Abbreviations
- CDC
center for disease control
- GT
Google Trend
TO THE EDITOR
Public health initiatives depend on timely data collection and dissemination of information. Recently, digital surveillance systems using “big data” such as internet search metrics, or online news stories, have predicted disease outbreaks such as severe acute respiratory syndrome 2 months before publication by World Health Organization and reported on a strange fever in Guinea 9 days before the official information release on the current Ebola epidemic in West Africa (Anema et al., 2014; Milinovich et al., 2015). Surveillance systems using search metric analyses such as Google Trends (GT) have shown promise in tracking influenza in real time, faster compared with traditional data collection on influenza, which typically lags 12–14 days behind (Ginsberg et al., 2009).
Epidemiological studies using search metrics assume that those falling ill with a particular disease will search for it online and the volume and geographical location of such searches can be interpreted as a proxy for disease incidence and location. Initial flaws in methodology resulted in an overestimation of influenza incidence due to search queries being overly influenced by media publicity rather than disease activity (Lazer et al., 2014). Newer algorithms are now being tested that take better account of such confounding factors (Santillana et al., 2014), and GT can now show major news stories on the same time line. Indeed, some emergency departments have demonstrated that such data may successfully be used to predict staffing and vaccine stocking needs (Araz et al., 2014; Thompson et al., 2014).
Although increasingly used in other fields of medicine, “big data” has so far seen little use in dermatology. In this study, we use GT to identify the geographical and seasonal trends in three tickborne diseases, (Lyme disease, ehrlichiosis, and Rocky Mountain spotted fever (RMSF)) and one fungal disease, (coccidioidomycosis). Such diseases are highly relevant to dermatologists who may be the first ones to diagnose them via their cutaneous manifestations (Supplementary Table S1 online ). We then compare this with traditional Center for Disease Control (CDC) data on actual disease events, which we hypothesized will correlate with search data and thereby demonstrate the utility of this resource for tracking and predicting these dermatologically relevant infectious diseases.
Supplementary Information.

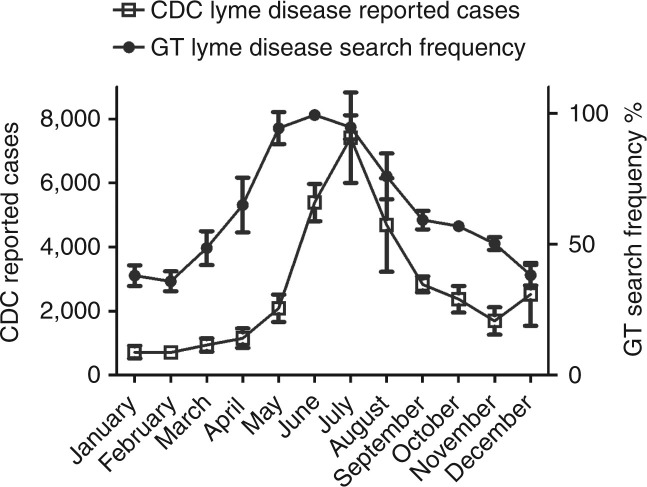
Tickborne diseases are most prevalent in the summer months (Figure 1 ) because of the life cycle of the tick vector and the increase in human outdoor activities (Dana, 2009; Shapiro, 2014). We demonstrated a correlation between monthly Google search frequency and the actual seasonal incidence of the tickborne diseases (Lyme r=0.69, P<0.0001; ehrlichiosis r=0.59, P<0.0001; RMSF r=0.46, P<0.0001; Table 1 and Supplementary Materials and Methods online). Unlike the tickborne diseases, coccidioidomycosis does not have a seasonal incidence peak according to the CDC data. Fittingly, our analysis showed only a weak seasonal correlation (r=0.4169) between GT and CDC data (Table 1). This result is likely due to the much larger data set we have analyzed, allowing even subtle correlations to be elicited. If we reduce our data to look at only 1 year, all of the tickborne seasonal data remain significant (P<0.05, for 2012 only), but coccidioidomycosis data then does not reach statistical significance (e.g., P=0.14; 2012 analyzed alone).
Figure 1.
Temporal correlation between Lyme disease search queries and Center for Disease Control (CDC) Morbidity And Mortality Weekly Report (MMWR) data. Open box plot shows averages and standard deviations of Lyme disease CDC reported cases each from 2007 to 2012. Solid circle plot shows Google search query average frequencies and standard deviations from 2007 to 2012 for the search topic Lyme disease. GT Search Frequency % denotes the format of GT data, which normalizes search frequency for each search term from 0 to 100%. GT, Google Trends.
Table 1.
Correlation between GT and CDC geographic and temporal data
| a. | Lyme Disease | Ehrlichiosis | RMSF | Coccidioidomycosis |
|---|---|---|---|---|
| Pearson’s r | 0.6912 | 0.5926 | 0.4572 | 0.4169 |
| 95% confidence interval | 0.5471–0.7955 | 0.4184–0.7248 | 0.2521–0.6229 | 0.1822–0.6066 |
| P-value (two-tailed) | <0.0001 | <0.0001 | <0.0001 | 0.0009 |
| b. | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 |
|---|---|---|---|---|---|---|
| Lyme disease | 0.7444 | 0.7505 | 0.6104 | 0.6855 | 0.6095 | 0.7194 |
| P-value (two-tailed) | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Ehrlichiosis | 0.3231 | |||||
| P-value (two-tailed) | 0.0346 | |||||
| RMSF | 0.6386 | 0.5938 | 0.3865 | 0.3184 | 0.2904 | 0.06475 |
| P-value (two-tailed) | <0.0001 | <0.0001 | 0.0061 | 0.0258 | 0.043 | 0.6654 |
| Coccidioidomycosis | 0.4813 | 0.4907 | ||||
| P-value (two-tailed) | 0.0173 | 0.0174 |
Abbreviations: CDC, Center for Disease Control; GT, Google Trends; MMWR, Morbidity And Mortality Weekly Report; RMSF, Rocky Mountain spotted fever.
Table 1a. Pearson’s correlation coefficients and P-values derived from the comparison of cumulative GT search data and CDC MMWR monthly reports for the listed diseases between 2007 and 2012. b. Spearman’s rank correlation coefficients and P-values derived from the comparison of state-based GT search data in the mainland United States to the CDC MMWR monthly reports by state for each individual year listed. Inadequate frequency of searches for state-based subanalysis for Ehrlichiosis from 2007 to 2011 and for Coccidioidomycosis from 2007 to 2010.
Tickborne diseases are restricted to the habitat of the tick vector—Lyme disease cases are most prevalent in the northeast and upper Midwest states corresponding to the habitat of the Lyme vector Ixodes scapularis. The soil-dwelling fungus coccidioidomycosis is prevalent in the southwestern United States (Welsh et al., 2012). Accordingly, we demonstrated a geographical correlation between the states with the most searches for the specific infectious disease and states having the most reported new infections (for year 2012 in order of decreasing correlation: Lyme r=0.74, P<0.0001; RMSF r=0.64, P<0.0001; coccidioidomycosis r=0.48, P=0.0173; ehrlichiosis r=0.32, P=0.03; Table 1 and Supplementary Materials and Methods online).
CDC infectious disease data have a typical 1–2 week reporting lag (Ginsberg et al., 2009; Lazer et al., 2014). GT has the potential to predict disease outbreaks closer to real time. In fact, when GT was dynamically recalibrated by combining it with CDC forward projected data (based on a 2-week lag), it was more predictive of influenza incidence than CDC or GT alone (Lazer et al., 2014).
As climate change alters the distribution of the Lyme disease vector, the black-legged tick (Feria-Arroyo et al., 2014; Ogden et al., 2014) or the host of the tick, the white-footed mouse, Peromyscus leucopus, (Roy-Dufresne et al., 2013) cases of Lyme disease are spreading to new locales (Robinson et al., 2014; Wang et al., 2014). In areas not normally affected by Lyme, “big data” may serve as a warning system that alerts physicians that disease may be extending into their area. Such clinical tips may allow earlier diagnosis and treatment and therefore lower morbidity in such diseases.
The methodology presented here has been subject to significant criticism (Lazer et al., 2014). For one, correlations do not indicate causality and the clinical relevance of weak correlations (such as some presented here) is subject to question. Confounding factors include search term selection and search algorithm updating by Google in accordance with their business model. Media publicity may explain the stronger correlations found with Lyme disease.
Correlations using search terms for uncommon conditions, such as the other diseases in this analysis, have not previously been reported in search metric analyses and may be a better representation of the true correlation rate. In fact, our findings may suggest a role for public health campaigns on less common conditions to facilitate following and tracking epidemics.
The correlation of this historical data suggests that big data mining using GT may be a useful resource in understanding the links between climate and infectious disease. In addition, it may prove useful in predicting disease outbreaks to help with emergency preparedness and resource distribution. In the future, we hope for more options in daily data extraction and more precise location information. We propose that a more ideal big data platform would be a research tool not tied to a company core business model and may allow for integration of traditional data sources such as CDC data.
Footnotes
The authors state no conflict of interest.
Supplementary Material
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
REFERENCES
- Anema A., Kluberg S., Wilson K. Digital surveillance for enhanced detection and response to outbreaks. Lancet Infect Dis. 2014;14:1035–1037. doi: 10.1016/S1473-3099(14)70953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araz O.M., Bentley D., Muelleman R.L. Using Google Flu Trends data in forecasting influenza-like-illness related ED visits in Omaha, Nebraska. Am J Emerg Med. 2014;32:1016–1023. doi: 10.1016/j.ajem.2014.05.052. [DOI] [PubMed] [Google Scholar]
- Dana A.N. Diagnosis and treatment of tick infestation and tick-borne diseases with cutaneous manifestations. Dermatol Ther. 2009;22:293–326. doi: 10.1111/j.1529-8019.2009.01244.x. [DOI] [PubMed] [Google Scholar]
- Feria-Arroyo T.P., Castro-Arellano I., Gordillo-Perez G. Implications of climate change on the distribution of the tick vector Ixodes scapularis and risk for Lyme disease in the Texas-Mexico transboundary region. Parasit Vectors. 2014;7:199. doi: 10.1186/1756-3305-7-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg J., Mohebbi M.H., Patel R.S. Detecting influenza epidemics using search engine query data. Nature. 2009;457:1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- Lazer D., Kennedy R., King G. Big data. The parable of Google Flu: traps in big data analysis. Science. 2014;343:1203–1205. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- Milinovich G.J., Magalhaes R.J., Hu W. Role of big data in the early detection of Ebola and other emerging infectious diseases. Lancet Glob Health. 2015;3:e20–e21. doi: 10.1016/S2214-109X(14)70356-0. [DOI] [PubMed] [Google Scholar]
- Ogden N.H., Radojevic M., Wu X. Estimated effects of projected climate change on the basic reproductive number of the Lyme disease vector Ixodes scapularis. Environ Health Perspect. 2014;122:631–638. doi: 10.1289/ehp.1307799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.J., Neitzel D.F., Moen R.A. Disease risk in a dynamic environment: the spread of tick-borne pathogens in Minnesota, USA. Ecohealth. 2014 doi: 10.1007/s10393-014-0979-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Dufresne E., Logan T., Simon J.A. Poleward expansion of the white-footed mouse (Peromyscus leucopus under climate change: implications for the spread of lyme disease. PLoS One. 2013;8:e80724. doi: 10.1371/journal.pone.0080724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santillana M., Zhang D.W., Althouse B.M. What can digital disease detection learn from (an external revision to) google flu trends? Am J Prev Med. 2014;47:341–347. doi: 10.1016/j.amepre.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Shapiro E.D. Clinical practice. Lyme disease. N Engl J Med. 2014;370:1724–1731. doi: 10.1056/NEJMcp1314325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.H., Malik M.T., Gumel A. Emergency department and “Google flu trends” data as syndromic surveillance indicators for seasonal influenza. Epidemiol Infect. 2014;142:2397–2405. doi: 10.1017/S0950268813003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Glowacki M.N., Hoet A.E. Emergence of Ixodes scapularis and Borrelia burgdorferi, the Lyme disease vector and agent, in Ohio. Front Cell Infect Microbiol. 2014;4:70. doi: 10.3389/fcimb.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh O., Vera-Cabrera L., Rendon A. Coccidioidomycosis. Clin Dermatol. 2012;30:573–591. doi: 10.1016/j.clindermatol.2012.01.003. [DOI] [PubMed] [Google Scholar]