surrounding residue 118, as seen from the unchanged conformations of the (truncated) catalytic triad, the S1 subsite residue His133, and the oxyanion hole (see Fig. 8b). The consistent conformations of these parts between the native and the mutant proteins make the mutant structure an appropriate model for enzyme–substrate interaction studies.
Fig. 8.
The structure of the S118A mutant 3CLSP. (a) Stereo view of the electron density map of the mutant residue Ala118 as well as the other two residues of the catalytic triad. The 1.0 σ contoured map confirmed the successful substitution of Ser118 by Ala. (b) Comparison of the native structure (light blue) with that of the mutant protein (magenta). The catalytic triad is shown as sticks. His133 is also shown because of its crucial role in substrate recognition. The missing loop and the oxyanion hole region are also highlighted. The consistent conformations of these parts between the native and the mutant proteins make the mutant structure an appropriate model for enzyme–substrate interaction studies.
Discussion
PRRSV nsp4 has been predicted and accepted as the main proteinase, with a pivotal role in the viral life cycle, but experimental proof had been lacking. Our work, for the first time, reports the crystal structure and in vitro proteolytic activity of this enzyme.
The S1 specificity pocket that accommodates the side chain of the substrate's P1 residue plays a vital role in substrate recognition and catalysis. In all the reported 3CLpros with known structures, a conserved histidine is found sitting at the bottom of the pocket.12, 22, 27, 28, 29, 30, 31, 32, 33 Mutational analysis found that any replacement of this residue both in human and feline coronavirus Mpro completely abolished their proteolytic activities.34, 35 As a member of the 3C-like protease family, it is not surprising that in PRRSV nsp4, His133 is localized appropriately to fulfill this function. However, besides this conserved His, the S1 subsite of both SGPE and EAV nsp4 includes two additional Ser/Thr residues as shown in Fig. 4a,12, 20 for which no counterparts can be found in our structure. We suspect that Ser136 must also be involved in P1 side-chain recognition based on two facts. First, this Ser is absolutely conserved in arterivirus nsp4 sequences. In EAV nsp4, the same Ser has been proposed to contact the side chain of the P1 glutamic acid (Fig. 4a). Furthermore, although we cannot define the position of Ser136 due to total lack of electron density, the two residues directly preceding it are clearly defined. Therefore, Ser136 is confined to a limited steric space close to the modeled P1 residue. A hydrogen bond between this disordered residue and the P1 amino acid could help stabilize both residues upon substrate binding. We also note that loop 136–140 lines one wall of the S1 subsite and is highly flexible. It may function like a lid that can partially cover the pocket after the entry of the P1 side chain and therefore help clinch the substrate to allow the occurrence of nucleophilic attack onto the scissile bond. This may also, to some extent, compensate the lack of a third partner in the S1 specificity pocket of PRRSV nsp4 (see Results). Snijder et al. reported that mutation of this third residue, Thr115 in EAV nsp4, to other amino acids can have various effects on catalysis, indicating its important role as part of the S1 subsite.11
It is noteworthy that our enzyme also shows weak proteolytic activity against SP8-M with a Gln at the P1 position (Fig. 6e; Table 2). This might reflect the flexibility of the substrate binding of the enzyme that can also accommodate Gln in addition to Glu. Recognition of both residues has been reported in other 3C proteinases, for example, 3C from members of the Picornaviridae.22, 36 Taking the similarity of the side chains of Glu and Gln into account, this might not be surprising. Furthermore, the flexibility of Ser136 might also be attributed to this partial compatibility.
We also report a rare form of oxyanion hole in our structure, which is totally lacking the signature helix-like turn. In serine proteinases, the catalytic cleavage of the substrate peptide bond is assumed to undergo a transformation from the ground planar state to a transition state that is characterized by the formation of a tetrahedral intermediate.37 The backbone NH groups (in the case of subtilisin also including the side chain of Asn155) can hydrogen-bond with the resultant carbonyl oxyanion of the substrate P1 residue during cleavage at the scissile bond and are therefore very important in catalysis.37, 38, 39, 40 Both in EAV nsp4 and SARS-CoV Mpro, two forms of the oxyanion hole were observed, an active form and a collapsed one.12, 28 Nevertheless, in both forms, the signature turn remains almost the same. In EAV nsp4, the different forms of the oxyanion hole are believed to arise mainly from the flipping of the peptide bond between Ser117 and Gly118, which is common in pairs of homologous protein structures.12, 41 However, as shown in Fig. 4b, the stretched form observed in our structure (lacking the helix-like turn) can only support the formation of one hydrogen bond with the oxyanion to stabilize the negative charge. We suspect that the hydrophobic strength of Phe112 might be the main cause for this atypical oxyanion hole. This is supported by our recent mutational work of Phe112Ala. The mutant enzyme shows some higher enzymatic activity, implying the role that Phe112 played in the formation of the atypical oxyanion hole, which might be attributed to the observed low enzymatic activity (data not shown). This needs to be verified by a clear crystal structure and further mutational work in the future. The unusual properties of both the S1 subsite and the oxyanion hole observed in our structure, as we discussed above, might well explain the low in vitro catalytic activity of PRRSV nsp4 compared to that of other 3C-like proteases in similar experiments.21, 22
Another interesting point is that in the neighborhood of the substrate-binding site, Cys111 is located in close proximity to Cys115 but has its side chain directed away from that of the latter, so that a disulfide bond cannot be formed. Disulfide bonds are rare in proteins that function in the cytosol, but for instance in coronavirus nonstructural proteins, several cases have been observed.42, 43, 44 We presume that the side chain of Cys111 can adopt a different conformation and form a disulfide bond with Cys115, perhaps after substrate binding to facilitate the proteolysis. This should be addressed in the future with a substrate–enzyme complex structure. With great efforts, we failed to obtain the complex crystals so far.
As the main proteinase in Arteriviridae, nsp4 must first release itself from the replicative polyprotein to fulfill its proteolytic function of processing the downstream domains into functional replicative subunits. Unlike EAV, whose nsp4/nsp5 cleavage site is reported to be cleaved in the major processing pathway with the presence of nsp2 as the cofactor,26 we have shown in our cis cleavage assay that a quick and thorough cleavage at the C-terminus of PRRSV nsp4 occurs automatically in vitro. Therefore, based on the results obtained in this study, we propose a possible nsp4 release model for PRRSV: Translation of ORF1a is followed by a rapid autocatalytic release of nsp1α, nsp1β, and nsp2;45, 46 subsequently, the nsp4/nsp5 junction site is quickly processed via cis cleavage by the 3CLSP to yield nsp3–nsp4 and nsp5–nsp8. Then, a trans cleavage of the nsp3/nsp4 joining sequence occurs to generate nsp3 and free nsp4, which can then fulfill its role as the main proteinase. Nevertheless, taking into account that the trans processing of the nsp3/nsp4 junction is very slow as shown by the peptide cleavage assay (see Results), we believe that cis cleavage at this site might also happen, especially in the early stage of translation of ORF1a. In support of this, a very long N-terminal loop was observed in our structure (Fig. 2a). This terminus can be brought into an appropriate position for cis cleavage by a conformational rearrangement upon breaking of the three hydrogen bonds mentioned above. Refinement of this cleavage model will require further studies in the future.
For coronaviral 3CLpros such as in SARS-CoV, there is plenty of evidence showing that the N-terminus of the protease is crucial for its proteolytic activity. Either addition of extra residues to the N-terminus or deletion of the N-terminal residues is deleterious to enzyme activity.47, 48, 49 There are also many reports showing that the homodimeric form of the SARS-CoV 3CLpro is the catalytically active form and the unique C-terminal extra domain plays a key role in controlling the dimer–monomer equilibrium.50, 51, 52, 53 In our previous report, we showed that the recombinant PRRSV nsp4 with nine extra amino acid residues at the N-terminus is monomeric in solution.16 These extra residues might also affect the enzymatic activity of PRRSV nsp4 and might be attributed to the high K m observed for the enzyme. Nevertheless, we confirm here that the recombinant enzyme displays obvious catalytic activity in vitro. Furthermore, the enzyme is actually active in vitro even with 21 extra N-terminal residues added as in the cis cleavage assay. Thus, the 3C-like proteases of Nidovirales likely evolved different proteolytic mechanisms for Coronaviridae such as SARS-CoV and Arteriviridae such as PRRSV. These aspects merit further study in the future.
Materials and Methods
Construction of the recombinant plasmids
The construction of the recombinant plasmid encoding wild-type 3CLSP has been described previously.16 Briefly, the coding sequence of the PRRSV nsp4 was amplified by reverse transcription-PCR reaction from the genomic RNA extracts of PRRSV isolate (GenBank accession no. EF112445) and then inserted into SmaI and XhoI sites of pGEX-6P-1 to yield plasmid pGEX-3CL. The S118A (Ser118 substituted by Ala) mutant construct was achieved by overlapping extension PCR. Using pGEX-3CL as template, with primer combinations of 3CL-F2/3CLM-R1 or 3CLM-F1/3CL-R2, two segments were obtained first and then they were mixed together at equal molar ratio as new templates in a second round of PCR by primer pairs 3CL-F2/3CL-R2. The resultant fragment that carries the S118A substitution was inserted into pGEX-6P-1 to generate pGEX-3CLM-S118A.
With primer pairs 3CL-EA-F1/3CL-EA-R1, using either pGEX-3CL or pGEX-3CLM-S118A as template, DNA fragments encoding translational fusion proteins his-nsp4-[nsp4/nsp5]-Myc-his and his-nsp4(S118A)-[nsp4/nsp5]-Myc-his were amplified respectively. The fragments were introduced into the pET-28b plasmid to create pET-3CL-EA and pET-3CL-S118A-EA (Fig. 1b).
All the constructs were verified by direct DNA sequencing, and the sequences of the primers used in this study are listed in Table 3 .
Table 3.
Primers used in this study
| Primer | Restriction site | Nucleic acid sequence (5′–3′) |
|---|---|---|
| 3CL-F2 | SmalI | cccgggGCGCTTTCAGAACTCAAAA |
| 3CL-R2 | XhoI | CCGctcgagTTATTCCAGTTCGGGTTTGGC |
| 3CLM-F1 | None | TGGCGATGCTGGATCCC |
| 3CLM-R1 | None | GGGATCCAGCATCGCCA |
| 3CL-EA-F1 | NdeI | CGCcatatgGGCGCTTTCAGAACTCAAAA |
| 3CL-EA-R1 | XhoI | CCGctcgagCAGATCCTCTTCTGAGATGAGTTTTTGTTCAAGTTGAACTGTGGAAAGGCCTCCTTCCAGTTCGGGTTTGGC |
Primers used in this study. Noncapitalized letters indicate the sequence of the restriction site inserted. Bold letters indicate nucleotide changes introduced. Italic letters represent the sequence encoding the Myc epitope. Underlined letters represent the coding sequence of the eight N-terminal residues of nsp5.
Protein purification and cystallization
The detailed production and purification procedure for the wild-type 3CLSP have been described previously.16 For the mutant enzyme, the same method was adopted. In brief, 1 μl of the plasmid was transformed into E. coli BL21 (DE3) competent cells for protein expression. The resultant GST fusion protein was purified by glutathione-Sepharose chromatography, cleaved with PreScission Protease, and the recombinant protein was further purified by size-exclusion chromatography.
SeMet-labeled 3CLSP was produced in BL21 (DE3) E. coli grown in SeMet minimal medium (0.65% YNB, 5% glucose, and 1 mM MgSO4 in M9 medium) supplemented with l-SeMet at 60 mg L− 1; lysine, threonine, and phenylalanine at 100 mg L− 1; and leucine, isoleucine, and valine at 50 mg L− 1.54 The expressed protein was purified as described above.
The purified proteins were concentrated, and their concentration was determined using BCA™ Protein Assay Kit (Pierce) according to the manufacturer's instructions.
Crystallization conditions were optimized, and the best crystals for both the wild-type and SeMet-labeled 3CLSP were obtained by vapor diffusion in hanging drops consisting of 1 μl of reservoir solution (0.1 M sodium citrate, pH 5.3, and 0.7 M monobasic ammonium phosphate) and 1 μl of concentrated protein solution (12 mg ml− 1 in 10 mM Tris–HCl and 10 mM NaCl, pH 8.0), followed by incubation at 18 °C for 7 days.
For the S118A mutant protein, the crystals were obtained under conditions similar to those for the wild-type 3CLSP, except that the pH value was elevated to 5.7 and the concentration of the mutant protein was 18 mg/ml. Crystals appeared after 10 days at 18 °C.
Data collection, phasing, and refinement
Crystals were flash-cooled in liquid nitrogen in cryoprotectant solution containing 15% glycerol and 50% reservoir solution. Multiwavelength X-ray diffraction data to 1.9 Å were collected from crystals of SeMet-labeled protein on beamline 3W1B of the Beijing Synchrotron Radiation Facility and processed with the HKL2000 suite of programs. Two selenium sites were located from data collected at peak and remote wavelengths using SOLVE.17 An initial model was built using RESOLVE55 followed by model refinement using REFMAC5 (CCP4 suite).56
X-ray diffraction data collected from crystals of the native protein on an in-house Rigaku MicroMax007 rotating-anode X-ray generator were phased using MR procedures in MOLREP as implemented in CCP4 Version 6.02. The initially determined structure was used as a search model. A series of iterative cycles of manual rebuilding were carried out in Coot and refinement with REFMAC5 (CCP4 suite).56
The S118A mutant structure was solved by MR method using the wild-type structure as the search model and refined to 2.0 Å.
The final statistics are summarized in Table 1. During the course of model building and refinement, the stereochemistry of the structures was checked by PROCHECK.18 All figures were generated using PyMOL57 and ESPript.58
In vitro peptide trans cleavage assay
The 16-mer substrate peptides (representing eight putative cleavage sites in PRRSV pp1a or pp1ab) for the in vitro 3CL protease peptide cleavage assay were purchased from Beijing Scilight Biotechnology Ltd. Co. Each peptide was dissolved in 100% dimethyl sulfoxide at a concentration of 50 mM and stored at − 80 °C. The nsp4 activity assay was performed in a reaction volume of 200 μl, using 5 μl nsp4 (or S118A mutant nsp4) (25 mg/ml) and 1 μl peptide (50 mM) in the reaction buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1 mM DTT, and 10% glycerol) to yield a final concentration of 25 μM enzyme and 250 μM peptide in the assay. Cleavage reactions were routinely incubated at 37 °C for 15 h. The reactions were terminated by the addition of an equal volume of 2% trifluoroacetic acid, and samples were frozen in liquid nitrogen immediately before being stored at − 70 °C. The samples were centrifuged for 10 min at 12,000 rpm before analysis by reverse-phase HPLC on a C18 column (4.6 mm × 250 mm). Cleavage products were resolved by using a 30-min, 0–40% linear gradient of acetonitrile in 0.1% trifluoroacetic acid. The absorbance was determined at 215 nm, and peak areas were calculated by integration. The identities of the product peptides were verified by quadrupole Fourier transform-MS analysis and/or comparison with HPLC traces of peptide standards.
Determination of the kinetic parameters of the recombinant PRRSV nsp4
A fluorescence-based assay with the fluorogenic peptide substrate Dabsyl-KTAYFQLE↓GRHFE-Edans (95% purity, Biosyntan GmbH, Berlin, Germany) was used to assess the activity of the PRRSV 3CLSP. This peptide contains the nsp11/nsp12 cleavage site (indicated by the arrow). The enhanced fluorescence due to the cleavage of this substrate as catalyzed by the enzyme was monitored at 490 nm with excitation at 340 nm, using a Cary Eclipse fluorescence spectrophotometer. The experiments were performed in a buffer consisting of 20 mM Tris–HCl (pH 7.3), 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid, and 5 mM DTT. Kinetic parameters K m and k cat were determined by initial-rate measurements at 25 °C. The reaction was initiated by adding proteinase (final concentration, 0.5 μM) to a solution containing different final concentrations of the fluorogenic peptide (10–40 μM). The initial rates were converted to enzyme activities (micromole substrate cleaved per second). Kinetic constants were derived by fitting the data to the Michaelis–Menten equation with the nonlinear regression analysis program Sigma Plot.
Analysis of the cis proteolytic activity of 3CLSP
To investigate the cis cleavage activity of 3CLSP, we designed two constructs bearing the nsp4/nsp5 scissile bond fused to a Myc epitope at the C-terminus of 3CLSP or 3CLSP (S118A), resulting in the expression vectors pET-3CL-EA and pET-3CL-S118A-EA. The N-terminal and C-terminal hexa-His tags were introduced into the recombinant proteins via vector pET-28b (Fig. 1b). The recombinant proteins were expressed and purified by using a Ni-NTA affinity chromatography column and a Superdex 75 Hiload 16/60 column (GE Healthcare). The uninduced cell sample, cell lysate after induction, and the purified protein were separated by 12% SDS-PAGE for Coomassie brilliant blue staining.
For Western blot analysis, 2 or 1 μg of the purified his-nsp4-[nsp4/nsp5]-Myc-his and his-nsp4(S118A)-[nsp4/nsp5]-Myc-his were loaded onto 12% SDS-PAGE for separation and then transferred onto Hybond nitrocellulose membranes (Amersham Biosciences). Membranes were blocked overnight in 10% milk powder dissolved in Tris-buffered saline. Blotted proteins were detected with the rabbit polyclonal anti-His primary antibody (Santa Cruz) or the mouse monoclonal anti-Myc antibody (Santa Cruz). A horseradish-peroxidase-labeled anti-rabbit or anti-mouse secondary antibody (Santa Cruz) was used. After each incubation step, membranes were washed four times with 0.05% Tween-20 in Tris-buffered saline. Detection was performed by using SuperSignal West Pico Chemiluminescent substrate solution (Pierce Biotechnology).
C-terminal sequencing of his-nsp4-[nsp4/nsp5]-Myc-his protein
The protein was prepared in 50 mM NH4HCO3 at a concentration of 1 mg/ml followed by digestion using trypsin (Roche) at 37 °C for 16 h with the mass ratio of trypsin to the protein set at 1:30. The digested products were further reduced by the addition of DTT to the digestion volume at 10 mM followed by incubation at 56 °C for 1 h. The resultant products were analyzed by electrospray ionization (ESI)-MS/MS. Then, a peptide fragment of 2117.06 Da was chosen for sequencing by ESI-MS/MS on a Q-TOF2 hybrid quadrupole/time-of-flight mass spectrometer (Micromass, UK).
PDB accession numbers
The coordinates and associated structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics PDB (PDB code 3FAN for the native protein; PDB code 3FAO for the S118A mutant structure).
Acknowledgements
We thank Stefan Anemüller for measuring the enzyme kinetics of PRRSV nsp4. This work was supported by National Key Technologies R&D Program (2006BAD06A04 and 2006BAD06A01) and National Basic Research Program (Project 973) of China (2005CB523001) and International Cooperation Project of the Ministry of Science and Technology (2006DFB32010 and 2007DFC30240). China–Japan Joint Laboratory of Molecular Immunology and Molecular Microbiology is, in part, supported by Japan Ministry of Education, Culture, Sports, Science and Technology. G.F.G. is a distinguished young investigator of the National Natural Science Foundation of China (Grant No. 30525010). R.H. acknowledges support by the Schleswig–Holstein Innovation Fund and the Fonds der Chemischen Industrie.
Edited by R. Huber
References
- 1.Nielsen H.S., Liu G., Nielsen J., Oleksiewicz M.B., Botner A., Storgaard T., Faaberg K.S. Generation of an infectious clone of VR-2332, a highly virulent North American-type isolate of porcine reproductive and respiratory syndrome virus. J. Virol. 2003;77:3702–3711. doi: 10.1128/JVI.77.6.3702-3711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paton D.J., Brown I.H., Edwards S., Wensvoort G. ‘Blue ear’ disease of pigs. Vet. Rec. 1991;128:617. doi: 10.1136/vr.128.26.617. [DOI] [PubMed] [Google Scholar]
- 3.A BVA Trust Symposium Porcine reproductive and respiratory syndrome (PRRS or blue-eared pig disease) Vet. Rec. 1992;130:87–89. doi: 10.1136/vr.130.5.87. [DOI] [PubMed] [Google Scholar]
- 4.Neumann E.J., Kliebenstein J.B., Johnson C.D., Mabry J.W., Bush E.J., Seitzinger A.H. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 2005;227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 5.Tian K., Yu X., Zhao T., Feng Y., Cao Z., Wang C. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE. 2007;2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y., Zhao T., Nguyen T., Inui K., Ma Y., Nguyen T.H. Porcine respiratory and reproductive syndrome virus variants, Vietnam and China, 2007. Emerg. Infect. Dis. 2008;14:1774–1776. doi: 10.3201/eid1411.071676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiss I., Sami L., Kecskemeti S., Hanada K. Genetic variation of the prevailing porcine respiratory and reproductive syndrome viruses occurring on a pig farm upon vaccination. Arch. Virol. 2006;151:2269–2276. doi: 10.1007/s00705-006-0805-0. [DOI] [PubMed] [Google Scholar]
- 8.Yuan S., Mickelson D., Murtaugh M.P., Faaberg K.S. Complete genome comparison of porcine reproductive and respiratory syndrome virus parental and attenuated strains. Virus Res. 2001;79:189–200. doi: 10.1016/S0168-1702(01)00295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 10.den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snijder E.J., Wassenaar A.L., van Dinten L.C., Spaan W.J., Gorbalenya A.E. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J. Biol. Chem. 1996;271:4864–4871. doi: 10.1074/jbc.271.9.4864. [DOI] [PubMed] [Google Scholar]
- 12.Barrette-Ng I.H., Ng K.K., Mark B.L., Van Aken D., Cherney M.M., Garen C. Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 2002;277:39960–39966. doi: 10.1074/jbc.M206978200. [DOI] [PubMed] [Google Scholar]
- 13.Blaha T. The “colorful” epidemiology of PRRS. Vet. Res. 2000;31:77–83. doi: 10.1051/vetres:2000109. [DOI] [PubMed] [Google Scholar]
- 14.Zimmerman J.J., Yoon K.J., Wills R.W., Swenson S.L. General overview of PRRSV: a perspective from the United States. Vet. Microbiol. 1997;55:187–196. doi: 10.1016/s0378-1135(96)01330-2. [DOI] [PubMed] [Google Scholar]
- 15.Wootton S., Yoo D., Rogan D. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch. Virol. 2000;145:2297–2323. doi: 10.1007/s007050070022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian X., Feng Y., Zhao T., Peng H., Yan J., Qi J. Molecular cloning, expression, purification and crystallographic analysis of PRRSV 3CL protease. Acta Crystallogr., Sect. F. 2007;63:720–722. doi: 10.1107/S1744309107033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 19.Holm L., Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 20.Nienaber V.L., Breddam K., Birktoft J.J. A glutamic acid specific serine protease utilizes a novel histidine triad in substrate binding. Biochemistry. 1993;32:11469–11475. doi: 10.1021/bi00094a001. [DOI] [PubMed] [Google Scholar]
- 21.van Aken D., Benckhuijsen W.E., Drijfhout J.W., Wassenaar A.L., Gorbalenya A.E., Snijder E.J. Expression, purification, and in vitro activity of an arterivirus main proteinase. Virus Res. 2006;120:97–106. doi: 10.1016/j.virusres.2006.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birtley J.R., Knox S.R., Jaulent A.M., Brick P., Leatherbarrow R.J., Curry S. Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J. Biol. Chem. 2005;280:11520–11527. doi: 10.1074/jbc.M413254200. [DOI] [PubMed] [Google Scholar]
- 23.Gao F., Ou H.Y., Chen L.L., Zheng W.X., Zhang C.T. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553:451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 25.van Dinten L.C., Rensen S., Gorbalenya A.E., Snijder E.J. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J. Virol. 1999;73:2027–2037. doi: 10.1128/jvi.73.3.2027-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wassenaar A.L., Spaan W.J., Gorbalenya A.E., Snijder E.J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J. Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Sisson W. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 30.Matthews D.A., Dragovich P.S., Webber S.E., Fuhrman S.A., Patick A.K., Zalman L.S. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl Acad. Sci. USA. 1999;96:11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosimann S.C., Cherney M.M., Sia S., Plotch S., James M.N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 32.Bergmann E.M., Mosimann S.C., Chernaia M.M., Malcolm B.A., James M.N. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 34.Ziebuhr J., Heusipp G., Siddell S.G. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 1997;71:3992–3997. doi: 10.1128/jvi.71.5.3992-3997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 36.Jewell D.A., Swietnicki W., Dunn B.M., Malcolm B.A. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry. 1992;31:7862–7869. doi: 10.1021/bi00149a017. [DOI] [PubMed] [Google Scholar]
- 37.Kraut J. Serine proteases: structure and mechanism of catalysis. Annu. Rev. Biochem. 1977;46:331–358. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- 38.Robertus J.D., Kraut J., Alden R.A., Birktoft J.J. Subtilisin; a stereochemical mechanism involving transition-state stabilization. Biochemistry. 1972;11:4293–4303. doi: 10.1021/bi00773a016. [DOI] [PubMed] [Google Scholar]
- 39.Henderson R. Structure of crystalline alpha-chymotrypsin. IV. The structure of indoleacryloyl-alpha-chyotrypsin and its relevance to the hydrolytic mechanism of the enzyme. J. Mol. Biol. 1970;54:341–354. doi: 10.1016/0022-2836(70)90434-1. [DOI] [PubMed] [Google Scholar]
- 40.Huber R., Bode W. Structural basis of the activation and action of trypsin. Acc. Chem. Res. 1978;11:114–122. [Google Scholar]
- 41.Hayward S. Peptide-plane flipping in proteins. Protein Sci. 2001;10:2219–2227. doi: 10.1110/ps.23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ponnusamy R., Moll R., Weimar T., Mesters J.R., Hilgenfeld R. Variable oligomerization modes in coronavirus non-structural protein 9. J. Mol. Biol. 2008;383:1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao J., Wu C.C., Lin T.L. Turkey coronavirus non-structure protein NSP15—an endoribonuclease. Intervirology. 2008;51:342–351. doi: 10.1159/000175837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, J., Vonrhein, C., Smart, O. S., Bricogne, G., Bollati, M., Kusov, Y. et al. (2009). The SARS-unique domain (SUD) of SARS coroanvirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. In press. [DOI] [PMC free article] [PubMed]
- 45.Kroese M.V., Zevenhoven-Dobbe J.C., Bos-de Ruijter J.N., Peeters B.P., Meulenberg J.J., Cornelissen L.A., Snijder E.J. The nsp1alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 2008;89:494–499. doi: 10.1099/vir.0.83253-0. [DOI] [PubMed] [Google Scholar]
- 46.den Boon J.A., Faaberg K.S., Meulenberg J.J., Wassenaar A.L., Plagemann P.G., Gorbalenya A.E., Snijder E.J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S., Chen L., Tan J., Chen J., Du L., Sun T. Severe acute respiratory syndrome coronavirus 3C-like proteinase N terminus is indispensable for proteolytic activity but not for enzyme dimerization. Biochemical and thermodynamic investigation in conjunction with molecular dynamics simulations. J. Biol. Chem. 2005;280:164–173. doi: 10.1074/jbc.M408211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verschueren K.H., Pumpor K., Anemuller S., Chen S., Mesters J.R., Hilgenfeld R. A structural view of the inactivation of the SARS coronavirus main proteinase by benzotriazole esters. Chem. Biol. 2008;15:597–606. doi: 10.1016/j.chembiol.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graziano V., McGrath W.J., Yang L., Mangel W.F. SARS CoV main proteinase: the monomer–dimer equilibrium dissociation constant. Biochemistry. 2006;45:14632–14641. doi: 10.1021/bi061746y. [DOI] [PubMed] [Google Scholar]
- 51.Hsu W.C., Chang H.C., Chou C.Y., Tsai P.J., Lin P.I., Chang G.G. Critical assessment of important regions in the subunit association and catalytic action of the severe acute respiratory syndrome coronavirus main protease. J. Biol. Chem. 2005;280:22741–22748. doi: 10.1074/jbc.M502556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273:1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 2004;279:24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Duyne G.D., Standaert R.F., Karplus P.A., Schreiber S.L., Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 55.Terwilliger T.C. Maximum-likelihood density modification. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collaborative Computing Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 57.DeLano W.L. DeLano Scientific; San Carlos, CA USA: 2002. The PyMOL Molecular Graphics System.http://www.pymol.org [Google Scholar]
- 58.Gouet P., Courcelle E., Stuart D.I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]