Abstract
Control of human African trypanosomiasis (HAT) is dependent on accurate diagnosis and treatment of infected patients. However, sensitivities of tests in routine use are unsatisfactory, due to the characteristically low parasitaemias in naturally infected individuals. We have identified a conserved sequence in the repetitive insertion mobile element (RIME) of the sub-genus Trypanozoon and used it to design primers for a highly specific loop-mediated isothermal amplification (LAMP) test. The test was used to analyse Trypanozoon isolates and clinical samples from HAT patients. The RIME LAMP assay was performed at 62 °C using real-time PCR and a water bath. DNA amplification was detectable within 25 min. All positive samples detected by gel electrophoresis or in real-time using SYTO-9 fluorescence dye could also be detected visually by addition of SYBR Green I to the product. The amplicon was unequivocally confirmed through restriction enzyme NdeI digestion, analysis of melt curves and sequencing. The analytical sensitivity of the RIME LAMP assay was equivalent to 0.001 trypanosomes/ml while that of classical PCR tests ranged from 0.1 to 1000 trypanosomes/ml. LAMP detected all 75 Trypanozoon isolates while TBR1 and two primers (specific for sub-genus Trypanozoon) showed a sensitivity of 86.9%. The SRA gene PCR detected 21 out of 40 Trypanosoma brucei rhodesiense isolates while Trypanosoma gambiense-specific glycoprotein primers (TgsGP) detected 11 out of 13 T. b. gambiense isolates. Using clinical samples, the LAMP test detected parasite DNA in 18 out of 20 samples which included using supernatant prepared from boiled blood, CSF and direct native serum. The sensitivity and reproducibility of the LAMP assay coupled with the ability to detect the results visually without the need for sophisticated equipment indicate that the technique has strong potential for detection of HAT in clinical settings. Since the LAMP test shows a high tolerance to different biological substances, determination of the appropriate protocols for processing the template to make it a user-friendly technique, prior to large scale evaluation, is needed.
Keywords: Loop-mediated isothermal amplification, Trypanozoon, Sleeping sickness, Diagnosis
1. Introduction
Human African trypanosomiasis (HAT) or sleeping sickness is endemic in sub-Saharan Africa where it is transmitted by tsetse flies of the Genus Glossina. The disease is caused by trypanosomes belonging to the sub-genus Trypanozoon namely Trypanosoma brucei rhodesiense and Trypanosoma brucei gambiense. T. b. rhodesiense causes disease in eastern, central and southern parts of Africa, while T. b. gambiense occurs in west and central Africa, extending from Angola to southern Sudan and Senegal. Other parasites of the sub-genus Trypanozoon are Trypanosoma brucei brucei and Trypanosoma evansi which cause diseases in livestock. However, a human case of T. evansi infection was recently reported in India (WHO, 2005, Truc et al., 2006), highlighting the need to develop definitive diagnostic tests for trypanosomes in humans.
Control of sleeping sickness relies on detection of the parasite and effective treatment of the patient. Routine diagnosis of the disease is based on direct visualisation of the parasite in blood, lymph node aspirates and CSF using a microscope (Van Meirvenne, 1999). This method has limited sensitivity due to fluctuating parasitaemia. In efforts to improve detection of trypanosomes, a number of diagnostic methods have been developed, including the mini-anion exchange centrifugation technique (mAECT; Lumsden et al., 1979), PCR (Welburn et al., 2001, Gibson et al., 2002, Radwanska et al., 2002, Jamonneau et al., 2003) and recently, a dipstick test (Deborggraeve et al., 2006) has been evaluated. Despite these advances, diagnosis of HAT remains unsatisfactory. The PCR-based tests have good sensitivity. However, the need for precision instruments and elaborate visualisation methods are obstacles to their wide application in clinical settings in Africa. Consequently, diagnosis of HAT involves a combination of parameters, such as origin of the patient, symptoms, demonstration of parasites by microscopy, or detection of specific antibodies using the Card Agglutination Test for Trypanosomiasis (CATT; Magnus et al., 1978). The paucity of definitive tests means that some patients go undetected and therefore become potential sources of infection to other people.
Early diagnosis of HAT is important in interrupting the transmission cycle of the parasite and progress of the disease to the late stage. Treatment of patients with late-stage disease, when parasites have invaded the CNS, is difficult due to the cost and long treatment schedules, which normally require hospital admission. Melarsoprol, the only drug that is effective for the late-stage T. b. rhodesiense form of disease, causes a post-treatment reactive encephalopathy in an estimated 10% of patients, half of which are fatal (Pépin and Milord, 1994, Kennedy, 2004). Modification of treatment regimes has not reduced mortality (Schmid et al., 2004) and treatment failure has been reported in the field (Legros et al., 1999, Matovu et al., 2001). Treatment of early stage HAT is much easier and safer, although some side effects have been reported when pentamidine or suramin are used (Lejon et al., 2003). Definitive diagnostic tests are therefore crucial for the early detection of cases. This would minimise false positives, and exposure of patients to drugs that are potentially dangerous (Inojosa et al., 2006), and whose efficacy may not be guaranteed.
Recently a rapid, simple and sensitive technique called loop-mediated isothermal amplification (LAMP) of DNA was developed by Notomi et al. (2000). LAMP is a novel strategy for gene amplification which relies on the auto-cycling strand displacement synthesis of DNA by Bst DNA polymerase under isothermal conditions (60–65 °C). The technique uses a set of six primers recognising eight sections of the target DNA (Nagamine et al., 2002). This increases amplification specificity, efficiency and rapidity. Moreover, the technique amplifies target DNA three-fold every half cycle, producing large amounts of product within 30–60 min (Notomi et al., 2000). The large amount of DNA formed allows visual detection of amplification through the addition of fluorescent dyes such as SYBR Green I (Poon et al., 2006) or Calcein (Boehme et al., 2007) and measurement of turbidity (Mori et al., 2001). LAMP has been used successfully in detection of human infectious agents for severe respiratory syndrome coronavirus (Hong et al., 2004), periodontis (Yoshida et al., 2005), malaria (Poon et al., 2006), peptic ulcers (Minami et al., 2006) and tuberculosis (Boehme et al., 2007). Further, Kuboki et al. (2003) demonstrated the potential of LAMP based on a single copy target (PFRA gene) for the detection of several T. brucei sp. More recently Thekisoe et al. (2007) have reported LAMP tests for T. evansi, Trypanosoma vivax, Trypanosoma congolense and T. b. gambiense. The LAMP test is attractive for diagnosis of HAT in sub-Saharan Africa where facilities are minimal, due to its speed, independence of specialised heating systems and results that can be visually inspected.
The availability of the sequenced genomes of several species of trypanosomes has provided information about genes that could be targeted as diagnostic markers. One of the targets is a non-autonomous retro-element, the repetitive insertion mobile element (RIME; Hasan et al., 1984). The RIME gene is ubiquitous, specific to the sub-genus Trypanozoon (Wuyts et al., 1994, Tilley et al., 2003) and constitutes the most common mobile element in the T. brucei genome with approximately 500 copies per haploid genome (Bhattacharya et al., 2002). In the present study, we have used the conserved region in the RIME gene to develop a sensitive and specific LAMP test for the subgenus Trypanozoon. The test was evaluated and compared with PCR using Trypanozoon isolates and clinical samples.
2. Materials and methods
Institutional Ethical Clearance for the collection of human samples was obtained from the Livestock Health Research Institute (LIRI) Tororo, Uganda, and the Uganda National Council of Science and Technology (UNCST), Kampala. The use of mice was approved by Murdoch University.
2.1. Parasites and preparation of templates
A total of 59 T. b. rhodesiense and T. b. gambiense isolates were used in this study (Table 1 ). They were isolated from humans, hyenas, tsetse flies, pigs and a dog between 1968 and 2005 in west, east, central and southern parts of Africa. Ten T. b. brucei and five T. evansi isolates were included in the analysis. Trypanosomes were amplified in laboratory mice and DNA prepared using either the published method (Sambrook and Russell, 2001) or the standard extraction commercial kits (Table 1). Stored infected mouse blood containing T. b. rhodesiense isolate ATCC 30027 was adjusted to achieve 1.0 × 106trypanosomes/ml, then 10-fold dilutions were prepared. One portion was used for DNA extraction and the other boiled for supernatant.
Table 1.
Trypanosome isolates used in the study
| Species/sub-speciesa | Identification code | Origin | Year of isolation | Original host |
|---|---|---|---|---|
| Trypanosoma brucei rhodesiense | LVH 56b | Lambwe valley, Kenya | 1978 | Human |
| T. b. rhodesiense | LVH 108b | Lambwe valley, Kenya | 1980 | Human |
| T. b. rhodesiense | KETRI 1883c | Lambwe valley, Kenya | 1970 | Reedbuck |
| T. b. rhodesiense | KETRI 1900c | Lambwe valley, Kenya | 1971 | Hyena |
| T. b. rhodesiense | KETRI 2544 | Lambwe valley, Kenya | 1981 | Human |
| T. b. rhodesiense | KETRI 2492c | Lambwe valley, Kenya | 1980 | Tsetse fly |
| T. b. rhodesiense | KETRI 2532c | Lambwe valley, Kenya | 1980 | Cow |
| T. b. rhodesiense | KETRI 3537c | Bugoma, Kenya | 1998 | Human |
| T. b. rhodesiense | KETRI 3624c | Busia, Kenya | 1998 | Human |
| T. b. rhodesiense | KETRI 3639c | Busia, Kenya | 1999 | Human |
| T. b. rhodesiense | KETRI 3739c | Busia, Kenya | 2001 | Dog |
| T. b. rhodesiense | KETRI 3007c | Busia, Kenya | 1987 | Pig |
| T. b. rhodesiense | EATRO 149c | Nyanza, Kenya | 1961 | Human |
| T. b. rhodesiense | KETRI 2473c | Nyanza, Kenya | 1970 | Human |
| T. b. rhodesiense | UTRO 2509b | Uganda | – | Human |
| T. b. rhodesiense | WB56b | Uganda | – | Human |
| T. b. rhodesiense | UTRO 2504c | Busoga, Uganda | 1979 | Dog |
| T. b. rhodesiense | KETRI 1911c | Busoga, Uganda | 1971 | Cow |
| T. b. rhodesiense | KETRI 2355b | Busoga, Uganda | 1977 | Human |
| T. b. rhodesiense | JE1 | Busoga, Uganda | 1990 | Human |
| T. b. rhodesiense | JE5 | Serere, Uganda | 2001 | Human |
| T. b. rhodesiense | JE6 | Serere, Uganda | 2001 | Human |
| T. b. rhodesiense | JE11 | Serere, Uganda | 1999 | Human |
| T. b. rhodesiense | JE12 | Serere, Uganda | 2003 | Human |
| T. b. rhodesiense | JE13 | Serere, Uganda | 2003 | Human |
| T. b. rhodesiense | JE14 | Serere, Uganda | 2001 | Human |
| T. b. rhodesiense | JE15 | Serere, Uganda | 2003 | Human |
| T. b. rhodesiense | TMRS 51a | Kibondo, Tanzania | 2004 | Human |
| T. b. rhodesiense | TMRS 51b | Kibondo, Tanzania | 2004 | Human |
| T. b. rhodesiense | TMRS 51c | Kibondo, Tanzania | 2005 | Human |
| T. b. rhodesiense | TMRS 52a | Urambo, Tanzania | 2005 | Human |
| T. b. rhodesiense | TMRS 52b | Urambo, Tanzania | 2004 | Human |
| T. b. rhodesiense | TMRS 52c | Urambo, Tanzania | 2006 | Human |
| T. b. rhodesiense | TMRS 53a | Mpanda, Tanzania | 2005 | Human |
| T. b. rhodesiense | TMRS 53b | Mpanda, Tanzania | 2005 | Human |
| T. b. rhodesiense | TMRS 53c | Mpanda, Tanzania | 2005 | Human |
| T. b. rhodesiense | TMRS JM | Kasulu, Tanzania | 2001 | Human |
| T. b. rhodesiense | TMRS 58 | Mpanda, Tanzania | 2006 | Human |
| T. b. rhodesiense | TMRS 4M | Urambo, Tanzania | 2006 | Human |
| T. b. rhodesiense | TMRS010b | Kasulu, Tanzania | 1991 | Human |
| T. b. rhodesiense | TMRS127b | Mpanda, Tanzania | 1994 | Human |
| T. b. rhodesiense | ATCC 30027 | Tanganyika | 1934 | Human |
| T. b. rhodesiense | Gambella IIb | Ethiopia | 1968 | Human |
| T. b. rhodesiense | 058b | Luangwa valley, Zambia | 1974 | Human |
| T. b. rhodesiense | TRPZ320b | Zambia | 1983 | Human |
| T. b. rhodesiense | EATRO 2636c | Mozambique | 1983 | Human |
| Trypanosoma brucei gambiense | MOSb | (Mbam) Cameroon | 1974 | Human |
| T. b. gambiense | Boulab | Bouenza, Congo | 1989 | Human |
| T. b. gambiense | NW2b | Uganda | 1992 | Human |
| T. b. gambiense | Dal 972b | Daloa, Ivory Coast | 1978 | Human |
| T. b. gambiense | Mbab | Daloa, Ivory Coast | 1978 | Human |
| T. b. gambiense | PT41b | Ivory Coast | 1992 | Human |
| T. b. gambiense | PT16b | Ivory Coast | 1992 | Human |
| T. b. gambiense | B014b | Fontem, Cameroon | 1988 | Human |
| T. b. gambiense | Font 1b | Fontem, Cameroon | 1993 | Human |
| T. b. gambiense | NW5b | Uganda | 1992 | Human |
| T. b. gambiense | JE16 | Adjuman, Uganda | 1992 | Human |
| T. b. gambiense | JE17 | Adjuman, Uganda | 1992 | Human |
| T. b. gambiense | KETRI 2565c | Sudan | 1982 | Human |
| Trypanosoma brucei brucei | LUMP 266b | Kiboko, Kenya | 1969 | Fly, Glossina pallidipes |
| T. b. brucei | KETRI 1814 | Kenya | 1970 | Rhino |
| T. b. brucei | KP2Nb | (Kouassi-Perita) Ivory coast | 1982 | Fly, G. palpalis |
| T. b. brucei | B8/18b | (Nsukka) Nigeria | 1962 | Pig |
| T. b. brucei | J10b | Luangwa valley, Zambia | 1973 | Hyena |
| T. b. brucei | STIB 215b | Serengeti, Tanzania | 1971 | Lion |
| T. b. brucei | Kateremab | Uganda | 1990 | Cow |
| T. b. brucei | TSW187/78Eb | Ivory coast | 1978 | Pig |
| T. b. brucei | LVBG 3Nb | Lambwe valley, Kenya | 1980 | Cow |
| T. b. brucei | H3b | Luangwa valley, Zambia | 1974 | Lion |
| Trypanosoma evansi | SA17c | Isiolo, Kenya | 2003 | Camel |
| T. evansi | KETRI 2426c | Ukunda, Kenya | 1978 | Camel |
| T. evansi | KETRI 3093c | Colombia, South America | 1979 | Horse |
| T. evansi | SA263c | Samburu, Kenya | 2003 | Camel |
| T. evansi | KETRI 2439c | Kulal, Kenya | 1979 | Camel |
| T. evansi | KETRI 3565c | Athi River, Kenya | 1994 | Camel |
| Trypanosoma congolense forest | Cam 22b | Mbetta, Cameroon | 1984 | Goat |
| T. congolense kilifi | WG5b | Kenya | 1980 | Sheep |
| T. congolensesavannah | KETRI 1869c | Kenya | – | – |
| Trypanosoma simiae | Ken 4b | Keneba, The Gambia | 1988 | Fly |
| T. simiae tsavo | KETRI 1864c | Kenya | – | Fly |
| Trypanosoma godfreyi | Ken 7b | Kenya | 1988 | Fly, Glossina morsitans |
| Trypanosoma vivax | Y58b | Nigeria | – | – |
The JE samples were processed using a Sigma Genomic DNA extraction kit, USA.
TMRS samples were processed using a Qiagen DNA extraction kit, Australia.
Ten picograms were used for each sample and the reactions were performed in triplicate and repeated after 2 weeks.
Wendy Gibson, University of Bristol, UK. DNA processed through the method of Sambrook and Russell (2001).
Trypanosomiasis Research Centre, Kenya. The DNA was prepared using Qiagen DNA extraction kit, Australia.
2.2. Clinical samples
Twenty archived human blood, CSF and serum samples collected between 1991 and 2007 from HAT patients in Uganda were used. Upon isolation, each sample was divided into two portions. The first portion was inoculated into mice and the second was processed for DNA either using a Sigma Genomic DNA extraction kit (St. Louis, MO USA) (JE samples) or using Gentra DNA purification Kit (Minneapolis, MN USA) (OM samples) (Table 2 ). In Tanzania, the blood was divided into two portions. The first portion was centrifuged and buffy coat collected. In the second portion, 15 μl of blood was mixed with 40 μl of ultra pure water (PCR grade) (Fisher Biotec), boiled for 3 min, centrifuged at 20,800g for 10 min and 10–15 μl of supernatant collected. CSF samples were boiled and centrifuged prior to addition into the reaction mixture while serum was added directly. Ten blood, CSF and serum samples from non-infected humans were used to check LAMP specificity. Two to 4 μl of supernatant was used in each LAMP reaction.
Table 2.
Analysis results for 20 human clinical samples from Uganda and Tanzania
| IDa | Source | Template | Origin | Year of isolation | Original host | Mouse inoculationb | Specific PCR tests |
RIME LAMP | Species/sub-speciesc | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TBR | SRA gene | TgsGP | |||||||||
| JE2 | Blood | DNA | Tororo, Uganda | 1991 | Human | + | − | − | − | + | Trypanosoma brucei rhodesiense |
| JE3 | Blood | DNA/supernatant | Tororo, Uganda | 2005 | Human | + | − | − | − | + | T. b. rhodesiense |
| JE4 | Blood | DNA | Tororo, Uganda | 2002 | Human | + | + | − | − | + | T. b. rhodesiense |
| TMRS10B | Blood | Supernatant/buffy coat | Tanzania | 2007 | Human | + | − | − | − | + | T. b. rhodesiense |
| TMRS11B | Blood | Supernatant/buffy coat | Tanzania | 2007 | Human | − | − | − | − | + | T. b. rhodesiense |
| JE8 | CSF | DNA | Tororo, Uganda | 2001 | Human | + | − | − | − | + | T. b. rhodesiense |
| JE9 | CSF | DNA/supernatant | Tororo, Uganda | 2001 | Human | + | + | − | − | + | T. b. rhodesiense |
| JE10 | CSF | DNA | Tororo, Uganda | 2001 | Human | + | + | − | − | + | T. b. rhodesiense |
| TMRS10C | CSF | supernatant | Tanzania | 2007 | Human | + | nd | nd | nd | + | T. b. rhodesiense |
| TMRS11C | CSF | Supernatant | Tanzania | 2007 | Human | − | nd | nd | nd | + | T. b. rhodesiense |
| TMRS10S | Serum | Directd | Tanzania | 2007 | Human | nd | nd | nd | nd | + | T. b. rhodesiense |
| TMRS11S | Serum | Directd | Tanzania | 2007 | Human | nd | nd | nd | nd | + | T. b. rhodesiense |
| OM55 | Blood | DNA | N.W Uganda | 2004 | Human | + | − | − | − | + | Trypanosoma brucei gambiense |
| OM56 | Blood | DNA | N.W Uganda | 2004 | Human | + | − | − | − | + | T. b. gambiense |
| OM66 | Blood | DNA | N.W Uganda | 2004 | Human | + | − | − | − | − | T. b. gambiense |
| OM62 | Blood | DNA | N.W Uganda | 2004 | Human | + | − | − | − | − | T. b. gambiense |
| OM54 | CSF | DNA | N.W Uganda | 2004 | Human | + | − | − | − | + | T. b. gambiense |
| OM64 | CSF | DNA/supernatant | N.W Uganda | 2004 | Human | + | + | − | − | + | T. b. gambiense |
| OM51 | Blood | DNA | N.W Uganda | 2004 | Human | + | + | − | − | + | T. b. gambiense |
| OM52 | Blood | DNA | N.W Uganda | 2004 | Human | + | − | − | − | + | T. b. gambiense |
TMRS11B was confirmed through a serum resistance-associated gene LAMP test (data not shown).
B, blood; C, CSF; and S, serum; thus TMR10B,C,S samples are from the same patient.
nd, not done; +, positive; and −, negative results.
Ethical clearance obtained from the Ugandan Council of Science and Technology (UNCST).
The first portion was inoculated into mice and the second portion was processed for DNA.
Identification was confirmed through specific PCR using the samples amplified in mice (first portion).
Three microliters of native serum was used for loop-mediated isothermal amplification (LAMP) test.
2.3. RIME sequencing and LAMP primer design
Primers RIME 1, 5′GTTCCCACCCCGTTGGCG and RIME 2, 5′CGTGGGCGCCCAGCCGTG were designed from the genetic databank sequence (Genbank Accession No. K01801) and used to amplify RIME monomer from members of the sub-genus Trypanozoon. After electrophoresis, a single band of ∼500 bp was excised and purified with a DNA purification kit (MO BIO Lab, Solana, USA). The products were cloned into a TOPO vector (Invitrogen, Australia) and transformed in Escherichia coli cells. Plasmid purification was performed using a miniprep column kit (Qiagen, Australia) and the target product sequenced using both M13 forward and reverse primers in an ABI automatic DNA 3730 analyser (Applied Biosystems). The resulting sequences (GenBank Accession Nos. EF567424 for T. b. brucei; EF567425 for T. evansi and EF567226 for T. b. rhodesiense) were used to design six LAMP primers targeting eight conserved regions within the RIME sequence. The outer forward primer (F3), outer backward primer (B3), forward inner primer (FIP) and backward inner primer (BIP) (Notomi et al., 2000) were designed using PrimerExplorer v3 software (http:/primerexplorer.jp/lamp) while the loop forward (LF) and backward (LB) (Nagamine et al., 2002) primers were manually designed (Table 3 ). The forward and backward primers were designed such that the restriction enzyme NdeI cuts in between primer F1c and B1 (Yamada et al., 2006).
Table 3.
Nucleotide sequences for the repetitive insertion mobile element (RIME) loop-mediated isothermal amplification (LAMP) primers
| Primer | Type | Sequence (5′–3′) | Length | Amplicon sizea | Target |
|---|---|---|---|---|---|
| RIME-F3 | F3 | CTGTCCGGTGATGTGGAAC | 19 | 179 | RIME |
| RIME-B3 | B3 | CGTGCCTTCGTGAGAGTTTC | 20 | ||
| RIME-FIP | FIP (F1c + F2) | GGAATACAGCAGATGGGGCGAGGCCAATTGGCATCTTTGGGA | 42 | ||
| RIME-BIP | BIP (B1c + B2) | AAGGGAGACTCTGCCACAGTCGTCAGCCATCACCGTAGAGC | 41 | ||
| RIME-LF | LF | GCCTCCCACCCTGGACTC | 18 | ||
| RIME-LB | LB | AGACCGATAGCATCTCAG | 18 |
F3 and B3 primers are only used for the initial strand displacement and are not involved in subsequent LAMP reaction (Notomi et al., 2000). Therefore the length between F2 and B2 is 179 bp. However, after amplification, the uppermost amplified amplicon size is 223 bp as the FIP and BIP primers consist of F1c = 22 bp and B1c = 22 bp sequences, respectively.
2.4. LAMP reaction
LAMP reactions of 25 μl were standardised for optimal reagent concentrations, temperature and time using T. b. rhodesiense isolate LVH 56 and T. b. gambiense isolate B014, following the Taguchi design (Cobb and Clarkson, 1994). Briefly, the FIP and BIP were varied from 0.8 to 2.4 μM, deoxynucleotides (dNTPs) from 100 to 400 μM, betaine from 0.2 to 0.8 M and MgSO4 from 0 to 4 mM. The reactions were optimised at 2.0 μM for each FIP and BIP primer, 0.8 μM for each loop primer (LF and LB), 0.2 μM for each of the F3 and B3 outer primers, 200 μM for each deoxynucleoside triphosphate, 0.8 M betaine (Sigma, St. Louis, Mo, USA), 20 mM Tris–HCl (pH8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100 and 8 U of BstDNA polymerase large fragment (New England Biolabs, MA, USA). For real-time reactions 3.34 μM SYTO-9 fluorescence dye (Molecular Probes, OR, USA) was added. The template was 1 μl [∼10 pg] for trypanosome DNA and 2–4 μl of processed supernatant, CSF and native serum. The LAMP test was carried out for 1 h at 58, 60, 62 and 64 °C using the Rotor-Gene 3000 thermocycler (Corbett Research, Sydney, Australia) or in a water bath, and terminated by increasing the temperature to 80 °C for 4 min.
2.5. Detection of LAMP product
Amplification of DNA in the LAMP reaction was monitored through electrophoresis in 1.0% agarose gels stained with ethidium bromide, direct visual inspection after addition of 1 μl of 1/10 dilution of SYBR Green I (Invitrogen, Australia), and by monitoring SYTO-9 dye fluorescence in the Rotor-Gene 3000. Real-time fluorescence data was obtained on the FAM channel (excitation at 470 nm and detection at 510 nm) as described by Monis et al. (2005). To confirm that the LAMP amplified the correct target: (i) the product was digested with specific restriction enzyme NdeI (New England Biolabs, MA, USA) which cuts between primers F1c and B1, (ii) melt curves were obtained and analysed as described by Monis et al. (2005), and (iii) a single LAMP band was cloned into a TOPO vector (Invitrogen, Australia), transformed into E. coliand sequenced. The resulting sequence was compared with target sequences using the DNAman software version 5.0 (Lynnon Biosoft, Canada).
2.6. Sensitivity and specificity of LAMP
To determine the analytical sensitivity of the LAMP test, 10-fold dilutions were made from ∼100 ng of DNA purified from T. b. rhodesiense isolate LVH 56 and T. b. gambiense isolate B014. The assay was carried out using both cold and pre-heated templates (Table 4 ). The LAMP test was compared with PCR tests specific for the Trypanozoon sub-genus (Masiga et al., 1992, Wuyts et al., 1994, Tilley et al., 2003). Specificity of the test was determined with approximately 1 ng of DNA from human, tsetse fly, bovine, camel, Plasmodium falciparum and trypanosomes belonging to subgenus Nannomonas(T. congolense savannah, T. congolense kilifi, T. congolense forest, Trypanosoma simiae, T. simiae tsavo, Trypanosoma godfreyi), T. vivax and Trypanosoma lewisi.
Table 4.
Analytical sensitivity of the repetitive insertion mobile element loop-mediated isothermal amplification (RIME LAMP) assay compared with other Trypanozoon sub-genus based tests using pre-heated templates from 10-fold serial dilution of Trypanosoma brucei rhodesiense LVH 56, ATCC 30027 and Trypanosoma brucei gambiense isolate B014 DNA
| Type of Test | Target sequence | Expected specificity | Ten-fold dilutionsa |
Reference | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | 10−7 | 10−8b | 10−9 | 10−10 | 10−11 | ||||
| RIME LAMP (WL) | RIME | Trypanozoon | + | + | + | + | + | + | + | + | + | − | − | This study |
| RIME LAMP (NL) | RIME | Trypanozoon | + | + | + | + | − | − | − | − | − | − | − | This study |
| PFRA LAMP | PFRA gene | Trypanozoon | + | + | + | + | − | − | − | − | − | − | − | Kuboki et al. (2003) |
| TBR1 & 2 | Repeatative region | Trypanozoon | + | + | + | + | + | + | + | − | − | − | − | Masiga et al. (1992) |
| pMUTEC | Retrotransposon | Trypanozoon | + | + | + | + | − | − | − | − | − | − | − | Wuyts et al. (1994) |
| RIME A & B | RIME | Trypanozoon | + | + | + | − | − | − | − | − | − | − | − | Tilley et al., 2003 |
WL, with loop primers; NL, without loop primers.
10−1 (∼1.0 × 105 tryps/ml), 10−2 (∼1.0 × 104 tryps/ml) and 10−9 dilution (∼0.001 tryp/ml).
Detection limit for ATCC 30027 DNA and cold templates.
3. Results
3.1. Detection and confirmation of LAMP product
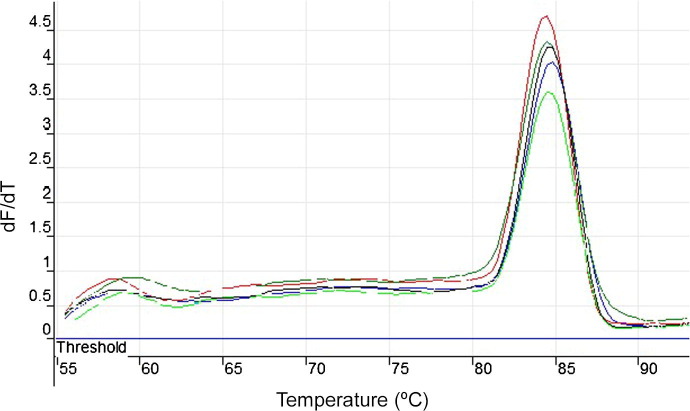
Optimum results were obtained when the reaction temperature was maintained at 62 °C. All positive LAMP reactions produced a characteristic ladder of multiple bands on an agarose gel, indicating that stem-loop DNA with inverted repeats was formed (Notomi et al., 2000). The developed LAMP test was reproducible in laboratories in Kenya, Uganda and Tanzania and no false positives were observed. Upon addition of SYBR Green I to the reaction products, all positive reactions turned green while the negative ones remained orange (Fig. 1 ). No colour change was recorded with non-infected human samples (blood, CSF or serum). Digestion of LAMP product with NdeI restriction enzyme gave the predicted sizes of 89 and 134 bp. The melting curves for RIME LAMP amplification of T. b. rhodesiense human samples JE8, TMRS10S and T. b. gambiense samples OM64, OM51 (Table 2) produced a single peak at 84.5 °C, suggesting amplicons of the same sequence (Fig. 2 ). The analysis of five clones obtained showed 100% identity with the target sequence, and revealed that the length varied with sequence repeats of primers F1c, F2, LFc, F1, B1c, LB, B2c, B1 and the sequence between F1c and B1 (Yamada et al., 2006).
Fig. 1.
The visual appearance of repetitive insertion mobile element loop-mediated isothermal amplification products from Trypanozoon isolates and human African trypanosomiasis clinical samples (Table 2) using SYBR Green I. The reactions were incubated in a water bath for 35 min at 62 °C. Positive samples turn green within 1 min and negative samples remain orange, enabling direct inspection of the results. 1. Trypanosoma brucei rhodesiense (Gambella II), 2. Trypanosoma brucei gambiense (MOS), 3. Trypanosoma vivax (Y58), 4. JE2, 5. TMRS10B, C – positive control [T. b. rhodesiense (LVH 56)], 6. JE9, 7. OM52, 8. OM56, 9. TMRS11C, 10. OM66 and NC – negative control (PCR water).
Fig. 2.
Melting curves for Trypanosoma brucei spp repetitive insertion mobile element loop-mediated isothermal amplification product as monitored in Rotor Gene 3000. The curves from top to bottom are: T. b. rhodesiense JE8, TMRS10S, Trypanosoma brucei gambiense OM64, OM51 and LVH 56 positive control. The curves were acquired after loop-mediated isothermal amplification amplification for 1 h at 62 °C and enzyme denaturing at 80 °C on the FAM channel using 1 °C steps and a hold of 30 s at each step from 60 to 96 °C. All isolates had a melting temperature (Tm) of 84.5 °C, indicating similar sequences, and hence similar amplicons. dF/dT = fluorescence.
3.2. Sensitivity and specificity of LAMP
Ten-fold serial dilutions of genomic DNA from T. b. rhodesiense, T. b. gambiense and supernatant prepared from infected mouse blood containing 1.0 × 106trypanosomes/ml of isolate ATCC 30027 were used to determine the lower detection limit for the LAMP assay at 62 °C. The RIME primer set without loop primers had a detection limit of 10−4 dilution (∼100 trypanosomes/ml). In contrast, the RIME reaction with loop primers had a detection limit of 10−9 dilution (∼0.001 trypanosomes/ml) for T. b. rhodesiense, T. b. gambiense and ATCC 30027 (Table 4). Amplification was detectable within 20–30 min for the test with loop primers, however the optimal time was set at 35 min to allow for very low DNA concentrations. A 10-fold increase in sensitivity was obtained when pre-heated templates (DNA and supernatant) were used. We observed inhibition of the LAMP reaction with DNA concentrations of ⩾200 ng. The RIME LAMP results were identical when either a Rotorgene 3000 thermocycler or a water bath was used to heat the reaction mixture. In all cases RIME LAMP showed a lower detection limit than PCR (Table 4). The RIME LAMP assay was specific and no reactivity was recorded with non-target DNA from other trypanosomes, hosts, vectors or P. faciparum.
3.3. Detection of sub-genus Trypanozoon isolates
RIME LAMP detected all 75 Trypanozoon isolates analysed (Table 5 ). The TBR primers detected 52/59 (88.1%) of T. b. rhodesiense and T. b. gambiense isolates while the SRA gene-specific PCR test (Gibson et al., 2002) detected 21/46 T. b. rhodesiense isolates. The TgsGP PCR (Radwanska et al., 2002) failed to detect two T. b. gambiense isolates (Table 5).
Table 5.
Summary of PCR and repetitive insertion mobile element loop-mediated isothermal amplification (RIME LAMP) results for samples used in the study
| DNA source | No. of isolates | Specific PCR tests |
RIME LAMPf | ||||
|---|---|---|---|---|---|---|---|
| TBRa | SRAb | TgsGPc | COX-1d | Minicirclee | |||
| Trypanosoma brucei rhodesiense | 46 | 40 (86.9%) | 21 (45.7%) | – | nd | nd | 46 (100%) |
| Trypanosoma brucei gambiense | 13 | 12 (92.3%) | – | 11 (84.6%) | nd | nd | 13 (100%) |
| Trypanosoma brucei brucei | 10 | 10 (100%) | – | – | 10 (100%) | nd | 10 (100%) |
| Trypanosoma evansi | 6 | 6 (100%) | – | – | – | 6 (100%) | 6 (100%) |
| Other trypanosomesg | 7 | – | – | – | – | – | – |
| Host and vectorh | 4 | – | – | – | – | nd | – |
| Plasmodium falciparum | 1 | – | – | – | – | – | – |
nd, not done; −, negative.
TBR 1 and 2 test (specific for subgenus Trypanozoon) (Masiga et al., 1992).
T. b. rhodesiense PCR (Gibson et al., 2002).
T. b. gambiense PCR (Radwanska et al., 2002).
T. b. brucei Maxicircle COX-1 PCR (Njiru et al., 2006).
T. evansi PCR (Masiga and Gibson, 1990).
The resulting amplicon was detected using SYTO-9 fluorescence dye in a real-time thermocycler; visual observation after the addition of SYBR Green I and by gel electrophoresis.
Trypanosoma congolense clade, Trypanosoma simiae clade, Trypanosoma godfreyi, Trypanosoma lewisi and Trypanosoma vivax.
Human, bovine, camel and tsetse fly.
3.4. Results for clinical samples
The results of the analysis of 20 archived human samples are shown in Table 2. The RIME LAMP assay detected all T. b. rhodesiense in blood, CSF supernatant and unprocessed serum, while it failed to detect two samples of T. b. gambiense DNA. There was 100% agreement in LAMP test replicates. The TBR PCR only detected 5/18 samples while specific PCR tests gave negative results. The RIME LAMP assay was specific and no amplification was recorded with DNA or supernatant prepared from CSF and blood of non-infected humans.
4. Discussion
Human African Trypanosomiasis (HAT) often presents non-specific clinical symptoms. Diagnosis is even more complicated as the disease progresses, since the clinical symptoms can mimic those of other diseases that are common in the endemic areas (Atouguia and Kennedy, 2000). Patients normally consult a health professional when the disease is advanced and irreversible brain damage has probably occurred (Robays et al., 2004). Development of a sensitive and reliable test for HAT is therefore a priority for early treatment and implementation of appropriate control measures. Furthermore, a test that is rapid and can give results at the point of care, would be ideal in the expansive and remote endemic areas of Africa. In the present study, we have demonstrated the use of LAMP technology in diagnosis of HAT using laboratory propagated isolates, as well as clinical samples. The RIME LAMP test that we have developed is rapid, and results are obtained within 35 min using a normal water bath to maintain the temperature at 62 °C. The analytical sensitivity was the equivalent of 0.001 trypanosomes/ml, indicating that it would be possible to detect very low parasitaemias in patients.
LAMP has inherent characteristics that make it advantageous as a diagnostic test for the rural endemic regions in Africa: (i) the Bst enzyme is active at relatively high temperatures (60–65 °C), reducing the prospect for non-specific priming, (ii) using six primers that recognise eight targets increases sensitivity and specificity (Nagamine et al., 2002), and (iii) the ability to read results visually eliminates the need for gel electrophoresis. In our hands, the RIME LAMP test could amplify DNA from clinical samples within 20–30 min. However, we optimised the tests for 35 min to ensure detection of DNA at very low concentrations. Purified DNA at ⩾200 ng (∼2.0 × 106 trypanosomes/ml) had an inhibitory effect on LAMP reaction as monitored in real time, and showed very weak bands on the agarose gel. It is, however, unlikely that such a high concentration of DNA would be found in a human host. The DNA concentrations in the 10−7 to 10−5 dilutions (∼0.1 to 10 trypanosomes/ml) gave the best results (efficient reaction in real time). Using the same dilutions, we observed an optimum concentration of ⩾10 pg (∼100 trypanosomes/ml) for LAMP tests based on maxicircle COX-1 and 18S genes, with detection limits below 10−7 (results not shown), a factor that is probably associated with the number of copies of each gene. In their studies to detect Trypanozoon sub-genus, Kuboki et al. (2003) used a lowcopy gene (PFRA) and four primers. Their detection limit was 1 pg (10 trypanosomes/ml), observations that were also confirmed in this study (Table 4). In the RIME LAMP test, we have used a target that is a multicopy gene and six primers, which may explain the higher sensitivity.
The robustness of RIME LAMP was further demonstrated when various templates were used. Heat-treated blood, serum and CSF supernatants were sufficient for amplification. Further, LAMP amplified DNA using 1–4 μl of largely native sera and buffy coat with no inhibitory effects in a 25 μl reaction. The ability to use heat-treated samples without compromising sensitivity eliminates the need for DNA extraction, and further shortens the LAMP reaction. Other studies have shown superior tolerance of LAMP for biological substances (Enomoto et al., 2005, Kaneko et al., 2007, Yamada et al., 2006), and heat-treated blood has been used successfully in the detection of malaria (Poon et al., 2006). During LAMP reactions, DNA separation is achieved through destabilisation of the DNA helix by betaine (Notomi et al., 2000). The increase in sensitivity associated with pre-heating of the template could therefore be due to faster DNA strand separation, translating into a more efficient LAMP test. Since HAT LAMP tests appear to be amiable to several templates, further work is needed in selecting and streamlining the best protocols for template preparation.
Comparisons of methods for detection of amplicons, including the addition of SYBR Green I, gel electrophoresis (ladder-like appearance) and real time monitoring, gave the same results, confirming the specificity of the three methods. The combination of SYBR Green I with the double-stranded DNA (amplicon) initiates a colour change from orange to green (Iwamoto et al., 2003). This colour change is rapid and eliminates the need to use ethidium bromide, which is potentially mutagenic, to visualise the products. In three laboratories (each in Kenya, Uganda and Tanzania), it was possible to process a template, perform the LAMP test and read results for 10 samples in 1 h, while the procedures for PCR took up to 4 h, excluding the time required for DNA isolation. The efficiency of amplification in a LAMP reaction is higher than that of PCR because there is no loss of time in the thermal change, since the reactions occur at a constant temperature. The test was reproducible in the field and no false positives were observed. Further, the usefulness of this assay was confirmed by its ability to detect both T. b. rhodesiense and T. b. gambiense directly from archived human blood and CSF samples (Table 2). The superior sensitivity demonstrated by detection of infections below the limits of other molecular techniques reported to date and negative results for P. falciparum, a co-endemic parasite in sub-saharan Africa, favours adaptation of the assay.
There is no current consensus on the diagnostic criteria for CNS involvement in HAT (Kennedy, 2007). Demonstration of trypanosomes in the CSF is the clearest indicator that CNS invasion has occurred. However this is always difficult to determine as the number of parasites in CSF is persistently low, leading to use of indirect and inconsistent markers such as white blood cell counts (WHO, 1998). Demonstration of CNS involvement is critical as it forms the grounds for the therapeutic choice, either early- or late-stage drugs. Some novel molecular tests such as proteomic signature analysis (Papadopoulos et al., 2004) and a PCR test (Jamonneau et al., 2003) have shown sensitivity and specificity of ⩾96%, however their adaptability in the endemic region is still a challenge. The high sensitivity and specificity of the RIME LAMP test recorded in this study and its ability to detect parasite DNA in the CSF samples (Table 2) could prove useful in confirming the presence or absence of parasites after treatment.
An equivocal confirmation that the LAMP test amplifies the target sequence is essential when the test is being developed. This is because LAMP yields a range of product sizes that appear as a ladder on agarose gels, unlike in PCR where a characteristic single band size is observed. Furthermore, the test kit should be developed with a focus on reading colour change, to limit post-reaction DNA manipulations. In the present study, amplification of the target sequence was confirmed with specific restriction enzyme digestion using NdeI, melting curves (Fig. 2), and unequivocally through sequence analysis. Since the technique uses six primers, higher specificity and sensitivity were achieved. Real time analysis forms an important component in diagnostic test development, since it allows the monitoring of the test instantaneously. It was possible to monitor the LAMP amplification, obtain the melt curves and cut-off point through monitoring fluorescence of the double-stranded DNA intercalating dye – SYTO-9. To our knowledge this is the first time that SYTO-9 has been used in LAMP studies. The data obtained was reproducible, robust and consistent. The SYTO-9 dye has an advantage over other intercalating dyes in that it has a less inhibitory effect, shows a broader working range of dye concentration and does not selectively bind to amplicons (Monis et al., 2005). This wider working flexibility makes SYTO-9 an effective option in LAMP studies.
In summary, this study shows that the RIME LAMP test is robust and has great potential as a test that can be deployed easily in endemic countries. The emerging information suggests: (i) that pre-heating of the template prior to its addition into the reaction mixture increases the test sensitivity by 10-fold, (ii) that amplification can easily be achieved using unprocessed template (buffy coat, supernatant and native serum) without inhibition or compromising RIME LAMP sensitivity and (iii) that a normal water bath is sufficient to reproduce results in endemic countries. This study provides direct evidence that addition of loop primers to the RIME LAMP test increased the test sensitivity and efficiency to a new level. It is apparent that a RIME LAMP test is promising as a diagnostic test and may be used as a back up to other tests in active or passive screening for HAT in endemic areas where diagnostic equipment are minimal. Determination and optimisation of protocols for processing the template to make the assay more user-friendly is a crucial next step. The data presented in this study will not only form an excellent comparator for further LAMP studies but will be useful towards development of a HAT test kit.
Acknowledgements
This project was funded by the Foundation for Innovative New Diagnostics (FIND), Geneva, Switzerland and Murdoch University, Australia. The views expressed by the authors do not necessarily reflect the views of the funding agencies. The authors acknowledge the provision of samples by Wendy Gibson (University of Bristol, UK) and informative and constructive comments from the anonymous reviewers.
References
- Atouguia J.L.M., Kennedy P.G.E. Neurological aspects of human African trypanosomiasis. In: Davis L.E., Kennedy P.G.E., editors. Infectious Diseases of the Nervous System. Butterworth–Heinemann; Oxford: 2000. pp. 321–372. [Google Scholar]
- Bhattacharya S., Barke A., Bhattacharya A. Mobile genetic elements in protozoan parasites. J. Genetics. 2002;81:73–86. doi: 10.1007/BF02715903. [DOI] [PubMed] [Google Scholar]
- Boehme C.C., Nabeta P., Henastroza G., Rubhana R., Rahim Z., Gerhardt M., Sanga E., Hoelscher M., Notomi T., Hase T., Perkins M.D. Operational feasibility of using loop-mediated isothermal amplification (LAMP) for the diagnosis of pulmonary TB in microscopy centres of developing countries. J. Clin. Microbiol. 2007;45:1936–1940. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb B., Clarkson JM. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids. Res. 1994;22:3301–3805. doi: 10.1093/nar/22.18.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deborggraeve S., Claes F., Laurent T., Mertens P., Leclipteux T., Dujardin J.C., Herdewijn P., Büscher P. Molecular dipstick test for diagnosis of sleeping sickness. J. Clin. Microbiol. 2006;44:2884–2889. doi: 10.1128/JCM.02594-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto Y., Yoshikawa T., Ihira M., Akimoto S., Miyake F., Usui C., Suga S., Suzuki K., Kawana T., Nishiyama Y., Asano Y. Rapid diagnosis of herpes simplex virus infection by a loop-mediated isothermal amplification method. J. Clin. Microbiol. 2005;43:951–955. doi: 10.1128/JCM.43.2.951-955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W., Backhouse T., Griffiths A. The human serum resistance associated gene is ubiquitous and conserved in Trypanosoma brucei rhodesiense throughout East Africa. Infect. Genet. Evol. 2002;25:1–8. doi: 10.1016/s1567-1348(02)00028-x. [DOI] [PubMed] [Google Scholar]
- Hasan G., Turner M.J., Cordingley J.S. Complete nucleotide sequence of an unusual mobile element from Trypanosoma brucei. Cell. 1984;37:333–341. doi: 10.1016/0092-8674(84)90329-5. [DOI] [PubMed] [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe respiratory syndrome Coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inojosa W.O., Augusto I., Bisoffi Z., Josenado T., Abel P.M., Stich A., Whitty C.J.M. Diagnosing human African trypanosomiasis in Angola using a card agglutination test: observational study of active and passive case finding strategies. BMJ. 2006;332:1479. doi: 10.1136/bmj.38859.531354.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T., Sonobe T., Hayashi K. Loop-mediated amplification for direct detection of Mycobacterium tuberculosis, M. avium and M. intracellulare in sputum samples. J. Clin. Microbiol. 2003;41:2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamonneau V., Solano P., Garcia A., Lejon V., Dje N., Miezan T.W., N’Guessan P., Cuny G., Buscher P. Stage determination and therapeutic decision in human African trypanosomiasis: value of polymerase chain reaction and immunoglobulin M quantification on the cerebrospinal fluid of sleeping sickness patients in Cote d’Ivoire. Trop. Med. Int. Health. 2003;8:589–594. doi: 10.1046/j.1365-3156.2003.01079.x. [DOI] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kennedy P.G.E. Human African trypanosomiasis of the CNS: current issues and challenges. J. Clin. Invest. 2004;113:496–504. doi: 10.1172/JCI21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy P.G.E. Diagnostic and neuropathogenesis issues in human African trypanosomiasis. Int. J. Parasitol. 2007;36:505–512. doi: 10.1016/j.ijpara.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Kuboki N., Inoue N., Sakurai T., Di Cello F., Grab D.J., Suzuki H., Sugimoto C., Igarashi I. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clinic. Microbiol. 2003;41:5517–5524. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros D., Fournier C., Gastellu E.M., Maiso F., Szumillin E. Therapeutic failure of melarsoprol among patients treated for late stage T. b. gambiense human African trypanosomiasis in Uganda. Bull. Soc. Pathol. Exot. 1999;92:171–172. [PubMed] [Google Scholar]
- Lejon V., Boelart M., Jannin J., Moore A., Buscher P. The challenge of Trypanosoma brucei gambiense sleeping sickness diagnosis outside Africa. Lancet. Infect. Dis. 2003;3:804–808. doi: 10.1016/s1473-3099(03)00834-x. [DOI] [PubMed] [Google Scholar]
- Lumsden W.H., Kimber C.D., Evans D.A., Doig S.J. Trypanosoma brucei: miniature anion-exchange centrifugation technique for detection of low parasitaemias: adaptation for field use. Trans. R Soc. Trop. Med. Hyg. 1979;73:312–317. doi: 10.1016/0035-9203(79)90092-0. [DOI] [PubMed] [Google Scholar]
- Magnus E., Vervoort T., Van Miervenne N. A card agglutination test with stained trypanosomes (CATT) for the serological diagnosis of T. b. gambiense trypanosomiasis. Ann. Soc. Belg. Med. Trop. 1978;58:169–176. [PubMed] [Google Scholar]
- Masiga D.K., Gibson C. Specific probes for Trypanosoma (Trypanozoon) evansi based on kinetoplast DNA minicircles. Parasitology. 1990;40:279–284. doi: 10.1016/0166-6851(90)90049-r. [DOI] [PubMed] [Google Scholar]
- Masiga D.K., Smyth A.J., Hayes P., Bromidge T.J., Gibson W.C. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int. J. Parasitol. 1992;22:909–918. doi: 10.1016/0020-7519(92)90047-o. [DOI] [PubMed] [Google Scholar]
- Matovu E., Enyaru J.C.K., Legros D., Schmid C., Seebeck T., Kaminsky R. Melarsoprol refractory T. b. gambiense from Omugo, North Western Uganda. Trop. Med. Int. Health. 2001;6:407–411. doi: 10.1046/j.1365-3156.2001.00712.x. [DOI] [PubMed] [Google Scholar]
- Minami M., Ohta M., Ohkura T., Ando T., Torii K., Hasegawa T., Goto H. Use of a combination of brushing technique and the loop-mediated isothermal amplification method as a novel, rapid, and safe system for detection of Helicobacter pylori. J. Clin. Microbiol. 2006;44:4032–4037. doi: 10.1128/JCM.00898-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monis P.T., Giglio S., Saint C.P. Comparison of SYTO9 and SYBR Green I for real-time polymerase chain and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal. Biochem. 2005;30:24–34. doi: 10.1016/j.ab.2005.01.046. [DOI] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Comm. 2001;289:15–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Prob. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Njiru Z.K., Constantine C.C., Masiga D.K., Reid S.A., Thompson R.C.A., Gibson W.C. Molecular characterization of Trypanosoma evansi type B. Infect. Genet. Evol. 2006;6:20–25. doi: 10.1016/j.meegid.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe, Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic. Acid. Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos M.C., Abel P.M., Agranoff D., Stitch A., Tarelli E., Bell B.A., Planche T., Loosemore A., Saadoun S., Wilkins P., Krishna S. A novel and accurate diagnostic test for human African trypanosomiasis. Lancet. 2004;363:1358–1363. doi: 10.1016/S0140-6736(04)16046-7. [DOI] [PubMed] [Google Scholar]
- Pépin J., Milord F. The treatment of human African trypanosomiasis. Adv. Parasitol. 1994;33:1–17. doi: 10.1016/s0065-308x(08)60410-8. [DOI] [PubMed] [Google Scholar]
- Poon L.L., Wong B.W., Ma E.H., Chan K.H., Chow L.M., Abeyewickreme W., Tangpukdee N., Yuen K.Y., Guan Y., Looareesuwan S., Peiris J.S. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clinic. Chem. 2006;52:303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- Radwanska M., Claes F., Magez S., Magnus E., Perez-Morga D., Pays E., Buscher P. Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am. J. Trop. Med. Hyg. 2002;67:289–295. doi: 10.4269/ajtmh.2002.67.289. [DOI] [PubMed] [Google Scholar]
- Robays J., Bilengue M.M., Van der Stuyft P., Boelaert M. The effectiveness of active population screening and treatment for sleeping sickness control in the Democratic Republic of Congo. Trop. Med. Int. Health. 2004;9:542–550. doi: 10.1111/j.1365-3156.2004.01240.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. third ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, US: 2001. Molecular Cloning. [Google Scholar]
- Schmid C., Nkunku S., Merolle A., Vounatso P., Burri C. Efficacy of 10-day melarsoprol schedule 2 years after treatment for late-stage gambiense sleeping sickness. Lancet. 2004;364:789–790. doi: 10.1016/S0140-6736(04)16940-7. [DOI] [PubMed] [Google Scholar]
- Thekisoe OM., Kuboki N., Nambota A., Fujisaki K., Sugimoto C., Igarashi I., Yasuda J., Inoue N. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Tropica. 2007;102:182–189. doi: 10.1016/j.actatropica.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Tilley A., Welburn S.C., Fevre E.M., Feil E.J., Hide G. Trypanosoma brucei: trypanosome strain typing using PCR analysis of mobile genetic elements (MGE-PCR) Exp. Parasitol. 2003;104:26–32. doi: 10.1016/s0014-4894(03)00114-0. [DOI] [PubMed] [Google Scholar]
- Truc P., Gibson W., Herder S. Genetic characterization of Trypanosoma evansi isolated from a patient in India. Infect. Genet. Evol. 2006;7:305–307. doi: 10.1016/j.meegid.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Van Meirvenne N. Biological diagnosis of human African trypanosomiasis. In: Dumas M., Boutelle B., Buguet A., editors. Progress in Human African Trypanosomiasis, Sleeping sickness. Springer; Paris: 1999. [Google Scholar]
- Welburn S.C., Picozzi K., Fevre E.M., Coleman P.G., Odiit M., Carrington M., Maudlin I. Identification of human-infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet. 2001;358:2017–2019. doi: 10.1016/s0140-6736(01)07096-9. [DOI] [PubMed] [Google Scholar]
- WHO A new form of human trypanosomiasis in India. Description of the first human case in the world caused by Trypanosoma evansi. Weekly Epidemiol. Rec. 2005;80:62–63. [PubMed] [Google Scholar]
- WHO, 1998. Control and surveillance of African trypanosomiasis. Report of a WHO expert committee. Technical Report Series, vol. 881, World Health Organisation, Geneva, Switzerland. [PubMed]
- Wuyts N., Chokesajjawatee N., Panyim S. A simplified and highly sensitive detection of Trypanosoma evansi by DNA amplification. S.E. Asian. J. Trop. Med. Publ. Health. 1994;25:266–271. [PubMed] [Google Scholar]
- Yamada Y., Itoh M., Yoshida M. Sensitive and rapid diagnosis of human parvovirus B19 infection by Loop-mediated isothermal amplification. Br. J. Dernatol. 2006;155:50–55. doi: 10.1111/j.1365-2133.2006.07379.x. [DOI] [PubMed] [Google Scholar]
- Yoshida A., Nagashima S., Ansai T., Tachibana M., Kato H., Watari, Notomi T., Takehara Loop-mediated isothermal amplification method for rapid detection of the periondontopathic bacteria Porphyromona gingivalis, Tannerella forsythia and Treponema denticola. J. Clin. Microbiol. 2005;43 doi: 10.1128/JCM.43.5.2418-2424.2005. 2418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]