Abstract
Peripheral blood mononuclear cells (PBMCs) represent an accessible tissue source for gene expression profiling in schizophrenia that could provide insight into the molecular basis of the disorder. This study used the Illumina HT_12 microarray platform and quantitative real time PCR (QPCR) to perform mRNA expression profiling on 114 patients with schizophrenia or schizoaffective disorder and 80 non-psychiatric controls from the Australian Schizophrenia Research Bank (ASRB). Differential expression analysis revealed altered expression of 164 genes (59 up-regulated and 105 down-regulated) in the PBMCs from patients with schizophrenia compared to controls. Bioinformatic analysis indicated significant enrichment of differentially expressed genes known to be involved or associated with immune function and regulating the immune response. The differential expression of 6 genes, EIF2C2 (Ago 2), MEF2D, EVL, PI3, S100A12 and DEFA4 was confirmed by QPCR. Genome-wide expression analysis of PBMCs from individuals with schizophrenia was characterized by the alteration of genes with immune system function, supporting the hypothesis that the disorder has a significant immunological component in its etiology.
Keywords: Schizophrenia, mRNA, Expression, PBMCs, Immune, Inflammation
1. Introduction
Schizophrenia is a heterogeneous disorder not characterized by a single gene or possibly even a single biological pathway and likely belongs within a “spectrum of psychosis” (Tienari et al., 2003). The existence of a continuum of psychiatric illness encompassing disorders displaying psychotic symptoms, such as schizophrenia and bipolar disorder, could manifest from altered gene expression, controlled at both transcriptional and post-transcriptional levels. Gene expression profiling may be useful for addressing the issue of heterogeneity by identifying genes related to the underlying biology of schizophrenia.
Gene expression studies have been conducted in several brain regions using post-mortem tissue (see Sequeira et al., 2012 for a recent review), but large sample sizes of post-mortem brain tissue are difficult to collect and gene expression profiling in brain tissue is impractical in living patients. Alternatively, the investigation of gene expression in peripheral blood mononuclear cells (PBMCs) that are easily obtained, offer the possibility of longitudinal follow-up, ensure excellent RNA quality and show a considerable degree of heritability and stability in gene expression levels (Meaburn et al., 2009) is a more feasible approach. Indeed, brain expressed genes are expressed in PBMCs and some show co-expression or similar expression levels in the same individuals, supporting their use as a surrogate tissue for gene expression profiling in schizophrenia (Bowden et al., 2006; Gladkevich et al., 2004; Liew et al., 2006; Rollins et al., 2010; Sullivan et al., 2006). In this regard, other groups have sought to identify blood-based expression profiles or develop panels of genes in small cohorts which may be useful for identifying functionally significant genetic and epigenetic changes in individuals with schizophrenia compared to healthy controls or even other related psychiatric disorders (Kurian et al., 2009; Maschietto et al., 2012; Middleton et al., 2005; Takahashi et al., 2010; Tsuang et al., 2005).
Gene expression profiling in PBMCs could also be used to identify functionally relevant pathways in schizophrenia as the lymphocyte constituents may act as a point of communication between the immune and nervous systems (Gladkevich et al., 2004; Sullivan et al., 2006). These cells have been shown to express a number of brain associated proteins including receptors for brain derived neurotrophic factor (BDNF), glucocorticoids, catecholamines, serotonin, dopamine and acetylcholine, and conversely many neurons express receptors for signaling molecules of the immune system (e.g. cytokines) (Guyon et al., 2008; Kronfol and Remick, 2000; McKenna et al., 2002; Muller and Ackenheil, 1998). That there may be an immunological link to the pathophysiology of schizophrenia is not a new concept. Linkage and GWAS support an association of a broad section of markers in the major histocompatability complex (MHC) region at 6p21.33 with schizophrenia (Consortium, 2008; Lewis et al., 2003; Li et al., 2010; O'Donovan et al., 2008; Ripke et al., 2011; Shi et al., 2009; Stefansson et al., 2009) and other reports suggest this locus is biologically relevant to schizophrenia (Laumbacher et al., 2003; Singh et al., 2008; Straub et al., 2002). Immune-associated genes, genetic variants and haplotypes are also implicated in schizophrenia (Lencz et al., 2007; Ozbey et al., 2009; Paul-Samojedny et al., 2011). A recent study combining Gene Set Enrichment Analysis (GSEA) and hypergeometric analysis of GWAS data showed pathways relating to apoptosis, inflammation or immunity were over-represented in schizophrenia (Jia et al., 2010).
Activation of the immune system is suggested by studies reporting altered levels of pro- and anti-inflammatory cytokines, acute phase proteins, complement components, antibodies and lymphocyte subset numbers, ratios, proliferation, activation and function in several tissue sources (serum, whole blood, cerebrospinal fluid (CSF)) from patients with schizophrenia (Maxeiner et al., 2009; Meyer et al., 2011; Miller et al., 2011; Potvin et al., 2008). Moreover, signs of inflammation and activation or increased densities of microglia have been observed in post-mortem brains and CSF from patients with schizophrenia (Doorduin et al., 2009; Drexhage et al., 2010; Monji et al., 2011; van Berckel et al., 2008). A wealth of evidence suggests diverse environmental risk factors impacting on immune function may play a role in schizophrenia including maternal infection and birth in the peak infection seasons of Winter/Spring (Brown and Derkits, 2010; Byrne et al., 2007; Meyer et al., 2006; Zuckerman and Weiner, 2005), maternal and neonatal deficiency in vitamin D which is critical for immunocompetency (McGrath et al., 2010; Schwalfenberg, 2011), malnutrition and psychological stress (reviewed in Markham and Koenig, 2011). Indeed, rodent models of maternal immune challenge results in schizophrenia-associated behavioral changes and cognitive deficits (Nawa and Takei, 2006; Shi et al., 2003).
In support of an immunological manifestation of the disorder we recently identified a microRNA (miRNA) signature associated with immune function in PBMCs from individuals with schizophrenia (Gardiner et al., 2011). Thus it is plausible that PBMCs reflect changes in the genome of patients with schizophrenia that define its character and etiology in those individuals. To further develop gene expression signatures that characterize the molecular background in individuals with schizophrenia we conducted gene or mRNA expression profiling in the largest PBMC cohort to date using the Australian Schizophrenia Research Bank (ASRB). The ASRB is a well characterized cohort of participants with a diagnosis of schizophrenia and carefully screened non-psychiatric controls (Loughland et al., 2010). Here we report differential expression of a large number of genes involved in the immune system.
2. Materials and methods
2.1. Participant recruitment and clinical assessment protocol
This study utilized participants, the majority of whom identified as Caucasian, from the Australian Schizophrenia Research Bank (ASRB) and the Hunter DNA Bank (HDB) (describe previously by Gardiner et al., 2011 and Loughland et al., 2010). Ethics approval was obtained from the Hunter Area Health Services Human Research Ethics Committee and written informed consent obtained from all participants. In this study, the cohort consisted of 114 participants with a lifetime diagnosis of schizophrenia or schizoaffective disorder (cases) as diagnosed by the World Health Organization's ICD-10 criteria and 80 non-psychiatric controls. Demographic and clinical variables of the cohort are summarized in Table 1 and detailed in Supplementary Table 1. There was a significant difference in the mean age of cases and controls (mean age cases 42.3 years, controls 38.7 years, 2-tailed t test p = 0.0498) that is of a small magnitude. Similarly, there was a difference in mean age between schizophrenia and schizoaffective cases (mean age schizophrenia 41 years, mean age schizoaffective disorder 46 years, 2-tailed t test p = 0.03). This small difference in age in mature adults was considered to be unlikely to have significantly impacted upon gene expression. There was a significant difference in gender distribution of cases and controls with more males in the patient group compared to the controls (60% male cases, 42% male controls, 2-tailed Pearson Chi-square p = 0.013) although this is similar to the gender distribution in population samples. There was no difference in gender distribution between schizophrenia and schizoaffective cases (Pearson Chi-square p = 0.286).
Table 1.
Summary of demographic and clinical characteristics of the participants.
| Demographic/clinical variable | Summary statistics |
|---|---|
| Non-psychiatric control | 80 |
| Mean age (years) | 38.7 |
| Gender: M/F | 34/46 |
| Mean RQI | 9.0 |
| Casesa | 114 |
| Schizophrenia | 77 |
| Schizoaffective disorder (manic type) | 16 |
| Schizoaffective disorder (depressed type) | 13 |
| Schizoaffective disorder (bipolar type) | 8 |
| Mean age (years) | 42.4 |
| Gender: M/F | 69/45 |
| Mean RQI | 9.1 |
| Mean age at onset of illness (years) | 23.96 |
| Family history schizophrenia (present/none/unknown) | 46/67/1 |
| Family history other psychosis (present/none/unknown) | 67/46/1 |
| Mean duration of illness (years) | 18.16 |
M – male; F – female; RQI – RNA quality indicator.
ICD-10 diagnosed schizophrenia or schizoaffective disorder (depressed, bipolar or manic subtype); family history other psychosis – reports any other psychiatric mental illness in any first or second degree relative.
2.2. Amplification and labeling of RNA
Whole blood was collected, followed by PBMC isolation, RNA extraction and integrity assessment as described previously (Gardiner et al., 2011). The mean (SD) RQI for this cohort was 9.1 (0.8) and the RQIs were considered to be within the range of acceptable RNA quality according to the manufacturer's instructions (Bio-Rad Laboratories). Contaminants including phenol–chloroform, salts and genomic DNA were removed from total RNA using the RNeasy minikit (Qiagen, VIC, Australia) according to the manufacturer's instructions. Each RNA sample was then amplified, biotinylated and column purified prior to hybridization to the array using the TotalPrep Amplification kit (Ambion, ABI, CA, USA) according to the manufacturer's protocol.
2.3. Differential gene expression profiling
Labeled RNA (750 ng) was hybridized to Illumina HT-12_V3 beadchips (∼48,000 probes) according to the manufacturer's protocol. Expression data underwent quality control analysis and background subtraction in GenomeStudio V3.0 (Illumina, CA, USA) and expression data was exported into R. Further quality control was conducted using the R with lumi packages (www.bioconductor.org) (Du et al., 2008), where the variance-stabilizing transformation (VST) (Lin et al., 2008) was applied. An average of 9624 transcripts were detected for the cohort representing 20% of the total number of transcripts present on the array (detection p value <0.05, before normalisation/background subtraction). Robust Quantile Normalization (RSN) was then applied to expression values for genes considered to be expressed (determined using the detection p value <0.05), followed by differential expression analysis using a linear empirical Bayes model (Smyth, 2004). Significantly differentially expressed genes were identified after p value correction for multiple testing by the Benjamini and Hochberg method. Initial analysis indicated 307 transcripts were differentially expressed in schizophrenia compared to non-psychiatric controls (Supplementary Table 2), which was refined to 164 altered transcripts after exclusion of genes with <10% fold change and discontinued or poorly annotated NCBI Entrez Gene Database records.
2.4. Quantitative real-time reverse transcription PCR (Q-PCR)
Q-PCR validation of differentially expressed mRNA was performed on a subset of the cohort (83 participants: 48 schizophrenia or schizoaffective patients, 35 non-psychiatric controls) as described previously (Santarelli et al., 2011). 10 genes were selected for Q-PCR validation based on strong differential expression of the array and/or biological significance. Both MEF2D (myocyte enhancer factor 2D) and EIF2C2 (or argonaute 2; AGO2) (eukaryotic translation initiation factor 2C, 2) are implicated in miRNA biogenesis which we have previously shown to be altered in post-mortem brain (Beveridge et al., 2010) as well as in PBMCs (Gardiner et al., 2011). In addition, MEF2D was altered in neuroblast culture in response to retinoic acid-induced differentiation suggesting it may be involved in neuronal differentiation, a process that is biologically relevant to schizophrenia. Several genes were also chosen for their involvement in immune function: EVL (Enah/Vasp-like), DEFA4 (defensin α4), PI3 (peptidase inhibitor 3, skin-derived), S100A12 (S100 calcium binding protein A12), CCR7 (Chemokine (C–C motif) receptor 7), CD6 molecule and HMHA1 (histocompatibility (minor) HA-1). VAMP5 (vesicle-associated membrane protein 5 (myobrevin)) was chosen, as it was one of the most strongly up-regulated genes. Primers were designed in Oligo Explorer V1.5 (Gene Link, NY, USA) (primer sequences are listed in Supplementary Table 3). Relative mRNA expression was calculated as the ratio of the gene and the geometric mean of the most stable and efficient housekeeping genes hydroxymethylbilane synthase (HMBS) and 18S ribosomal RNA (18S). Outliers (expression >3 standard deviations from the mean) were excluded from further analysis. Statistical significance of differential mRNA expression between schizophrenia and control groups was determined by Student's t-test (one-tailed p < 0.05).
2.5. Effects of demographic variables on gene expression
The effect of demographic variables was tested by correlation analysis where Pearson's Correlation was used for normally distributed data and Spearman Correlation was used for data that did not follow a normal distribution. Expression values from the microarray (for all 194 differentially expressed genes) and Q-PCR (validated genes) were tested for correlation with age. Additionally, for the validated genes, microarray and Q-PCR expression were analyzed for covariance with gender, RQI and diagnosis (schizophrenia compared to schizoaffective disorder) using a 2-tailed Mann–Whitney-U test and one-way ANOVA.
2.6. Bioinformatic functional analyses
To determine the most significant biological functions and pathways represented by the differentially expressed genes, a list of these genes and their corresponding fold changes were uploaded into Ingenuity Pathway Analysis (IPA) knowledge base v6.3 (Ingenuity Systems, USA, www.ingenuity.com). Of the 166 differentially expressed genes, 164 unique transcripts mapped to annotated gene IDs of which 140 were included in network analysis and 118 were eligible for functions annotation and pathways analysis. Functional Annotation Analysis of the differentially expressed genes was applied to determine the significant Biological Functions and Functions Annotation (p < 0.05 after Benjamini–Hochberg correction for multiple testing) with at least 2 or more genes representing each annotation. Networks showing relationships and interactions between differentially expressed genes and others that functionally interact with them, were generated and ranked in terms of their relevance i.e. the number of participating genes, degree of connectivity and size relative to the total number of network eligible genes.
IPA also allowed the integration of mRNA expression data with miRNA expression data previously collected in an overlapping cohort in which 134 participants were common to both studies (61 controls and 73 cases) (Gardiner et al., 2011). Expression data for the 83 miRNA that were identified as significantly differentially expressed in schizophrenia (FDR <5) was uploaded to IPA where 60 had target prediction information. IPA identifies putative mRNA targets for the miRNA using experimentally validated interactions (TarBase and miRecords) as well as predicted miRNA–mRNA interactions (TargetScan Human Release 6.2; http://www.targetscan.org/; Lewis et al., 2005) and miRNA-related findings from the peer-reviewed literature. The putative miRNA:mRNA pairs were then filtered with respect to fold change to identify inverse miRNA:mRNA target pairs (where the expression of the miRNA is the opposite of it's mRNA target).
3. Results
3.1. Gene expression profiling
mRNA expression was measured in 114 participants with schizophrenia or schizoaffective disorder (cases) compared to 80 non-psychiatric controls. A total of 164 genes displayed differential expression with changes ≥10% and p < 0.05; 105 were down-regulated and 59 up-regulated in the cases compared to controls (Fig. 1 , Table 2 ). Ten genes were chosen for Q-PCR validation based on strong differential expression on the array and/or biological significance. Six genes were confirmed to have significant alterations in expression in the cases; myocyte enhancer factor 2D (MEF2D), eukaryotic translation initiation factor 2C, 2 (EIF2C2; or argonaute 2 (AGO2)) and Enah/Vasp-like (EVL) were down-regulated and defensin α4 (DEFA4), peptidase inhibitor 3, skin-derived (PI3) and S100 calcium binding protein A12 (S100A12) were up-regulated, validating the results of the microarray (Fig. 2 and Table 3 ). Validation of four additional genes was conducted. Chemokine (C–C motif) receptor 7 (CCR7; p = 0.054) and CD6 molecule (p = 0.053), showed a strong trend toward down-regulation in the cases, whilst histocompatibility (minor) HA-1 (HMHA1; p = 0.089) and vesicle-associated membrane protein 5 (myobrevin) (VAMP5; p = 0.101) showed general trends toward down and up-regulation respectively (Fig. 2A and B), all of which were consistent with the microarray analysis. The fold changes detected by the microarray and Q-PCR were significantly correlated (Pearson r = 0.976, p < 0.0001, data not shown) and in all but one instance, fold changes detected by Q-PCR were of greater magnitude compared to those detected on the microarray.
Fig. 1.
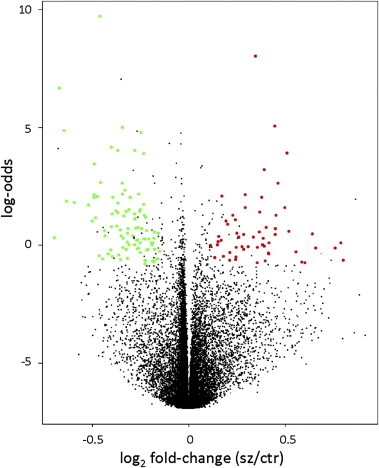
Volcano plot of differentially expressed genes. A scatter-plot of the log odds (probability) against the log2 fold change in expression in PBMCs (schizophrenia/control). Genes with statistically significant differential expression in schizophrenia (Benjamini–Hochberg corrected p < 0.05, fold change >10% up or down-regulation) with current NCBI Entrez gene records are depicted in the upper quadrants; 105 genes down-regulated on the left (green) and 59 genes up-regulated on the right (red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Differentially expressed genes in PBMCs in schizophrenia compared to non-psychiatric controls.
| Gene symbol | Gene name | Chromosomal location | p Value | Fold change |
|---|---|---|---|---|
| Up-regulated | ||||
| ASGR2 | Asialoglycoprotein receptor 2 | 17p | 0.0008 | 1.27 |
| RNASE3 | Ribonuclease, RNase A family, 3 | 14q24–q31 | 0.0049 | 1.36 |
| RNASE2 | Ribonuclease, RNase A family, 2 | 14q24–q31 | 0.0069 | 1.43 |
| ARPC4 | Actin related protein 2/3 complex, subunit 4 | 3p25.3 | 0.0106 | 1.31 |
| TCN1 | Transcobalamin I (vitamin B12 binding protein, R binder family) | 11q11–q12 | 0.0147 | 1.38 |
| SLPI | Secretory leukocyte peptidase inhibitor | 20q12 | 0.0192 | 1.3 |
| RPL34 | Ribosomal protein L34 | 4q25 | 0.0192 | 1.23 |
| ATP5S | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit S (factor B) | 14q21.3 | 0.0192 | 1.13 |
| C9ORF16 | Chromosome 9 open reading frame 16 | 16p11.2 | 0.0229 | 1.23 |
| VAMP5 | Vesicle-associated membrane protein 5 (myobrevin) | 2p11.2 | 0.0229 | 1.41 |
| RNASE6 | Ribonuclease, RNase A family, 6 | 14q11.2 | 0.0246 | 1.29 |
| CKLF | Chemokine-like factor | 16q21 | 0.0247 | 1.37 |
| EXOSC1 | Exosome component 1 | 10q24 | 0.0247 | 1.17 |
| WLS | Wntless homolog (drosophila) | 1p31.3 | 0.0247 | 1.13 |
| GTF2H5 | General transcription factor IIH, polypeptide 5 | 6q25.3 | 0.0255 | 1.18 |
| CYBRD1 | Cytochrome b reductase 1 | 2q31.1 | 0.0259 | 1.14 |
| HIBADH | 3-hydroxyisobutyrate dehydrogenase | 7p15.2 | 0.0277 | 1.15 |
| NTPCR | Nucleoside-triphosphatase, cancer-related | 1q42.2 | 0.0290 | 1.2 |
| CFP | Complement factor properdin | Xp11.4 | 0.0296 | 1.32 |
| DYNLL1 | Dynein, light chain, LC8-type 1 | 12q24.23 | 0.0297 | 1.37 |
| CTSD | Cathepsin D | 11p15.5 | 0.0303 | 1.44 |
| PPPDE2 | PPPDE peptidase domain containing 2 | 22q13.2 | 0.0309 | 1.29 |
| ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | 10q24 | 0.0309 | 1.19 |
| DEFA4 | Defensin, alpha 4, corticostatin | 8p23.1 | 0.0309 | 1.56 |
| NME1 | Non-metastatic cells 1, protein (NM23A) expressed in | 17q21.3 | 0.0314 | 1.22 |
| LSM1 | LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 8p11.2 | 0.0314 | 1.37 |
| TFF3 | Trefoil factor 3 (intestinal) | 21q22.3 | 0.0322 | 1.11 |
| CSTB | Cystatin B (stefin B) | 21q22.3 | 0.0326 | 1.31 |
| RAB32 | RAB32, member RAS oncogene family | 6q24.3 | 0.0326 | 1.26 |
| MRPS18C | Mitochondrial ribosomal protein S18C | 4q21.23 | 0.0326 | 1.2 |
| CNPY2 | Canopy 2 homolog (zebrafish) | 12q15 | 0.0326 | 1.19 |
| UQCRB | Ubiquinol-cytochrome c reductase binding protein | 8q22 | 0.0331 | 1.12 |
| C18ORF10 | Chromosome 18 open reading frame 10 | 18q12.2 | 0.0336 | 1.11 |
| DEFA1B | Defensin, alpha 1B | 8p23.1 | 0.0339 | 1.73 |
| HIST1H2BK | Histone cluster 1, H2bk | 6p21.33 | 0.0339 | 1.34 |
| AZU1 | Azurocidin 1 | 19p13.3 | 0.0348 | 1.31 |
| ACPP | Acid phosphatase, prostate | 3q21–q23 | 0.0348 | 1.11 |
| RNF130 | Ring finger protein 130 | 5q35.3 | 0.0354 | 1.31 |
| LGALS3 | Lectin, galactoside-binding, soluble, 3 | 14q22.3 | 0.0356 | 1.28 |
| RAB5C | RAB5C, member RAS oncogene family | 17q21.2 | 0.0356 | 1.24 |
| HMGN2 | High mobility group nucleosomal binding domain 2 | 1p36.1 | 0.0356 | 1.21 |
| RPS4Y2 | Ribosomal protein S4, Y-linked 2 | Yq11.223 | 0.0356 | 1.19 |
| S100A12 | S100 calcium binding protein A12 | 1q21 | 0.0357 | 1.7 |
| HBD | Hemoglobin, delta | 11p15.5 | 0.0357 | 1.58 |
| POLR2C | Polymerase (RNA) II (DNA directed) polypeptide C, 33 kDa | 16q13–q21 | 0.0366 | 1.22 |
| LCN2 | Lipocalin 2 | 9q34 | 0.0392 | 1.47 |
| MS4A3 | Membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) | 11q12.1 | 0.0402 | 1.16 |
| DBI | Diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) | 2q12–q21 | 0.0412 | 1.33 |
| C19ORF70 | Chromosome 19 open reading frame 70 | 19p13.3 | 0.0418 | 1.33 |
| TM2D1 | TM2 domain containing 1 | 1p31.3 | 0.0440 | 1.13 |
| GMPR2 | Guanosine monophosphate reductase 2 | 14q12 | 0.0448 | 1.19 |
| CHMP3 (VPS24) | Vacuolar protein sorting 24 homolog (S. cerevisiae) | 2p11.2 | 0.0453 | 1.1 |
| CDC42SE1 | CDC42 small effector 1 | 1q21.3 | 0.0454 | 1.18 |
| MRPS33 | Mitochondrial ribosomal protein S33 | 7q34 | 0.0454 | 1.16 |
| PI3 | Peptidase inhibitor 3, skin-derived | 20q13.12 | 0.0457 | 1.74 |
| PRDX5 | Peroxiredoxin 5 | 11q13 | 0.0460 | 1.32 |
| RPL23 | Ribosomal protein L23 | 17q | 0.0478 | 1.5 |
| UBTD1 | Ubiquitin domain containing 1 | 10q24.2 | 0.0487 | 1.28 |
| RBX1 | Ring-box 1, E3 ubiquitin protein ligase | 22q13.2 | 0.0488 | 1.52 |
| Down-regulated | ||||
| TRRAP | Transformation/transcription domain-associated protein | 7q21.2–q22.1 | 0.0003 | 0.73 |
| CCR7 | Chemokine (C–C motif) receptor 7 | 17q12–q21.2 | 0.0017 | 0.63 |
| EVL | Enah/Vasp-like | 14q32.2 | 0.0049 | 0.64 |
| ZNF827 | Zinc finger protein 827 | 4q31.22 | 0.0049 | 0.79 |
| BRAT1 | BRCA1-associated ATM activator 1 | 7p22.3 | 0.0049 | 0.83 |
| LEMD2 | LEM domain containing 2 | 6p21.31 | 0.0049 | 0.84 |
| C16ORF58 | Chromosome 16 open reading frame 58 | 16p11.2 | 0.0067 | 0.76 |
| E4F1 | E4F transcription factor 1 | 16p13.3 | 0.0067 | 0.77 |
| RNF216 | Ring finger protein 216 | 7p22.1 | 0.0067 | 0.82 |
| IL16 | Interleukin 16 (lymphocyte chemoattractant factor) | 15q26.3 | 0.0069 | 0.85 |
| HMHA1 | Histocompatibility (minor) HA-1 | 19p13.3 | 0.0097 | 0.71 |
| CHTOP | Chromatin target of PRMT1 | 1q21.3 | 0.0133 | 0.88 |
| IQSEC1 | IQ motif and Sec7 domain 1 | 3p25.2 | 0.0147 | 0.73 |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 3q13.11 | 0.0147 | 0.79 |
| LRP5L | Low density lipoprotein receptor-related protein 5-like | 22q11.23 | 0.0189 | 0.79 |
| SAFB | Scaffold attachment factor B | 19p13.3–p13.2 | 0.0192 | 0.71 |
| PLA2G4B | Phospholipase A2, group IVB (cytosolic) | 15q11.2–q21.3 | 0.0192 | 0.71 |
| SGSM2 | Small G protein signaling modulator 2 | 17p13.3 | 0.0192 | 0.72 |
| SEC16A | SEC16 homolog A (S. cerevisiae) | 9q34.3 | 0.0192 | 0.76 |
| LONP1 | Lon peptidase 1, mitochondrial | 19p13.2 | 0.0192 | 0.79 |
| SEC24C | SEC24 family, member C (S. cerevisiae) | 10q22.2 | 0.0192 | 0.81 |
| MTMR14 | Myotubularin related protein 14 | 3p26 | 0.0192 | 0.84 |
| CD6 | CD6 molecule | 11q13 | 0.0205 | 0.64 |
| TYK2 | Tyrosine kinase 2 | 19p13.2 | 0.0211 | 0.66 |
| CBX6 | Chromobox homolog 6 | 22q13.1 | 0.0212 | 0.77 |
| RASAL3 | RAS protein activator like 3 | 19p13.12 | 0.0212 | 0.7 |
| STK10 | Serine/threonine kinase 10 | 5q35.1 | 0.0212 | 0.85 |
| FAM153B | Family with sequence similarity 153, member B | 5q35.2 | 0.0237 | 0.81 |
| C7ORF54 | Chromosome 7 open reading frame 54 | 7q31 | 0.0237 | 0.84 |
| CLUAP1 | Clusterin associated protein 1 | 16p13.3 | 0.0241 | 0.77 |
| BTBD11 | BTB (POZ) domain containing 11 | 12q23.3 | 0.0241 | 0.8 |
| P2RY11 | Purinergic receptor P2Y, G-protein coupled, 11 | 19p13.2 | 0.0241 | 0.84 |
| TJAP1 | Tight junction associated protein 1 (peripheral) | 6p21.1 | 0.0246 | 0.77 |
| ITPR3 | Inositol 1,4,5-trisphosphate receptor, type 3 | 6p21 | 0.0247 | 0.72 |
| TMEM175 | Transmembrane protein 175 | 4p16.3 | 0.0247 | 0.76 |
| TGFBRAP1 | Transforming growth factor, beta receptor associated protein 1 | 2q12.1 | 0.0247 | 0.79 |
| SURF6 | Surfeit 6 | 9q34.2 | 0.0247 | 0.82 |
| NOP2 | NOP2 nucleolar protein homolog (yeast) | 12p13 | 0.0247 | 0.82 |
| PMPCA | Peptidase (mitochondrial processing) alpha | 9q34.3 | 0.0247 | 0.83 |
| CTC1 | CTS telomere maintenance complex component 1 | 17p13.1 | 0.0247 | 0.83 |
| ZNF212 | Zinc finger protein 212 | 7q36.1 | 0.0247 | 0.85 |
| ASXL1 | Additional sex combs like 1 (drosophila) | 20q11.1 | 0.0247 | 0.86 |
| CDK12 | Cyclin-dependent kinase 12 | 17q12 | 0.0247 | 0.88 |
| MEF2D | Myocyte enhancer factor 2D | 1q12–q23 | 0.0259 | 0.71 |
| COX19 | COX19 cytochrome c oxidase assembly homolog (S. cerevisiae) | 7p22.3 | 0.0290 | 0.77 |
| UBN1 | Ubinuclein 1 | 16p13.3 | 0.0295 | 0.76 |
| SNX29 | Sorting nexin 29 | 16p13.13–p13.12 | 0.0296 | 0.73 |
| REC8 | REC8 homolog (yeast) | 14q11.2–q12 | 0.0297 | 0.8 |
| RHBDF2 | Rhomboid 5 homolog 2 (drosophila) | 17q25.1 | 0.0297 | 0.83 |
| MOV10 | Mov10, moloney leukemia virus 10, homolog (mouse) | 1p13.2 | 0.0297 | 0.83 |
| QSOX2 | Quiescin Q6 sulfhydryl oxidase 2 | 9q34.3 | 0.0297 | 0.84 |
| URGCP | Upregulator of cell proliferation | 7p13 | 0.0297 | 0.88 |
| C19ORF6 | Chromosome 19 open reading frame 6 | 19p13.3 | 0.0297 | 0.78 |
| NAT10 | N-acetyltransferase 10 (GCN5-related) | 11p13 | 0.0297 | 0.74 |
| YY1AP1 | YY1 associated protein 1 | 1q22 | 0.0297 | 0.86 |
| EIF2C2 (AGO2) | Eukaryotic translation initiation factor 2C, 2 | 8q24 | 0.0303 | 0.72 |
| PPP1R3E | Protein phosphatase 1, regulatory (inhibitor) subunit 3E | 14q11.2 | 0.0304 | 0.88 |
| INTS1 | Integrator complex subunit 1 | 7p22.3 | 0.0309 | 0.8 |
| ISYNA1 | Inositol-3-phosphate synthase 1 | 19p13.11 | 0.0309 | 0.9 |
| ANP32A-IT1 | ANP32A intronic transcript 1 (non-protein coding) | 15q23 | 0.0309 | 0.89 |
| FCGBP | Fc fragment of IgG binding protein | 19q13.1 | 0.0319 | 0.79 |
| SBF1 | SET binding factor 1 | 22q13.33 | 0.0319 | 0.82 |
| SPG7 | Spastic paraplegia 7 (pure and complicated autosomal recessive) | 16q24.3 | 0.0322 | 0.74 |
| EDC4 | Enhancer of mRNA decapping 4 | 16q22.1 | 0.0326 | 0.62 |
| REXO1 | REX1, RNA exonuclease 1 homolog (S. cerevisiae) | 19p13.3 | 0.0326 | 0.79 |
| METTL16 | Methyltransferase like 16 | 17p13.3 | 0.0326 | 0.82 |
| C9ORF91 | Chromosome 9 open reading frame 91 | 9q32 | 0.0326 | 0.85 |
| ZSCAN18 | Zinc finger and SCAN domain containing 18 | 19q13.43 | 0.0326 | 0.83 |
| CACNA1I | Calcium channel, voltage-dependent, T type, alpha 1I subunit | 22q13.1 | 0.0326 | 0.85 |
| HIC2 | Hypermethylated in cancer 2 | 22q11.21 | 0.0326 | 0.87 |
| RASA3 | RAS p21 protein activator 3 | 13q34 | 0.0326 | 0.88 |
| BCL11B | B-cell CLL/lymphoma 11B (zinc finger protein) | 14q32.2 | 0.0331 | 0.83 |
| HDC | Histidine decarboxylase | 15q21–q22 | 0.0339 | 0.8 |
| TSHZ1 | Teashirt zinc finger homeobox 1 | 18q22.3 | 0.0339 | 0.84 |
| MED29 | Mediator complex subunit 29 | 19q13.2 | 0.0339 | 0.87 |
| FBXO32 | F-box protein 32 | 8q24.13 | 0.0339 | 0.88 |
| LRWD1 | Leucine-rich repeats and WD repeat domain containing 1 | 7q22.1 | 0.0348 | 0.81 |
| SFMBT2 | Scm-like with four mbt domains 2 | 10p14 | 0.0348 | 0.82 |
| ATF5 | Activating transcription factor 5 | 19q13.3 | 0.0348 | 0.88 |
| TNPO2 | Transportin 2 | 19p13.2 | 0.0356 | 0.79 |
| CREBBP | CREB binding protein | 16p13.3 | 0.0357 | 0.84 |
| CTRL | Chymotrypsin-like | 16q22.1 | 0.0360 | 0.83 |
| C17ORF63 | Chromosome 17 open reading frame 63 | 17q11.2 | 0.0366 | 0.86 |
| ZNF672 | Zinc finger protein 672 | 1q44 | 0.0367 | 0.8 |
| POM121 | POM121 membrane glycoprotein | 7q11.23 | 0.0367 | 0.88 |
| ULK1 | Unc-51-like kinase 1 (C. elegans) | 12q24.3 | 0.0370 | 0.76 |
| TELO2 | TEL2, telomere maintenance 2, homolog (S. cerevisiae) | 16p13.3 | 0.0370 | 0.86 |
| ZNF446 | Zinc finger protein 446 | 19q13.43 | 0.0384 | 0.9 |
| ELP2 | Elongation protein 2 homolog (S. cerevisiae) | 18q12.2 | 0.0406 | 0.84 |
| ZNF746 | Zinc finger protein 746 | 7q36.1 | 0.0416 | 0.75 |
| ZNF395 | Zinc finger protein 395 | 8p21.1 | 0.0429 | 0.78 |
| TRABD | TraB domain containing | 22q13.33 | 0.0435 | 0.72 |
| CCDC130 | Coiled-coil domain containing 130 | 19p13.2 | 0.0436 | 0.76 |
| MAP7D1 | MAP7 domain containing 1 | 1p34.3 | 0.0453 | 0.76 |
| TNK2 | Tyrosine kinase, non-receptor, 2 | 3q29 | 0.0453 | 0.8 |
| ZNF828 | Zinc finger protein 828 | 13q34 | 0.0453 | 0.8 |
| EIF2B5 | Eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82 kDa | 3q27.1 | 0.0454 | 0.89 |
| EML3 | Echinoderm microtubule associated protein like 3 | 11q12.3 | 0.0454 | 0.73 |
| ABCF1 | ATP-binding cassette, sub-family F (GCN20), member 1 | 6p21.33 | 0.0454 | 0.78 |
| CCNT1 | Cyclin T1 | 12q13.11 | 0.0454 | 0.89 |
| MYO9B | Myosin IXB | 19p13.1 | 0.0457 | 0.85 |
| ZNF362 | Zinc finger protein 362 | 1p35.1 | 0.0460 | 0.86 |
| ZSWIM4 | Zinc finger, SWIM-type containing 4 | 19p13.13 | 0.0460 | 0.88 |
| KIAA0182 | KIAA0182 | 16q24.1 | 0.0461 | 0.88 |
| DMAP1 | DNA methyltransferase 1 associated protein 1 | 1p34 | 0.0497 | 0.85 |
Gene symbols for significantly up-regulated (n = 59) and down-regulated (n = 105) genes are shown (p value represents the corrected p value, after Benjamini–Hochberg multiple testing correction <0.05).
Fig. 2.
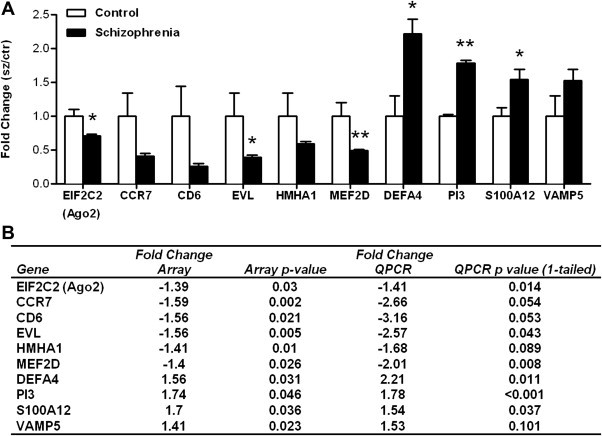
A QPCR validation of differentially expressed genes. The expression of ten genes highlighted by the microarray was analyzed by QPCR. Bars indicate mean fold change + SEM for 83 participants: 48 schizophrenia or schizoaffective patients and 35 non-psychiatric controls. The control cohort is set at 1. Differential expression of EIF2C2, EVL, DEFA4, S100A12 (*p < 0.05) and PI3 and MEF2D (**p < 0.01) was validated, with a further two genes (CCR7 p = 0.054; CD6 p = 0.053) showing a non-significant trend in the same direction as the microarray data. B Fold changes and p values for each gene on the microarray and the QPCR (one tailed student's t-test) are shown.
Table 3.
Top biological functions enriched for differentially expressed genes in schizophrenia using IPA (Benjamini–Hochberg corrected p value range).
| p-Value range | # Molecules | |
|---|---|---|
| Diseases and disorders | ||
| Infectious disease | 7.02E-08–3.35E-02 | 22 |
| Respiratory disease | 7.02E-08–3.35E-02 | 12 |
| Inflammatory response | 3.52E-05–4.63E-02 | 18 |
| Dermatological diseases and conditions | 1.13E-04–4.72E-02 | 20 |
| Genetic disorder | 5.46E-04–4.98E-02 | 24 |
| Molecular and cellular functions | ||
| Antigen presentation | 3.52E-05–4.98E-02 | 8 |
| Cellular movement | 3.52E-05–4.98E-02 | 14 |
| Cell–cell signaling and interaction | 7.13E-05–4.98E-02 | 15 |
| RNA damage and repair | 9.34E-05–1.69E-02 | 3 |
| Cell death | 1.29E-04–4.98E-02 | 15 |
| Physiological system development and function | ||
| Hematological system development and function | 3.52E-05–4.98E-02 | 10 |
| Immune cell trafficking | 3.52E-05–4.98E-02 | 8 |
| Tissue development | 2.94E-03–4.17E-02 | 6 |
| Cell-mediated immune response | 8.48E-03–4.98E-02 | 4 |
| Connective tissue development and function | 8.48E-03–4.98E-02 | 4 |
3.2. Effects of demographic variables on gene expression
Neither microarray nor Q-PCR data for MEF2D, EIF2C2 (AGO2), EVL, PI3, S100A12 and DEFA4 showed a correlation with age or RQI (microarray: 2 tailed Spearman p > 0.086; Q-PCR: Pearson correlation, p > 0.304). There was no difference in expression of these genes between males and females (2-tailed Mann–Whitney-U test p > 0.154) with the exception of DEFA4 where the expression was greater in males than females by microarray (2-tailed Mann–Whitney-U test, p = 0.001). Indeed, this up-regulation of DEFA4 was significant in male cases compared to male controls but was also up-regulated in female cases compared to female controls (2-tailed Mann–Whitney-U test p = 0.003 and p = 0.004 respectively) and this trend was confirmed by the Q-PCR data (not shown). One-way ANOVA revealed no difference in expression between schizophrenia and schizoaffective cases with the exception of PI3, in which mean expression in schizophrenia was higher than schizoaffective disorder on the microarray (p = 0.013) that was not supported by the Q-PCR data (p = 0.485). There was a significant correlation between the expression of CCR7 (microarray data) and age (Spearman r = −0.2792, two-tailed p = 0.0001).
3.3. Functional annotation and bioinformatic analysis
The list of differentially expressed genes and fold changes was submitted for Ingenuity Pathways Analysis (IPA). This analysis revealed the strong presence of genes involved in various aspects of immune function with ∼37% of the total functional annotations categorized as being immune/inflammation-associated (Fig. 3 ). Top ranked biological functions included Infectious Disease, Inflammatory Response, Antigen Presentation, Immune Cell Trafficking and Cell-mediated Immune Response (Table 3). Immune/Inflammation related Functional Annotations included Severe Acute Respiratory Syndrome, Chemotaxis/Recruitment of various immune cells, Replication of a Virus, Respiratory/Infectious Disorder, Antimicrobial Response, Inflammatory/Immune Response (Table 4 ). The full Functional Annotation Analysis is provided in Supplementary Table 4. IPA identifies molecular relationships of differentially expressed genes in the context of biological pathways and indicated an enrichment of differentially expressed Immune/Inflammation-related genes in top scoring networks and canonical pathways (Supplementary Tables 5 and 6). Network 1, enriched with functions including Cell-to-Cell Signaling and Interaction, Infectious Disease and Respiratory Disease is illustrated in Supplementary Fig. 1.
Fig. 3.
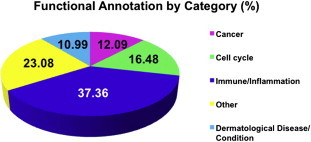
Functional annotations of differentially expressed genes by category. Assortment of functional annotations by broad functional categories revealed over-representation of genes with immune and inflammation-related functions (∼37%) in the list of genes differentially expressed between schizophrenia and controls.
Table 4.
Immune and inflammation related functions enriched with differentially expressed genes in schizophrenia using IPA (Benjamini–Hochberg corrected p value).
| Category | Functions annotation | p-Value | # Molecules | Genes |
|---|---|---|---|---|
| Infectious disease; respiratory disease | Severe acute respiratory syndrome | <0.001 | 10 | CCR7, DEFA1 (includes others), DEFA4, HIST1H2BJ/HIST1H2BK, LCN2, RAB32, RNASE2, S100A12, SLPI, TCN1 |
| Antigen presentation; cellular movement; hematological system development and function; immune cell trafficking; inflammatory response | Chemotaxis of antigen presenting cells | <0.001 | 6 | AZU1, CCR7, CKLF, DEFA1 (includes others), IL16, RNASE2 |
| Cell death | Killing of cells | <0.001 | 6 | AZU1, CTSD, DEFA1 (includes others), LGALS3, PI3, SLPI |
| Antigen presentation; cellular movement; hematological system development and function; immune cell trafficking; inflammatory response | Chemotaxis of phagocytes | <0.001 | 7 | AZU1, CCR7, CKLF, DEFA1 (includes others), IL16, RNASE2, SLPI |
| Infection mechanism | Replication of HIV | 0.00121 | 5 | CCNT1, DEFA1 (includes others), IL16, MOV10, S100A12 |
| Inflammatory response | Inflammation of tissue | 0.0015 | 3 | AZU1, CTSD, SLPI |
| Antigen presentation; cellular movement; hematological system development and function; immune cell trafficking; inflammatory response | Chemotaxis of macrophages | 0.00319 | 3 | AZU1, CKLF, DEFA1 (includes others) |
| Infection mechanism | Replication of RNA virus | 0.00443 | 10 | CCNT1, DEFA1 (includes others), DMAP1, EIF2C2, IL16, LONP1, MOV10, S100A12, SAFB, TNK2 |
| Inflammatory response | Immune response | 0.00451 | 17 | ABCF1, AZU1, CCR7, CFP, CKLF, CTSD, DEFA1 (includes others), ENTPD1, IL16, IQSEC1, LCN2, PRDX5, RNASE2, RNF216, S100A12, SLPI, TYK2 |
| Infection mechanism | Replication of HIV-1 | 0.00474 | 4 | CCNT1, DEFA1 (includes others), MOV10, S100A12 |
| Cellular movement; hematological system development and function; immune cell trafficking | Cell movement of leukocytes | 0.00612 | 8 | AZU1, CCR7, CKLF, DEFA1 (includes others), IL16, LGALS3, RNASE2, SLPI |
| Antigen presentation; cellular movement; hematological system development and function; immune cell trafficking; inflammatory response | Chemotaxis of dendritic cells | 0.00779 | 3 | CCR7, IL16, RNASE2 |
| Cellular movement | Chemotaxis of eukaryotic cells | 0.00784 | 8 | AZU1, CCR7, CKLF, DEFA1 (includes others), IL16, RNASE2, SLPI, TFF3 |
| Antimicrobial response | Inhibition of HIV | 0.00892 | 2 | DEFA1 (includes others), SLPI |
| Cellular movement; hematological system development and function; immune cell trafficking | Homing of mononuclear leukocytes | 0.01 | 5 | AZU1, CCR7, CKLF, DEFA1 (includes others), IL16 |
| Inflammatory response; antimicrobial response | Antimicrobial response | 0.0107 | 4 | AZU1, CFP, RNF216, S100A12 |
| Cellular movement; hematological system development and function; immune cell trafficking | Homing of lymphocytes | 0.0118 | 4 | CCR7, CKLF, DEFA1 (includes others), IL16 |
| Inflammatory response | Inflammatory response | 0.0124 | 9 | ABCF1, AZU1, CCR7, CKLF, DEFA1 (includes others), IL16, PRDX5, RNASE2, SLPI |
| Infection mechanism | Production of virus | 0.0125 | 3 | CREBBP, MOV10, ULK1 |
| Inflammatory response | Inflammation | 0.0129 | 5 | AZU1, CTSD, ENTPD1, S100A12, SLPI |
| Cellular movement; immune cell trafficking | Migration of leukocytes | 0.0163 | 8 | AZU1, CCR7, CKLF, DEFA1 (includes others), IL16, LGALS3, RNASE2, SLPI |
| Cellular movement; hematological system development and function; immune cell trafficking | Chemotaxis of leukocyte cell lines | 0.0188 | 2 | CCR7, DEFA1 (includes others) |
| Inflammatory response; antimicrobial response | Antibacterial response of organism | 0.0233 | 2 | AZU1, CFP |
| Infectious disease | Infectious disorder | 0.0252 | 22 | ABCF1, ASGR2, CBLB, CCNT1, CCR7, DEFA1 (includes others), DEFA4, EIF2B5, ELP2, HIST1H2BJ/HIST1H2BK, IL16, LCN2, POLR2C, RAB32, RNASE2, RNASE3, RNF216, S100A12, SLPI, SPG7, TCN1, TYK2 |
| Cellular movement; hematological system development and function; immune cell trafficking; cell-mediated immune response | Homing of T lymphocytes | 0.0258 | 3 | CCR7, DEFA1 (includes others), IL16 |
| Cellular movement; hematological system development and function; immune cell trafficking | Cell rolling of leukocytes | 0.0282 | 2 | CCR7, LGALS3 |
| Cellular movement; immune cell trafficking | Migration of antigen presenting cells | 0.0286 | 3 | CCR7, IL16, LGALS3 |
| Antigen presentation; cellular movement; hematological system development and function; immune cell trafficking; inflammatory response; lymphoid tissue structure and development | Chemotaxis of neutrophils | 0.0335 | 3 | AZU1, CKLF, SLPI |
| Immunological disease; hematological disease | Hypereosinophilia | 0.0354 | 2 | RNASE2, RNASE3 |
| Cellular movement; hematological system development and function; immune cell trafficking; inflammatory response | Chemotaxis of mononuclear leukocytes | 0.038 | 4 | AZU1, CKLF, DEFA1 (includes others), IL16 |
| Cellular movement; hematological system development and function; immune cell trafficking | Cell movement of granulocytes | 0.0404 | 4 | AZU1, CKLF, LGALS3, SLPI |
| Infection mechanism | Binding of virus | 0.0431 | 2 | ASGR2, CCNT1 |
| Cellular movement; immune cell trafficking; inflammatory response | Migration of phagocytes | 0.0463 | 4 | CCR7, IL16, LGALS3, SLPI |
| Cell-to-cell signaling and interaction | Recruitment of cells | 0.0481 | 3 | CCR7, ENTPD1, SLPI |
Comparison of the mRNA expression data with the previously described miRNA expression data (Gardiner et al., 2011) revealed 102 miRNA:mRNA pairings (in all cases the miRNA was down-regulated while the mRNA was up-regulated), consisting of 42 unique miRNA targeting 37 unique mRNA (Supplementary Table 7).
4. Discussion
In this study, we conducted differential mRNA expression profiling in the largest cohort of patients with schizophrenia or schizoaffective disorder compared with non-psychiatric controls reported to date. This revealed 164 differentially expressed genes (≥10%) after correction for multiple testing, supported by Q-PCR analysis of gene expression. Bioinformatic analysis of differentially expressed genes indicated enrichment of immune/inflammation-related functions providing supporting evidence for immune dysfunction in schizophrenia.
Interestingly, when we considered up-regulated genes in this enriched group separately, the innate immune response in particular was featured. For example, Secretory Leukocyte Peptidase Inhibitor (SLPI) and Azurocidin 1 (AZU1) are chemotactic for cells of the innate immune system and modulate the inflammatory response (Sallenave, 2002; Soehnlein et al., 2008; Subramaniyam et al., 2011). Lipocalin 2 (LCN2) is up-regulated during inflammation and in the plasma of patients with mild cognitive impairment (Choi et al., 2011), and is induced by IL-1β, a protein elevated in the CSF of first episode schizophrenia patients (Cowland et al., 2003; Soderlund et al., 2009). Chemokine-like factor (CKLF), has roles in dendritic cell maturation (Han et al., 2001; Ke et al., 2002; Shao et al., 2010) and is chemotactic for immune cells and possibly has roles in brain development (Han et al., 2001; Wang et al., 2010). Two α-defensin family members, DEFA4 (Q-PCR validated) and DEFA1β were up-regulated, consistent with increased α-defensin protein levels in T cell lysates from treated and antipsychotic free schizophrenia patients (Craddock et al., 2008). Defensins (endogenous peptide antibiotics) are abundant in neutrophil granules, natural killer cells and T lymphocyte subsets and act as immunomodulatory factors regulating acute inflammation, phagocytosis, cell migration/maturation and cytokine secretion (Klotman and Chang, 2006; Rodriguez-Garcia et al., 2007; Selsted and Ouellette, 2005). DEFA4 inhibits synthesis of anti-inflammatory glucocorticoids known to have significant influence on developing synaptic structure and function in the adult brain (Owen et al., 2005). Many α-defensin genes cluster at 8p23, near a schizophrenia linkage site (Fallin et al., 2011; Suarez et al., 2006) known to be a hot spot for copy number variation (CNV) in normal individuals (Aldred et al., 2005).
Alternatively, when we considered the down-regulated genes, we observed a more even distribution of genes with function in both innate and cell-mediated immunity. Indeed, Surfeit 6 (SURF6), a marker of lymphocyte activation for proliferation (Moraleva et al., 2009) and Interleukin 16, a dendritic cell chemo-attractant and modulator of T cell activation and inflammation (Cruikshank and Little, 2008; Kaser et al., 1999) were down-regulated in schizophrenia. CCR7 controls memory T cell migration to sites of inflammation and stimulates dendritic cell maturation (Dieu et al., 1998; Forster et al., 1999). Down-regulation of CD6, a gene involved in T-cell activation promoting their commitment to a Th1 subtype and enhancing their sensitivity to the pro-inflammatory IL-2 (Nair et al., 2010), might be expected to have anti-inflammatory consequences.
This immune-related gene expression signature is in agreement with other blood-based studies in schizophrenia. Glatt et al. (2005) identified several down-regulated genes from the MHC region in PBMCs and we identified changes to ABCF1, LEMD2, TJAP1, ITPR3 and HIST1H2BK that reside at or near this locus. Interestingly, the major histocompatibility complex, class II, DR beta 1 (HLA-DRB1) was also down-regulated in the prefrontal cortex (Glatt et al., 2005). Three genes up-regulated in our study and in Glatt's study include the Galectin family member lectin, galactoside-binding, soluble 3 (LGALS3), a negative regulator of T lymphocyte activation (Yang et al., 2008); the antiprotease, antibacterial and possibly anti-inflammatory Peptidase inhibitor 3, skin-derived (PI3) (Sallenave, 2002) and the pro-inflammatory S100 calcium binding protein A12, calgranulin C (S100A12) (Glatt et al., 2005). S100A12 was also up-regulated in leukocytes obtained from discordant schizophrenic sibling pairs with known linkage to 5q (Middleton et al., 2005). Related S100 genes (S1200A1 and S100A6) were also up-regulated in our post-mortem study of the superior temporal gyrus (STG) in schizophrenia (Bowden et al., 2008) and S100A9 was up-regulated in whole blood from schizophrenia patients (Tsuang et al., 2005).
Similarly, Kurian et al. (2011) identified differentially expressed genes in whole blood associated with “high hallucination state” or “high delusion state” groups, with ‘Inflammatory Response’, ‘IL-8 Signaling’ and ‘Chemokine Signalling’ as well as ‘IL-15 production’ among the top Diseases/Disorders or canonical pathways (Kurian et al., 2011). Takahashi et al. (2010) reported up-regulated genes in whole blood from patients with schizophrenia using the supervised classifier Artificial Neural Networks that featured in Gene Ontology (GO) biological processes such as ‘Inflammatory Response’, ‘Lymphocyte Homeostasis’, ‘Defense Response’, ‘Immune System Process’, ‘Cytokine Biosynthetic Process’ and ‘Cytokine Metabolic Process’ (Takahashi et al., 2010).
The immune-associated mRNA expression signature also agrees with genes predicted to be targeted by a cluster of differentially expressed miRNA identified in an overlapping cohort to this study at chr14q32 that have roles in a range of immune-related pathways such as ‘T Cell Receptor Signaling’, ‘Chemokine Signaling’, ‘Natural Killer Cell-Mediated Cytotoxicity’ and ‘Cytokine–Cytokine Receptor Interaction’ (Gardiner et al., 2011). Down-regulation of EVL is interesting since it is a host gene for miR-342 (Grady et al., 2008), a member of the down-regulated 14q32 miRNA cluster. EVL is involved in remodeling of the actin cytoskeleton which is essential for processes in the CNS (such as axon guidance) and in the immune system (interaction between T lymphocytes and antigen-presenting cells, phagocytosis and chemotaxis of immune cells) (Krause et al., 2003). Down-regulation of EIF2C2 (encoding the endonuclease argonaute 2; AGO2) which functions in small non-coding RNA mediated gene silencing (Cenik and Zamore, 2011) is interesting considering that members of the 14q32 miRNA cluster down-regulated in the cases in this cohort (Gardiner et al., 2011) were over-represented among miRNA that were down-regulated in dopaminergic neurons from the striatum of Ago2-deficient mice (Schaefer et al., 2010). Members of this cluster also contain structural features associated with dicer-independent/Ago2-slicer activity-dependent processing (Diederichs and Haber, 2007; Frank et al., 2010; O'Carroll et al., 2007). This suggests that EIF2C2 is especially important for 14q32 cluster biogenesis and may be related to their down-regulation in PBMCs in these cases (Gardiner et al., 2011) and supports our previous observations of altered miRNA biogenesis in schizophrenia in the cortex (Beveridge et al., 2010). Similarly, down-regulation of MEF2D, a calcium-activated transcription factor, may also provide a mechanism driving the schizophrenia-associated down-regulation of the 14q32 miRNA cluster observed in this cohort since this transcription factor was shown to be a positive regulator of some of these miRNA in rat neurons (Fiore et al., 2009). Moreover, MEF2s have been shown to regulate immune cells (Aude-Garcia et al., 2010; Potthoff and Olson, 2007) and MEF2D has important roles in the brain in neuro-development, neuronal survival and synaptic plasticity (Heidenreich and Linseman, 2004; Lam and Chawla, 2007; She et al., 2011). In addition, we have previously reported up-regulation of MEF2D in response to retinoic acid-induced neuronal differentiation (Beveridge et al., 2009). The distinct roles this transcription factor has in immune function and in the brain makes it an attractive candidate for future investigation.
To further investigate the relationship between mRNA expression and differentially expressed miRNA reported in the overlapping cases (Gardiner et al., 2011), the expression data was integrated revealing 102 predicted inverse miRNA:mRNA target pairings (where the miRNA was down-regulated and would be expected to lead to de-repression of the expression of the target mRNA, which was up-regulated), suggesting that miRNA and post transcriptional gene silencing has a significant influence on schizophrenia-associated changes in gene expression and regulatory networks.
Could this schizophrenia-associated change in the expression of genes with immune function in PBMCs have implications for brain development or function? One possibility is that this peripheral immune-related expression signature could be reflective of immune-dysfunction that is also manifested in the brain and may be indicative of underlying neuropathology. Indeed, the peripheral expression signature we observed was consistent to some extent with gene expression in the CNS which also show aberrant expression of immune and inflammation-associated genes, proteins and pathways, suggestive of an immune component in schizophrenia (Arion et al., 2007; Fillman et al., 2012; Levin et al., 2009; Martins-de-Souza et al., 2010; Matigian et al., 2010; Mistry et al., 2012; Saetre et al., 2007; Schmitt et al., 2011).
Despite these connections, the biological significance of altered expression of genes associated with immune function in schizophrenia is unknown. Therefore it is difficult to determine whether the immune-related expression signature reflects abnormality central and specific to the underlying pathogenesis of schizophrenia or is an indirect consequence of its pathophysiology or co-morbid environmental factor(s). The immune-related signature could reflect the state of illness. Narayan et al. (2008) report that differentially expressed genes in post mortem brain from schizophrenia subjects are involved in inflammation, stimulus-response and immune-related pathways and were more strongly associated with long-term chronic schizophrenia (Narayan et al., 2008). The immune-related expression signature in our cohort, perhaps more representative of chronic schizophrenia is consistent with this observation. The molecular basis of the disorder may change with duration of illness and therefore whether these immune signatures are persistent through exacerbation and remission requires longitudinal studies. Additionally, whether these changes can be detected at the onset of illness could be assessed using first-episode psychosis cohorts. Another possibility is that medications are contributing toward the immune-associated gene expression signature. Indeed, a number of antipsychotics and antidepressants have been shown to display immunosuppressive and anti-inflammatory effects (Chen et al., 2012; Drzyzga et al., 2006; Schmitt et al., 2005; Tynan et al., 2012). Nevertheless, the influence of antipsychotic medication on gene expression could not be determined due to the nature of this information being self-reported by the participants with schizophrenia rather than through medical records. Similarly, obesity could also foster a pro-inflammatory state and may be affecting gene expression. However, since measures of current weight status e.g. body mass index (BMI) or waist circumference were not available, this possibility could not be further investigated.
Alternatively, the immune-related expression pattern may be indicative of a generalized immune disturbance apparent in many psychiatric and neurological disorders. In support of this, immune-related changes or abnormalities have been identified in schizophrenia and bipolar disorder (Bousman et al., 2010; Shao and Vawter, 2008), depression (Maes et al., 2009; Wager-Smith and Markou, 2011), anxiety (Hou and Baldwin, 2012) and Alzheimer's disease (Deretic, 2005; Horesh et al., 2011). This hints at some common elements and suggests the immune system is vitally important for the development and function of the brain.
In summary, we have conducted a large genome-wide survey of gene expression in PBMCs from individuals with schizophrenia and schizoaffective disorder and identified a significant over-representation of genes associated with the immune system. While this has immediate functional implications for the relationship between the brain and the immune system, it may also be reflecting genetic, environmental or developmentally significant insults that are relevant to the pathogenesis of the disorder (Bilbo and Schwarz, 2009; Kinney et al., 2010). In other words the immune expression signature in blood may be a residual image of this disturbance and provide insight into the etiopathogenesis of schizophrenia.
Role of the funding source
This research was supported by the Schizophrenia Research Institute, the Hunter Medical Research Institute and the Neurobehavioral Genetics Unit, utilizing infrastructure funding from NSW Ministry of Health. It was supported by a MC Ainsworth Research Fellowship in Epigenetics (MC); a NARSAD Young Investigator Award Samples and clinical and demographic data for this study were provided by the Australian Schizophrenia Research Bank (Chief Investigators: Carr V, Schall U, Scott R, Jablensky A, Mowry B, Michie P, Catts S, Henskens F, Pantelis C), which is supported by the National Health and Medical Research Council of Australia (Enabling Grant No. 386500), the Pratt Foundation, Ramsay Health Care, the Viertel Charitable Foundation and the Schizophrenia Research Institute.
Contributors
E. Gardiner contributed to the study design, experimental work, data analysis and preparation of the manuscript.
M. Cairns contributed to the study design, manuscript preparation and sourcing of funding.
B. Liu contributed to data analysis.
N. Beveridge contributed to data analysis.
V. Carr contributed to the study design, manuscript preparation and sourcing of funding.
B. Kelly contributed to the study design, manuscript preparation and sourcing of funding.
R. Scott contributed to the study design, manuscript preparation and sourcing of funding.
P. Tooney contributed to the study design, manuscript preparation and sourcing of funding.
Conflict of interest
None of the authors have conflicts of interest regarding this manuscript.
Acknowledgments
PBMCs were processed by Melissa Tooney, Trish Collinson, Amy Martin, and Janelle Collins-Langworthy.
Footnotes
Supplementary material related to this article can be found at http://dx.doi.org/10.1016/j.jpsychires.2012.11.007.
Appendix A. Supplementary material
References
- Aldred P.M., Hollox E.J., Armour J.A. Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Human Molecular Genetics. 2005;14:2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- Arion D., Unger T., Lewis D.A., Levitt P., Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biological Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aude-Garcia C., Collin-Faure V., Bausinger H., Hanau D., Rabilloud T., Lemercier C. Dual roles for MEF2A and MEF2D during human macrophage terminal differentiation and c-Jun expression. The Biochemical Journal. 2010;430:237–244. doi: 10.1042/BJ20100131. [DOI] [PubMed] [Google Scholar]
- van Berckel B.N., Bossong M.G., Boellaard R., Kloet R., Schuitemaker A., Caspers E. Microglia activation in recent-onset schizophrenia: a quantitative (R)-[11C]PK11195 positron emission tomography study. Biological Psychiatry. 2008;64:820–822. doi: 10.1016/j.biopsych.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Beveridge N.J., Gardiner E., Carroll A.P., Tooney P.A., Cairns M.J. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Molecular Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge N.J., Tooney P.A., Carroll A.P., Tran N., Cairns M.J. Down-regulation of miR-17 family expression in response to retinoic acid induced neuronal differentiation. Cellular Signalling. 2009;21:1837–1845. doi: 10.1016/j.cellsig.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Bilbo S.D., Schwarz J.M. Early-life programming of later-life brain and behavior: a critical role for the immune system. Frontiers in Behavioral Neuroscience. 2009;3:14. doi: 10.3389/neuro.08.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousman C.A., Chana G., Glatt S.J., Chandler S.D., Lucero G.R., Tatro E. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2010;153B:494–502. doi: 10.1002/ajmg.b.31006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden N.A., Scott R.J., Tooney P.A. Altered gene expression in the superior temporal gyrus in schizophrenia. BMC Genomics. 2008;9:199. doi: 10.1186/1471-2164-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden N.A., Weidenhofer J., Scott R.J., Schall U., Todd J., Michie P.T. Preliminary investigation of gene expression profiles in peripheral blood lymphocytes in schizophrenia. Schizophrenia Research. 2006;82:175–183. doi: 10.1016/j.schres.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Brown A.S., Derkits E.J. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. American Journal of Psychiatry. 2010;167:261–280. doi: 10.1176/appi.ajp.2009.09030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M., Agerbo E., Bennedsen B., Eaton W.W., Mortensen P.B. Obstetric conditions and risk of first admission with schizophrenia: a Danish national register based study. Schizophrenia Research. 2007;97:51–59. doi: 10.1016/j.schres.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Cenik E.S., Zamore P.D. Argonaute proteins. Current Biology: CB. 2011;21:R446–R449. doi: 10.1016/j.cub.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Chen M.L., Tsai T.C., Wang L.K., Lin Y.Y., Tsai Y.M., Lee M.C. Clozapine inhibits Th1 cell differentiation and causes the suppression of IFN-gamma production in peripheral blood mononuclear cells. Immunopharmacology and Immunotoxicology. 2012;34:686–694. doi: 10.3109/08923973.2011.651535. [DOI] [PubMed] [Google Scholar]
- Choi J., Lee H.W., Suk K. Increased plasma levels of lipocalin 2 in mild cognitive impairment. Journal of the Neurological Sciences. 2011;305:28–33. doi: 10.1016/j.jns.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Consortium I.S. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowland J.B., Sorensen O.E., Sehested M., Borregaard N. Neutrophil gelatinase-associated lipocalin is up-regulated in human epithelial cells by IL-1 beta, but not by TNF-alpha. Journal of Immunology. 2003;171:6630–6639. doi: 10.4049/jimmunol.171.12.6630. [DOI] [PubMed] [Google Scholar]
- Craddock R.M., Huang J.T., Jackson E., Harris N., Torrey E.F., Herberth M. Increased alpha-defensins as a blood marker for schizophrenia susceptibility. Molecular & Cellular Proteomics: MCP. 2008;7:1204–1213. doi: 10.1074/mcp.M700459-MCP200. [DOI] [PubMed] [Google Scholar]
- Cruikshank W., Little F. Interleukin-16: the ins and outs of regulating T-cell activation. Critical Reviews in Immunology. 2008;28:467–483. doi: 10.1615/critrevimmunol.v28.i6.10. [DOI] [PubMed] [Google Scholar]
- Deretic V. Autophagy in innate and adaptive immunity. Trends in Immunology. 2005;26:523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Diederichs S., Haber D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Dieu M.C., Vanbervliet B., Vicari A., Bridon J.M., Oldham E., Ait-Yahia S. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. Journal of Experimental Medicine. 1998;188:373–386. doi: 10.1084/jem.188.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduin J., de Vries E.F., Willemsen A.T., de Groot J.C., Dierckx R.A., Klein H.C. Neuroinflammation in schizophrenia-related psychosis: a PET study. Journal of Nuclear Medicine: Official Publication, Society of Nuclear Medicine. 2009;50:1801–1807. doi: 10.2967/jnumed.109.066647. [DOI] [PubMed] [Google Scholar]
- Drexhage R.C., Knijff E.M., Padmos R.C., Heul-Nieuwenhuijzen L., Beumer W., Versnel M.A. The mononuclear phagocyte system and its cytokine inflammatory networks in schizophrenia and bipolar disorder. Expert Review of Neurotherapeutics. 2010;10:59–76. doi: 10.1586/ern.09.144. [DOI] [PubMed] [Google Scholar]
- Drzyzga L., Obuchowicz E., Marcinowska A., Herman Z.S. Cytokines in schizophrenia and the effects of antipsychotic drugs. Brain, Behavior, and Immunity. 2006;20:532–545. doi: 10.1016/j.bbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Fallin M.D., Lasseter V.K., Liu Y., Avramopoulos D., McGrath J., Wolyniec P.S. Linkage and association on 8p21.2-p21.1 in schizophrenia. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2011;156:188–197. doi: 10.1002/ajmg.b.31154. [DOI] [PubMed] [Google Scholar]
- Fillman S.G., Cloonan N., Catts V.S., Miller L.C., Wong J., McCrossin T. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Molecular Psychiatry. 2012 doi: 10.1038/mp.2012.110. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Fiore R., Khudayberdiev S., Christensen M., Siegel G., Flavell S.W., Kim T.K. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO Journal. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R., Schubel A., Breitfeld D., Kremmer E., Renner-Muller I., Wolf E. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- Frank F., Sonenberg N., Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Gardiner E., Beveridge N.J., Wu J.Q., Carr V., Scott R.J., Tooney P.A. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Molecular Psychiatry. 2011;18:827–840. doi: 10.1038/mp.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladkevich A., Kauffman H.F., Korf J. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Progress in Neuropsychopharmacology & Biological Psychiatry. 2004;28:559–576. doi: 10.1016/j.pnpbp.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Glatt S.J., Everall I.P., Kremen W.S., Corbeil J., Sasik R., Khanlou N. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady W.M., Parkin R.K., Mitchell P.S., Lee J.H., Kim Y.H., Tsuchiya K.D. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880–3888. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- Guyon A., Massa F., Rovere C., Nahon J.L. How cytokines can influence the brain: a role for chemokines? Journal of Neuroimmunology. 2008;198:46–55. doi: 10.1016/j.jneuroim.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Han W., Lou Y., Tang J., Zhang Y., Chen Y., Li Y. Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. The Biochemical Journal. 2001;357:127–135. doi: 10.1042/0264-6021:3570127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich K.A., Linseman D.A. Myocyte enhancer factor-2 transcription factors in neuronal differentiation and survival. Molecular Neurobiology. 2004;29:155–166. doi: 10.1385/MN:29:2:155. [DOI] [PubMed] [Google Scholar]
- Horesh Y., Katsel P., Haroutunian V., Domany E. Gene expression signature is shared by patients with Alzheimer's disease and schizophrenia at the superior temporal gyrus. European Journal of Neurology: The Official Journal of the European Federation of Neurological Societies. 2011;18:410–424. doi: 10.1111/j.1468-1331.2010.03166.x. [DOI] [PubMed] [Google Scholar]
- Hou R., Baldwin D.S. A neuroimmunological perspective on anxiety disorders. Human Psychopharmacology. 2012;27:6–14. doi: 10.1002/hup.1259. [DOI] [PubMed] [Google Scholar]
- Jia P., Wang L., Meltzer H.Y., Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophrenia Research. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A., Dunzendorfer S., Offner F.A., Ryan T., Schwabegger A., Cruikshank W.W. A role for IL-16 in the cross-talk between dendritic cells and T cells. Journal of Immunology. 1999;163:3232–3238. [PubMed] [Google Scholar]
- Ke X., Jia L., Jing H., Liu Y., Zhang Y., Di C. Effects of novel human chemokine-like factor 1 (CKLF1) on bone marrow hematopoietic stem cell/progenitor cell in vitro. Zhonghua Xue Ye Xue Za Zhi = Zhonghua Xueyexue Zazhi. 2002;23:301–303. [PubMed] [Google Scholar]
- Kinney D.K., Hintz K., Shearer E.M., Barch D.H., Riffin C., Whitley K. A unifying hypothesis of schizophrenia: abnormal immune system development may help explain roles of prenatal hazards, post-pubertal onset, stress, genes, climate, infections, and brain dysfunction. Medical Hypotheses. 2010;74:555–563. doi: 10.1016/j.mehy.2009.09.040. [DOI] [PubMed] [Google Scholar]
- Klotman M.E., Chang T.L. Defensins in innate antiviral immunity. Nature Reviews. Immunology. 2006;6:447–456. doi: 10.1038/nri1860. [DOI] [PubMed] [Google Scholar]
- Krause M., Dent E.W., Bear J.E., Loureiro J.J., Gertler F.B. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annual Review of Cell and Developmental Biology. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- Kronfol Z., Remick D.G. Cytokines and the brain: implications for clinical psychiatry. American Journal of Psychiatry. 2000;157:683–694. doi: 10.1176/appi.ajp.157.5.683. [DOI] [PubMed] [Google Scholar]
- Kurian S.M., Heilman R., Mondala T.S., Nakorchevsky A., Hewel J.A., Campbell D. Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS One. 2009;4:e6212. doi: 10.1371/journal.pone.0006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian S.M., Le-Niculescu H., Patel S.D., Bertram D., Davis J., Dike C. Identification of blood biomarkers for psychosis using convergent functional genomics. Molecular Psychiatry. 2011;16:37–58. doi: 10.1038/mp.2009.117. [DOI] [PubMed] [Google Scholar]
- Lam B.Y., Chawla S. MEF2D expression increases during neuronal differentiation of neural progenitor cells and correlates with neurite length. Neuroscience Letters. 2007;427:153–158. doi: 10.1016/j.neulet.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Laumbacher B., Muller N., Bondy B., Schlesinger B., Gu S., Fellerhoff B. Significant frequency deviation of the class I polymorphism HLA-A10 in schizophrenic patients. Journal of Medical Genetics. 2003;40:217–219. doi: 10.1136/jmg.40.3.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T., Morgan T.V., Athanasiou M., Dain B., Reed C.R., Kane J.M. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Molecular Psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- Levin Y., Wang L., Schwarz E., Koethe D., Leweke F.M., Bahn S. Global proteomic profiling reveals altered proteomic signature in schizophrenia serum. Molecular Psychiatry. 2009;15:1088–1100. doi: 10.1038/mp.2009.54. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis C.M., Levinson D.F., Wise L.H., DeLisi L.E., Straub R.E., Hovatta I. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. American Journal of Human Genetics. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Li Z., Chen P., Zhao Q., Wang T., Huang K. Common variants in major histocompatibility complex region and TCF4 gene are significantly associated with schizophrenia in Han Chinese. Biological Psychiatry. 2010;68:671–673. doi: 10.1016/j.biopsych.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Liew C.C., Ma J., Tang H.C., Zheng R., Dempsey A.A. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. The Journal of Laboratory and Clinical Medicine. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Lin S.M., Du P., Huber W., Kibbe W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Research. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughland C., Draganic D., McCabe K., Richards J., Nasir A., Allen J. Australian Schizophrenia Research Bank: a database of comprehensive clinical, endophenotypic and genetic data for aetiological studies of schizophrenia. Australian and New Zealand Journal of Psychiatry. 2010;44:1029–1035. doi: 10.3109/00048674.2010.501758. [DOI] [PubMed] [Google Scholar]
- Maes M., Yirmyia R., Noraberg J., Brene S., Hibbeln J., Perini G. The inflammatory & neurodegenerative (I&ND) hypothesis of depression: leads for future research and new drug developments in depression. Metabolic Brain Disease. 2009;24:27–53. doi: 10.1007/s11011-008-9118-1. [DOI] [PubMed] [Google Scholar]
- Markham J.A., Koenig J.I. Prenatal stress: role in psychotic and depressive diseases. Psychopharmacology. 2011;214:89–106. doi: 10.1007/s00213-010-2035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-de-Souza D., Maccarrone G., Wobrock T., Zerr I., Gormanns P., Reckow S. Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. Journal of Psychiatric Research. 2010;44:1176–1189. doi: 10.1016/j.jpsychires.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Maschietto M., Silva A.R., Puga R.D., Lima L., Pereira C.B., Nakano E.Y. Gene expression of peripheral blood lymphocytes may discriminate patients with schizophrenia from controls. Psychiatry Research. 2012 doi: 10.1016/j.psychres.2012.04.030. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Matigian N., Abrahamsen G., Sutharsan R., Cook A.L., Vitale A.M., Nouwens A. Disease-specific, neurosphere-derived cells as models for brain disorders. Disease Models & Mechanisms. 2010;3:785–798. doi: 10.1242/dmm.005447. [DOI] [PubMed] [Google Scholar]
- Maxeiner H.G., Rojewski M.T., Schmitt A., Tumani H., Bechter K., Schmitt M. Flow cytometric analysis of T cell subsets in paired samples of cerebrospinal fluid and peripheral blood from patients with neurological and psychiatric disorders. Brain, Behavior, and Immunity. 2009;23:134–142. doi: 10.1016/j.bbi.2008.08.003. [DOI] [PubMed] [Google Scholar]
- McGrath J.J., Burne T.H., Feron F., Mackay-Sim A., Eyles D.W. Developmental vitamin D deficiency and risk of schizophrenia: a 10-year update. Schizophrenia Bulletin. 2010;36:1073–1078. doi: 10.1093/schbul/sbq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna F., McLaughlin P.J., Lewis B.J., Sibbring G.C., Cummerson J.A., Bowen-Jones D. Dopamine receptor expression on human T- and B-lymphocytes, monocytes, neutrophils, eosinophils and NK cells: a flow cytometric study. Journal of Neuroimmunology. 2002;132:34–40. doi: 10.1016/s0165-5728(02)00280-1. [DOI] [PubMed] [Google Scholar]
- Meaburn E.L., Fernandes C., Craig I.W., Plomin R., Schalkwyk L.C. Assessing individual differences in genome-wide gene expression in human whole blood: reliability over four hours and stability over 10 months. Twin Research and Human Genetics: The Official Journal of the International Society for Twin Studies. 2009;12:372–380. doi: 10.1375/twin.12.4.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U., Feldon J., Schedlowski M., Yee B.K. Immunological stress at the maternal-foetal interface: a link between neurodevelopment and adult psychopathology. Brain, Behavior, and Immunity. 2006;20:378–388. doi: 10.1016/j.bbi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Meyer U., Schwarz M.J., Muller N. Inflammatory processes in schizophrenia: a promising neuroimmunological target for the treatment of negative/cognitive symptoms and beyond. Pharmacology & Therapeutics. 2011;132:96–110. doi: 10.1016/j.pharmthera.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Middleton F.A., Pato C.N., Gentile K.L., McGann L., Brown A.M., Trauzzi M. Gene expression analysis of peripheral blood leukocytes from discordant sib-pairs with schizophrenia and bipolar disorder reveals points of convergence between genetic and functional genomic approaches. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2005;136B:12–25. doi: 10.1002/ajmg.b.30171. [DOI] [PubMed] [Google Scholar]
- Miller B.J., Buckley P., Seabolt W., Mellor A., Kirkpatrick B. Meta-analysis of cytokine alterations in schizophrenia: clinical status and antipsychotic effects. Biological Psychiatry. 2011;70:663–671. doi: 10.1016/j.biopsych.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry M., Gillis J., Pavlidis P. Genome-wide expression profiling of schizophrenia using a large combined cohort. Molecular Psychiatry. 2012 doi: 10.1038/mp.2011.172. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monji A., Kato T.A., Mizoguchi Y., Horikawa H., Seki Y., Kasai M. Neuroinflammation in schizophrenia especially focused on the role of microglia. Progress in Neuropsychopharmacology & Biological Psychiatry. 2012 doi: 10.1016/j.pnpbp.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Moraleva A.A., Malysheva M.V., Magoulas C., Polzikov M.A., Zatsepina O.V. Early expression of nucleolar SURF-6 protein in mouse spleen lymphocytes activated for proliferation in vitro. Bulletin of Experimental Biology and Medicine. 2009;147:578–582. doi: 10.1007/s10517-009-0578-z. [DOI] [PubMed] [Google Scholar]
- Muller N., Ackenheil M. Psychoneuroimmunology and the cytokine action in the CNS: implications for psychiatric disorders. Progress in Neuropsychopharmacology & Biological Psychiatry. 1998;22:1–33. doi: 10.1016/s0278-5846(97)00179-6. [DOI] [PubMed] [Google Scholar]
- Nair P., Melarkode R., Rajkumar D., Montero E. CD6 synergistic co-stimulation promoting proinflammatory response is modulated without interfering with the activated leucocyte cell adhesion molecule interaction. Clinical and Experimental Immunology. 2010;162:116–130. doi: 10.1111/j.1365-2249.2010.04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan S., Tang B., Head S.R., Gilmartin T.J., Sutcliffe J.G., Dean B. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Research. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawa H., Takei N. Recent progress in animal modeling of immune inflammatory processes in schizophrenia: implication of specific cytokines. Neuroscience Research. 2006;56:2–13. doi: 10.1016/j.neures.2006.06.002. [DOI] [PubMed] [Google Scholar]
- O'Carroll D., Mecklenbrauker I., Das P.P., Santana A., Koenig U., Enright A.J. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes & Development. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan M.C., Craddock N., Norton N., Williams H., Peirce T., Moskvina V. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nature Genetics. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Owen D., Andrews M.H., Matthews S.G. Maternal adversity, glucocorticoids and programming of neuroendocrine function and behaviour. Neuroscience and Biobehavioral Reviews. 2005;29:209–226. doi: 10.1016/j.neubiorev.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Ozbey U., Tug E., Namli M. Interleukin-10 gene promoter polymorphism in patients with schizophrenia in a region of East Turkey. The World Journal of Biological Psychiatry: The Official Journal of the World Federation of Societies of Biological Psychiatry. 2009;10:461–468. doi: 10.1080/15622970802626580. [DOI] [PubMed] [Google Scholar]
- Paul-Samojedny M., Owczarek A., Suchanek R., Kowalczyk M., Fila-Danilow A., Borkowska P. Association study of interferon gamma (IFN-gamma) +874T/A gene polymorphism in patients with paranoid schizophrenia. Journal of Molecular Neuroscience: MN. 2011;43:309–315. doi: 10.1007/s12031-010-9442-x. [DOI] [PubMed] [Google Scholar]
- Potthoff M.J., Olson E.N. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Potvin S., Stip E., Sepehry A.A., Gendron A., Bah R., Kouassi E. Inflammatory cytokine alterations in schizophrenia: a systematic quantitative review. Biological Psychiatry. 2008;63:801–808. doi: 10.1016/j.biopsych.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Ripke S., Sanders A.R., Kendler K.S., Levinson D.F., Sklar P., Holmans P.A. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Garcia M., Oliva H., Climent N., Garcia F., Gatell J.M., Gallart T. Human immature monocyte-derived dendritic cells produce and secrete alpha-defensins 1-3. Journal of Leukocyte Biology. 2007;82:1143–1146. doi: 10.1189/jlb.0507295. [DOI] [PubMed] [Google Scholar]
- Rollins B., Martin M.V., Morgan L., Vawter M.P. Analysis of whole genome biomarker expression in blood and brain. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2010;153B:919–936. doi: 10.1002/ajmg.b.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetre P., Emilsson L., Axelsson E., Kreuger J., Lindholm E., Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallenave J.M. Antimicrobial activity of antiproteinases. Biochemical Society Transactions. 2002;30:111–115. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- Santarelli D.M., Beveridge N.J., Tooney P.A., Cairns M.J. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biological Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- Schaefer A., Im H.I., Veno M.T., Fowler C.D., Min A., Intrator A. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. Journal of Experimental Medicine. 2010;207:1843–1851. doi: 10.1084/jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A., Bertsch T., Tost H., Bergmann A., Henning U., Klimke A. Increased serum interleukin-1beta and interleukin-6 in elderly, chronic schizophrenic patients on stable antipsychotic medication. Neuropsychiatric Disease and Treatment. 2005;1:171–177. doi: 10.2147/nedt.1.2.171.61048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A., Leonardi-Essmann F., Durrenberger P.F., Parlapani E., Schneider-Axmann T., Spanagel R. Regulation of immune-modulatory genes in left superior temporal cortex of schizophrenia patients: a genome-wide microarray study. The World Journal of Biological Psychiatry: The Official Journal of the World Federation of Societies of Biological Psychiatry. 2011;12:201–215. doi: 10.3109/15622975.2010.530690. [DOI] [PubMed] [Google Scholar]
- Schwalfenberg G.K. A review of the critical role of vitamin D in the functioning of the immune system and the clinical implications of vitamin D deficiency. Molecular Nutrition & Food Research. 2011;55:96–108. doi: 10.1002/mnfr.201000174. [DOI] [PubMed] [Google Scholar]
- Selsted M.E., Ouellette A.J. Mammalian defensins in the antimicrobial immune response. Nature Immunology. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- Sequeira P.A., Martin M.V., Vawter M.P. The first decade and beyond of transcriptional profiling in schizophrenia. Neurobiology of Disease. 2012;45:23–36. doi: 10.1016/j.nbd.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Li T., Mo X., Majdic O., Zhang Y., Seyerl M. Expressional and functional studies of CKLF1 during dendritic cell maturation. Cellular Immunology. 2010;263:188–195. doi: 10.1016/j.cellimm.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Shao L., Vawter M.P. Shared gene expression alterations in schizophrenia and bipolar disorder. Biological Psychiatry. 2008;64:89–97. doi: 10.1016/j.biopsych.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She H., Yang Q., Shepherd K., Smith Y., Miller G., Testa C. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. The Journal of Clinical Investigation. 2011;121:930–940. doi: 10.1172/JCI43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Levinson D.F., Duan J., Sanders A.R., Zheng Y., Pe'er I. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Fatemi S.H., Sidwell R.W., Patterson P.H. Maternal influenza infection causes marked behavioral and pharmacological changes in the offspring. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2003;23:297–302. doi: 10.1523/JNEUROSCI.23-01-00297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Banerjee S., Bera N.K., Nayak C.R., Chaudhuri T.K. Analysis of the role of human leukocyte antigen class-I genes to understand the etiopathology of schizophrenia. Indian Journal of Psychiatry. 2008;50:166–170. doi: 10.4103/0019-5545.43625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Soderlund J., Schroder J., Nordin C., Samuelsson M., Walther-Jallow L., Karlsson H. Activation of brain interleukin-1beta in schizophrenia. Molecular Psychiatry. 2009;14:1069–1071. doi: 10.1038/mp.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soehnlein O., Kenne E., Rotzius P., Eriksson E.E., Lindbom L. Neutrophil secretion products regulate anti-bacterial activity in monocytes and macrophages. Clinical and Experimental Immunology. 2008;151:139–145. doi: 10.1111/j.1365-2249.2007.03532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H., Ophoff R.A., Steinberg S., Andreassen O.A., Cichon S., Rujescu D. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub R.E., Jiang Y., MacLean C.J., Ma Y., Webb B.T., Myakishev M.V. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. American Journal of Human Genetics. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez B.K., Duan J., Sanders A.R., Hinrichs A.L., Jin C.H., Hou C. Genomewide linkage scan of 409 European-ancestry and African American families with schizophrenia: suggestive evidence of linkage at 8p23.3-p21.2 and 11p13.1-q14.1 in the combined sample. American Journal of Human Genetics. 2006;78:315–333. doi: 10.1086/500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniyam D., Hollander C., Westin U., Erjefalt J., Stevens T., Janciauskiene S. Secretory leukocyte protease inhibitor inhibits neutrophil apoptosis. Respirology. 2011;16:300–307. doi: 10.1111/j.1440-1843.2010.01901.x. [DOI] [PubMed] [Google Scholar]
- Sullivan P.F., Fan C., Perou C.M. Evaluating the comparability of gene expression in blood and brain. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Hayashi H., Watanabe Y., Sawamura K., Fukui N., Watanabe J. Diagnostic classification of schizophrenia by neural network analysis of blood-based gene expression signatures. Schizophrenia Research. 2010;119:210–218. doi: 10.1016/j.schres.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Tienari P., Wynne L.C., Laksy K., Moring J., Nieminen P., Sorri A. Genetic boundaries of the schizophrenia spectrum: evidence from the Finnish Adoptive Family Study of Schizophrenia. American Journal of Psychiatry. 2003;160:1587–1594. doi: 10.1176/appi.ajp.160.9.1587. [DOI] [PubMed] [Google Scholar]
- Tsuang M.T., Nossova N., Yager T., Tsuang M.M., Guo S.C., Shyu K.G. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2005;133B:1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- Tynan R.J., Weidenhofer J., Hinwood M., Cairns M.J., Day T.A., Walker F.R. A comparative examination of the anti-inflammatory effects of SSRI and SNRI antidepressants on LPS stimulated microglia. Brain, Behavior, and Immunity. 2012;26:469–479. doi: 10.1016/j.bbi.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Wager-Smith K., Markou A. Depression: a repair response to stress-induced neuronal microdamage that can grade into a chronic neuroinflammatory condition? Neuroscience and Biobehavioral Reviews. 2011;35:742–764. doi: 10.1016/j.neubiorev.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Z., Li G., Chen X.Y., Zhao M., Yuan Y.H., Wang X.L. Chemokine-like factor 1, a novel cytokine, induces nerve cell migration through the non-extracellular Ca2+-dependent tyrosine kinases pathway. Brain Research. 2010;1308:24–34. doi: 10.1016/j.brainres.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Yang R.Y., Rabinovich G.A., Liu F.T. Galectins: structure, function and therapeutic potential. Expert Reviews in Molecular Medicine. 2008;10:e17. doi: 10.1017/S1462399408000719. [DOI] [PubMed] [Google Scholar]
- Zuckerman L., Weiner I. Maternal immune activation leads to behavioral and pharmacological changes in the adult offspring. Journal of Psychiatric Research. 2005;39:311–323. doi: 10.1016/j.jpsychires.2004.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


