Abstract
Background
Since 2008, severe cases of emerging human adenovirus (HAdV) type 55 (HAdV-55) were reported sporadically in China. But no comparative studies had been conducted to discern the differences in epidemiologic and clinical abnormalities between HAdV-55 and other types (HAdV-7, HAdV-3, HAdV-14, HAdV-50, and HAdV-C).
Methods
A multicenter surveillance study for adult and adolescent community-acquired pneumonia (CAP) was conducted prospectively in Beijing and Yan Tai between November 2010 and April 2012. A standardized data form was used to record clinical information. The viral DNA extracted from the clinical samples or adenovirus viral isolates was sequenced.
Results
Among 969 cases, 48 (5%) were identified as adenovirus pneumonia. Six branches were clustered: HAdV-55 in 21, HAdV-7 in 11, HAdV-3 in nine, HAdV-14 in four, HAdV-50 in two, and HAdV-C in one. Most HAdV-55 cases were identified during February and March. All the hypervariable regions of the hexon genes of the 21 HAdV-55 strains were completely identical. Patients who had HAdV-55 were about 10 years older (P = .027) and had higher pneumonia severity index scores (P = .030) compared with those with other types (HAdV-7, HAdV-3, HAdV-14, HAdV-50, and HAdV-C). Systemic BP was also higher among patients in the HAdV-55 group (P = .006). Unilateral or bilateral consolidations were the most common radiologic findings in both patients with HAdV-55 and those with other types (57.9% vs 36%). More than one-half of the patients were admitted to hospital; oxygen therapy was given to 29.2% of the 48 patients, and two needed mechanical ventilation.
Conclusions
HAdV-55 has established itself as a major pneumonia pathogen in the Chinese population, and further surveillance and monitoring of this agent as a cause of CAP is warranted.
Abbreviations: BNACAP, Beijing Network for Adult Community-Acquired Pneumonia; CAP, community-acquired pneumonia; CPE, cytopathic effect; HAdV, human adenovirus; PCR, polymerase chain reaction; PSI, pneumonia severity index
Community-acquired pneumonia (CAP) refers to pneumonia acquired outside of hospital or long-term care facilities. The overall annual incidence of CAP ranges from five to 20 per 1,000 adults.1 Many microbial pathogens can cause CAP, and the role of viruses may have been underestimated thus far because of a lack of appropriate diagnostic methods.2, 3 Modern molecular techniques have revealed that respiratory viruses account for about 22% of adult CAP cases.4, 5, 6, 7, 8, 9, 10 The most common viruses are influenza, parainfluenza, respiratory syncytial virus, metapneumovirus, and adenovirus.
We previously reported 18 sporadic CAP cases caused by human adenovirus (HAdV) from our single center between August 2008 and April 2011. Polymerase chain reaction (PCR) analysis using type-specific primers targeting the hexon gene revealed that they all belonged to species B (HAdV-11, HAdV-7, HAdV-3, and HAdV-14),11 and HAdV-11 accounted for 58.8% (10 of 17) of them. However, further genome sequence analysis proved that these 10 HAdV-11 strains were actually HAdV type 55 (HAdV-55). HAdV-55, an intertypic recombinant described originally as genome type 11a, was identified from an outbreak of acute respiratory tract infection in Shanxi Province, China, in 2006.12 It exhibited a neutralizing antigen epitope of HAdV-11 and the pathogenic properties of HAdV-14.13, 14 The whole-genome sequencing analysis showed that HAdV-55 had an HAdV-14 chassis with a partial HAdV-11 in the hexon gene.15, 16 For this reason, it was renamed HAdV-55.13
Our previous case series11 indicated that HAdV-55 apparently emerged in Beijing.12 Adenovirus 14 is an emerging agent of concern that has been causing outbreaks of pneumonia not just in China, but worldwide. Adenovirus 55, which is related to adenovirus 14, is now also emerging as an agent of concern. We investigated whether HAdV-55 has a different clinical profile from the profiles of other adenovirus types circulating in China.
Materials and Methods
Beijing Network for Adult CAP
The Beijing Network for Adult Community-Acquired Pneumonia (BNACAP), which consists of 11 general hospitals from nine different districts in Beijing and one teaching hospital in Yan Tai, is a clinic-based, multicenter, prospective surveillance system for adults and adolescents with CAP. Yan Tai is a city by the sea in Shan Dong Province, located about 770 km southeast of Beijing. The institutional review board of Beijing Chao-Yang Hospital approved the study (project approval number 10-KE-49). All patients gave their written informed consent.
Study Population
Between November 2010 and April 2012, all adolescent and adult patients (aged 14 years or older) from 12 general hospitals who met the inclusion criteria of CAP were prospectively enrolled during daytime 7 days a week.3 Patients with HIV infection or neutropenia, those receiving immunosuppressive chemotherapy or prednisone steroids equivalent to 15 mg/d for 30 days, pregnant or breast-feeding women, and those with known or suspected active TB were excluded.
Clinical Data Collection
Clinical information collected by investigators with a standardized data form included the following: age, sex, comorbidities, smoking history, vaccination against influenza and Streptococcus pneumoniae in the past year, symptoms (fever, cough, sputum, dyspnea, chest pain), GI symptoms (nausea, vomiting, diarrhea, and abdominal pain), and neurologic symptoms (headache, dizziness). Clinical signs (body temperature, heart rate, respiratory frequency, BP, and crackles) and treatments (antibiotics, antiviral therapy, or oxygen use) were also recorded. The pneumonia severity index (PSI) was used to assess the severity of illness on the day of enrollment.17
Symptoms and signs of all patients were followed up, either during their hospitalization or after discharge, until all symptoms disappeared. For outpatients, the same information was gathered. All the information collected from the patients was input into a computerized database.
Microbiologic Diagnostic Tests Undertaken
The nasal or throat swab specimens collected by the attending physicians were collected in 2-mL viral transport media, transported at 2°C to 8°C, and preserved at −80°C. The viral RNA was extracted from the clinical samples using a QIAamp RNA mini kit (QIAGEN). Following this, a commercially available Seeplex RV 15 ACE Detection kit (Seegene Inc), a multiplex, one-step, reverse transcriptase PCR, was used to screen for 15 different viruses as the cause of the respiratory illness. The kit included assays for adenovirus, influenza A and B viruses, human metapneumovirus, rhinovirus, respiratory syncytial virus (groups A and B), coronavirus (229E, NL63, OC43, and HKU1), parainfluenza virus (type 1, 2, 3, 4), bocavirus, and enterovirus.
Blood cultures were performed for patients presenting with chills and shivering. If pleural fluid and sputum samples were available, Gram stain and culture were performed. Urinary antigen tests for Legionella pneumophila and S pneumoniae (Binax) were also performed on all urine specimens. Acute sera (1-3 days after onset) and convalescent sera (2-4 weeks after onset) were collected for testing of the antibody for HAdV or other respiratory viruses.
Criteria for Viral Pneumonia
Viral pneumonia was diagnosed based on one of the following criteria: (1) the presence of HAdV or other respiratory viruses detected in sputum or throat swab samples by molecular methods or (2) seroconversion, defined as a fourfold or greater increase in titers of antibodies to HAdV or other respiratory viruses.
Cell Culture and Virus Isolation
Nasal or throat swab specimens were inoculated onto Hep-2 cells and cultured in a maintenance medium for detection of a cytopathic effect (CPE). Cells were observed for CPE every 7 days. Cultures exhibiting adenovirus-like CPE were processed again to confirm the presence of the virus.
Extraction of Viral DNA and PCR
Viral DNA was extracted from the clinical samples and adenovirus viral isolates using a QIAamp DNA mini kit (QIAGEN). The hexon and fiber genes were both amplified using primers described previously by Zhu et al.12 PCR was performed with a 25-μL reaction mixture containing 2 μL of template DNA. Reaction conditions were determined as described previously by Zhu et al.12
Sequence Analysis
The PCR products were purified (QIAGEN) and sequenced by a dye terminator method (BigDye Terminator, version 3.1, cycle sequencing kit; Applied Biosystems) with an ABI Prism 3100 genetic analyzer (Applied Biosystems). Sequence data were stored as standard chromatogram format files (.abl) and were analyzed with Sequencher software (version 4.0.5; Gene Codes Corp), the Basic Local Alignment Search Tool program (National Center for Biotechnology Information), BioEdit sequence alignment editor software (version 5.0.9; Tom Hall, North Carolina State University, Raleigh, North Carolina), and the Molecular Evolutionary Genetics Analysis (MEGA) program (Sudhir Kumar, Arizona State University, Phoenix, Arizona).
Statistical Analysis
Data analysis was performed using SPSS 15.0 (IBM). A two-tailed independent-samples t test or a Mann-Whitney U test (in the case of nonnormal distributions) was used to compare continuous variables between the two groups. For the categorical data, univariate analysis was carried out using the χ2 test or Fisher exact test. Significance was fixed at P < .05.
Results
Epidemiology
Between November 2010 and April 2012, 1,013 cases with CAP were enrolled in the BNACAP study. Forty-four cases were ruled out: In 30, no throat/nasal specimen was obtained, and clinical information was missing in 14. Therefore, 969 cases were available for the etiology study. Among them, 393 were positive for at least one pathogen: respiratory viruses in 262, Mycoplasma pneumoniae in 168, typical bacteria in 47, Mycobacterium tuberculosis in 15, and Legionella pneumoniae in four. Dual causes were found in 65 patients (e-Table 1).
Types of HAdV
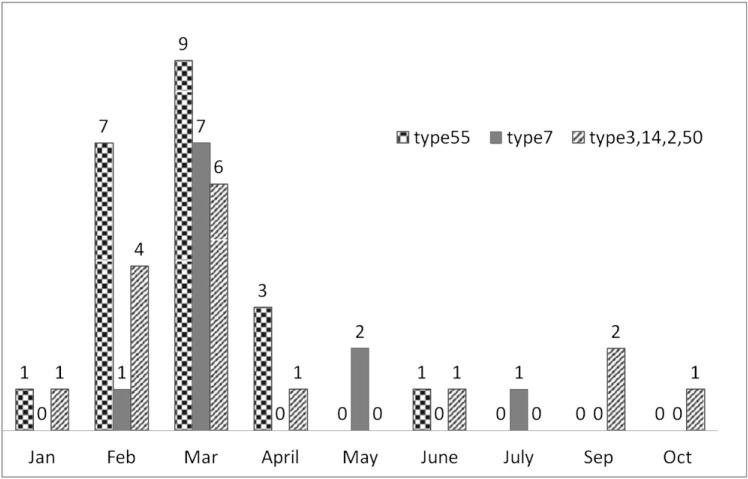
Forty-eight patients (48 of 969 [5%]) were identified as having adenovirus pneumonia, and 26 of the 48 adenovirus-positive samples showed characteristic adenovirus-like CPE. Basic Local Alignment Search Tool analysis based on the hypervariable region of the hexon genes from all 48 adenovirus-positive samples was performed. Among the 48 samples, 21 (43.8%) were HAdV-55, 11 (22.9%) were HAdV-7, nine (18.8%) were HAdV-3, four (8.3%) were HAdV-14, two (4.2%) were HAdV-50, and one (2.1%) was HAdV-C. Most HAdVs were identified in February and March. No adenovirus was found in November or December (Fig 1 ). The type distribution was similar between Beijing and Yan Tai City (Table 1 ).
Figure 1.
Epidemiologic distribution of different types of human adenoviruses. Most human adenovirus type 55 was identified during February and March, and it had epidemiologic characteristics similar to other types. No adenovirus pneumonia was found in November and December, the typical influenza season months.
Table 1.
—Epidemiologic and Clinical Characteristics of Patients With CAP Caused by Adenoviruses (Comparison Between HAdV-55 and Other Types)
| Characteristic | Total (N = 48) | HAdV-55 (n = 21) | Other Types (n = 27) | P Value |
|---|---|---|---|---|
| Age, y | 38.0 ± 18.5 | 44.7 ± 21.8 | 32.9 ± 13.8 | .027a |
| Male (female), No. | 33 (15) | 16 (5) | 17 (10) | .366 |
| From Beijing | 29 (60.4) | 11 (52.4) | 18 (66.7) | .38 |
| Underlying diseases | 5 (10.4) | 4 (19.1) | 1 (3.7) | .153 |
| Current smokers | 15 (31.3) | 8 (38.1) | 7 (25.9) | .531 |
| Influenza vaccination within 1 y, No. | 3 | 1 | 2 | 1.0 |
| Streptococcus pneumoniae vaccination within 1 y, No. | 1 | 0 | 1 | 1.0 |
| Antibiotics before enrollment, No. | 32 | 13 | 19 | .555 |
| Clinical features | ||||
| PSI score | 41.0 ± 22.8 | 49.1 ± 27.0 | 34.8 ± 16.8 | .030a |
| Fever | 43 (89.6) | 19 (90.5) | 24 (88.9) | 1.0 |
| Tmax, °C | 39.3 ± 0.9 | 38.9 ± 0.7 | 39.4 ± 1.0 | .070 |
| Cough | 45 (93.8) | 20 (95.2) | 25 (92.6) | 1.0 |
| Sputum | 33 (68.8) | 16 (76.2) | 17 (63.0) | .366 |
| Purulent sputum | 18 (37.5) | 10 (47.6) | 8 (29.6) | .380 |
| Dyspnea | 9 (18.8) | 3 (14.3) | 6 (22.2) | .712 |
| Chest pain | 4 (8.3) | 1 (4.8) | 3 (11.1) | .621 |
| GI symptoms | 6 (12.5) | 4 (19.1) | 2 (7.4) | .383 |
| Neurologic symptoms | 6 (12.5) | 3 (14.3) | 3 (11.1) | 1.0 |
| Systemic BP, mm Hg | 118.3 ± 11.3 | 123.4 ± 11.7 | 114.5 ± 9.5 | .006a |
| Heart rate, beats/min | 86.6 ± 11.5 | 84.2 ± 10.1 | 88.4 ± 12.4 | .208 |
| Respiratory rate, breaths/min | 20.1 ± 3.2 | 20.8 ± 4.2 | 19.6 ± 2.3 | .235 |
| Moist rales | 17 (35.4) | 8 (38.1) | 9 (33.3) | .377 |
| Dry rales | 2 (4.2) | 2 (9.5) | 0 (0) | .186 |
| Conjunctival congestion | 1 (2.1) | 0 (0) | 1 (3.7) | 1.0 |
| Rashes | 2 (4.2) | 1 (4.8) | 1 (3.7) | 1.0 |
Data are presented as mean ± SD or No. (%) unless indicated otherwise. CAP = community-acquired pneumonia; HAdV = human adenovirus; PSI = pneumonia severity index; Tmax = maximal temperature.
P < .05.
Demographic Characteristics
The mean age of the 48 cases was 38 years; 12 of the 48 (25%) were aged > 50 years. Patients infected by HAdV-55 were about 10 years older than those infected by other types (P = .027). Men predominated over women, with a sex ratio of about 2:1. More patients infected by HAdV-55 had underlying diseases (19.7% vs 3.7%), although the difference was not significant (Table 1).
Clinical Features: Comparison Between HAdV-55 and Other Types
Most clinical symptoms and signs between patients infected by HAdV-55 and those infected by other types did not differ, except for PSI score and systemic BP; these were significantly higher in patients infected by HAdV-55 (P = .030 and P = .006, respectively) (Table 1).
There was no difference in laboratory findings between HAdV-55-infected cases and those infected by other types (Table 2 ). Eight HAdV cases involved coinfections, including HAdV-55 with M pneumoniae in three, HAdV-55 with parainfluenza virus 3 and influenza virus B in one, HAdV-7 with M pneumoniae in one, HAdV-2 with respiratory syncytial virus A in one, HAdV-14 and parainfluenza virus 4 in one, and HAdV-3 with human coronavirus in one (Table 2).
Table 2.
—Laboratory Findings and Chest Radiologic Characteristics of Patients With CAP Caused by Adenoviruses (Comparison Between HAdV-55 and Other Types)
| Characteristic | Total (N = 48) | HAdV-55 (n = 21) | Other Types (n = 27) | P Value |
|---|---|---|---|---|
| WBC, 109/L | 7.19 ± 3.59 | 6.70 ± 3.31 | 7.33 ± 3.84 | .749 |
| Leukocyte < 4,000/mm3, % | 4 (8.3) | 2 (9.5) | 2 (7.4) | 1.0 |
| Leukocyte > 10,000/mm3, % | 8 (12.5) | 3 (14.3) | 4 (14.8) | .715 |
| Neutrophil, % | 68.7 ± 12.3 | 69.9 ± 11.8 | 67.7 ± 12.9 | .553 |
| Lymphocyte, % | 21.9 ± 9.3 | 20.9 ± 9.0 | 22.8 ± 9.8 | .484 |
| Hemoglubulin, g/L | 141.4 ± 14.8 | 139.0 ± 15.0 | 143.3 ± 14.7 | .327 |
| Platelet, 109/L | 186.9 ± 77.4 | 196.2 ± 87.6 | 179.7 ± 69.3 | .469 |
| AST, μ/L | 26.5 (14-176) | 28 (14-130) | 26 (17-176) | .975 |
| AST > 40 μ/L | 10 (20.8) | 4 (19.1) | 6 (22.2) | 1.0 |
| ALT, μ/L | 23 (6-122) | 26.5 (9-122) | 22 (6-109) | .511 |
| ALT > 40 μ/L | 8 (16.7) | 3 (14.3) | 5 (18.5) | 1.0 |
| ALB, g/L | 36 (24.5-45.9) | 34.6 (30.7-43.7) | 36.4 (24.5-45.9) | .600 |
| LDH, μ/L | 201 (120-794) | 193 (120-467) | 217 (129-794) | .771 |
| LDH > 250 μ/L | 13 (27.1) | 4 (19) | 9 (33.3) | .338 |
| CK, μ/L | 47 (34-1994) | 70.5 (41-345) | 87 (34-1994) | .659 |
| CK > 200 μ/L | 5 (10.4) | 1 (4.8) | 4 (14.8) | .369 |
| Tbil, μmol/L | 9.9 (4.3-39.4) | 11.8 (6.0-39.4) | 8.9 (4.3-38.4) | .063 |
| Tbil > 17.1 μmol/L | 8 (16.7) | 5 (23.8) | 3 (11.1) | .272 |
| Cr, μmol/L | 68.9 (1.9-148.7) | 77.5 (1.9-148.7) | 62 (3.6-110.2) | .372 |
| K, mmol/L | 3.92 ± 0.42 | 3.91 ± 0.46 | 3.93 ± 0.40 | .845 |
| Na, mmol/L | 136.2 ± 4.1 | 135.1 ± 4.2 | 137.0 ± 3.9 | .102 |
| Pao2, mm Hg | 83.4 ± 23.8 | 91.8 ± 31.3 | 77.1 ± 14.7 | .167 |
| Paco2, mm Hg | 32.6 ± 6.5 | 32.3 ± 7.3 | 32.9 ± 6.1 | .827 |
| ESR,a mm/h | 27 (8-70) | 32 (8-66) | 23.5 (9-70) | .212 |
| CRP,b mg/L | 11.1 (2-147) | 11.1 (2-147) | 12 (2-104) | .728 |
| CRP > 20 mg/L | 10 (43.5) | 3 (25) | 7 (46.7) | |
| PCT,c ng/mL | 0.32 (0.02-2.65) | 0.31 (0.05-0.87) | 0.32 (0.02-2.65) | .876 |
| PCT > 0.5 ng/mL | 4 (33.3) | 1 (20) | 3 (42.8) | 1.0 |
| PCT > 1 ng/mL | 2 (16.7) | 0 (0) | 2 (28.6) | 1.0 |
| Chest radiographyd | ||||
| Bilateral involvement | 19 (39.6) | 9 (47.4) | 10 (40) | .761 |
| Consolidation | 20 (45.5) | 11 (57.9) | 9 (36) | .223 |
| Patchy infiltration | 18 (40.9) | 7 (36.8) | 11 (44) | .760 |
| Ground-grass opacity | 12 (27.3) | 5 (26.3) | 7 (28) | 1.0 |
| Pleural effusion | 3 (6.8) | 1 (5.3) | 2 (8) | 1.0 |
| Coinfectionse | 8 (16.7) | 4 (19.1) | 4 (14.8) | .715 |
Data are presented as mean ± SD, No. (%), or median (range). ALB = albumin; ALT = alanine aminotransferase; AST = aspartate aminotransferase; CK = creatine kinase; Cr = creatinine; CRP = C reactive protein; ESR = erythrocyte sedimentation rate; LDH = lactate dehydrogenase; PCT = procalcitonin; Tbil = total bilirubin. See Table 1 legend for expansion of other abbreviations.
n = 27.
n = 23.
n = 12.
Total (N = 44), HAdV-55 (n = 19), and other types (n = 25).
Eight HAdV cases involved coinfections, including HAdV-55 with Mycoplasma pneumoniae in three, HAdV-55 with parainfluenza virus 3 and influenza virus B in one, HAdV-7 with Mycoplasma pneumoniae in one, HAdV-2 with respiratory syncytial virus A in one, HAdV-14 and parainfluenza virus 4 in one, and HAdV-3 with human coronavirus in one.
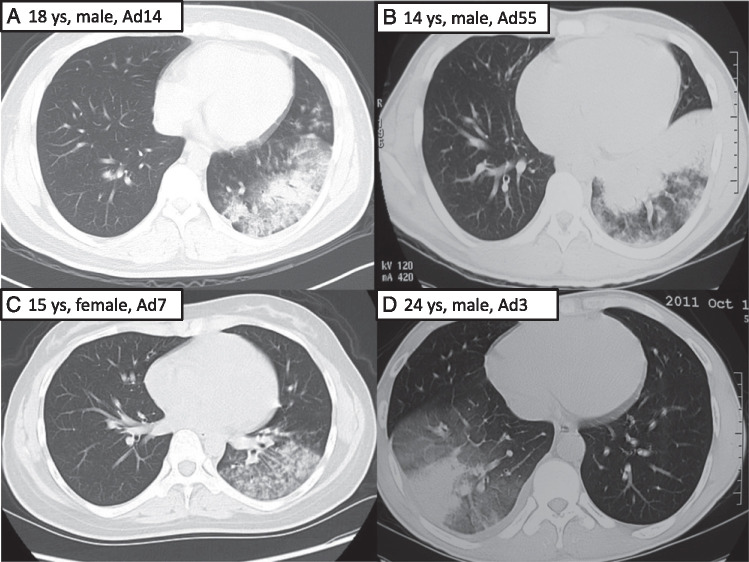
Forty percent of the patients had bilateral involvement on chest radiography (Table 2). Consolidation, patchy infiltrate, and ground-grass opacity were the most common findings in pneumonia caused by HAdV. Patients infected by HAdV-55 presented consolidation more commonly than did those infected by other types (57.9% vs 36%) (Fig 2 ), but the difference was not significant.
Figure 2.
Radiographic findings of four patients infected with different types of adenoviruses. A, An 18-year-old young man infected with human adenovirus (HAdV)-14. Chest CT scan showed patchy infiltrate, ground-grass opacity, and partial consolidation of left lower lobe. B, A 14-year-old boy infected with HAdV-55. Chest CT scan showed consolidation and ground-grass opacity of left lower lobe. C, A 15-year-old girl infected with HAdV-7. Chest CT scan showed patchy infiltrate and ground-grass opacity of left lower lobe. D, A 24-year-old man infected with HAdV-3. Chest CT scan showed consolidation and ground-grass opacity of right lower lobe. ys = years old.
Complications, Management, and Prognosis of Patients With HAdV Pneumonia
More than one-half of the patients were admitted to hospital, but there was no difference between HAdV-55 and other types (Table 3 ). No case was proved to have a coinfection with bacteria, but coinfections with other respiratory viruses (25%) or M pneumoniae (12.5%) were common. Oxygen therapy was given to 29.2% of the patients, and only two needed mechanical ventilation. Antibiotics were given to all the patients, but only four were prescribed antiviral drugs (all from Beijing Chao-Yang Hospital). The clinical outcomes, including duration of fever and other respiratory symptoms, length of stay in hospital, and hospitalization expenses, were similar between HAdV-55 and other types (Table 3).
Table 3.
—Complications, Management, and Prognosis of Patients With Adenoviral Pneumonia
| Characteristics | Total (N = 48) | HAdV-55 (n = 21) | Other Types (n = 27) | P Value |
|---|---|---|---|---|
| Hospitalization | 26 (54.2) | 11 (52.4) | 15 (55.6) | 1.0 |
| Complications | ||||
| Coinfection with bacteria | 0 (0) | 0 (0) | 0 (0) | … |
| Coinfection with Mycoplasma pneumoniae | 6 (12.5) | 3 (14.3) | 3 (11.1) | 1.0 |
| Coinfection with other respiratory virus | 12 (25) | 4 (19.1) | 7 (25.9) | .726 |
| ARDS | 2 (4.2) | 0 (0) | 2 (7.4) | .497 |
| Management | ||||
| Oxygen therapy | 14 (29.2) | 7 (33.3) | 7 (25.9) | .750 |
| Mechanical ventilation | 2 (4.2) | 0 (0) | 2 (7.4) | .497 |
| Antibiotics | 48 (100) | 21 (100) | 27 (100) | 1.0 |
| Antivirala | 4 (8.3) | 4 (19.1) | 0 (0) | .031 |
| Outcomes | ||||
| Duration of fever, d | 6 (2-20) | 7 (2-20) | 5 (2-9) | .201 |
| Duration of cough, d | 14.5 (4-37) | 14.5 (5-37) | 14 (4-28) | .810 |
| Duration of sputum, d | 13.5 (4-37) | 13.5 (4-37) | 12 (6-29) | 1.0 |
| Duration of dyspnea, d | 6 (1-21) | 6 (2-11) | 6 (1-21) | .905 |
| Duration of chest pain, d | 5 (5-7) | 5 (5-5) | 6 (5-7) | .667 |
| LOS in hospital, d | 7.5 (3-26) | 9 (5-21) | 6 (3-26) | .087 |
| Hospitalization expenses, RMB | 6,020 (1,200-49,947) | 7,608 (4,229-32,684) | 4,865 (1,200-49,947) | .234 |
Data are presented as No. (%) or median (range). LOS = length of stay; RMB = Chinese Yuan. See Table 1 legend for expansion of other abbreviations.
Three of the patients were prescribed acyclovir and the fourth was given ribavirin. All the patients were from Beijing Chao-Yang Hospital.
Genome Sequence and Analysis of HAdV-55
A phylogenetic analysis was conducted based on the hypervariable region of the hexon gene to demonstrate the genetic relationship between HAdVs strains and the other seven HAdVs species (A-G); 47 of 48 strains belonged to HAdV species B and only one was HAdV-C (e-Fig 1A). Further partial hexon gene of HAdV strains were compared with the sequences of HAdV-B species in GenBank (e-Fig 1B); 21 of the 47 HAdV-Bs formed a dependent branch and revealed 100%, 96.7%, and 80% homologies with HAdV-55 (FJ643676), HAdV-11 (AF532578), and HAdV-14 (AY803294), respectively. Another phylogenetic tree was then conducted based on the partial fiber gene of the 15 HAdVs, hexon genes which were homologous to HAdV-55 (e-Fig 1C). The partial fiber gene had 99.8% to 100%, 99.4% to 99.5%, and 94.2% to 94.3% nucleotide identity with HAdV-55 (FJ643676), HAdV-14 (AY803294), and HAdV-11 (AF532578), respectively. (The amplification or sequencing of the other six strains failed, and the real-time PCR indicated that their hexon genes were HAdV-14 and their fiber genes were HAdV-14 [data not show]).
All the hypervariable regions of the hexon genes (18281-19247nt, 967bp) of the 21 HAdV-55 strains were completely identical. Two of the 15 partial fiber genes (30689-31818nt, 1130bp) had one nucleotide difference from the others. The nucleotide sequences data from these 48 HAdVs were submitted to GenBank, and accession numbers were allocated as KC510700 to KC510747 (Hexon genes) and KC510748 to KC510762 (fiber genes).
Discussion
To our knowledge, this study is the first large cohort on the epidemiology and clinical features of CAP associated with HAdV-55, the emerging pathogen among immunocompetent adolescents and adults. Our data showed clearly that HAdV-55 has established itself as a major pneumonia pathogen in the Chinese population and that further surveillance and monitoring of this agent as a cause of CAP is warranted.
HAdV has been recognized as an important viral cause of ARDS. The HAdV types most frequently associated with ARDSs include subspecies B1 HAdV-3, HAdV-7, and HAdV-21 and species E HAdV-4. The association of subspecies B2 HAdV (HAdV-11, HAdV-14, HAdV-34, HAdV-35) infection with ARDS has been rarely reported historically, with some of that documentation covering military trainees.15, 18, 19
In China, HAdV-3 and HAdV-7 were the most common types of pathogens.20, 21, 22 HAdV-11a associated with ARDSs can be traced back to the 1980s23 and it reemerged as an ARDS pathogen in 2006.12, 16 HAdV-55 (formerly known as HAdV-11a) was renamed HAdV-55 based on complete genomic sequence data,13 which clearly showed that HAdV-55 was a recombinant between the HAdV-11 and HAdV-14 ancestral strains.
Adenovirus 14 is an emerging agent of concern that has been causing outbreaks of pneumonia not just in China but worldwide.24, 25 Tate et al26 reported an outbreak of severe respiratory disease associated with HAdV-14 in a US Air Force training facility. Five hundred fifty-one of 1,147 trainees (48%) with febrile respiratory illness were infected with HAdV-14; 23 trainees were hospitalized with pneumonia; four of those required admission to an ICU, and one died. Subsequently, outbreaks associated with HAdV-14 were reported in other states, such as Oregon, Alaska, and so forth.27, 28
Today, adenovirus 55, which is related to adenovirus 14, is also emerging as an agent of concern. Because of the absence of cases caused by other HAdV types, Vento et al29 could only compare pneumonia caused by HAdV-14 with HAdV-14-negative cases. Our study had the advantage of being able to compare the differences in clinical, laboratory, or radiographic abnormalities caused by HAdV-55 and other types of pathogens. We proved that patients with diseases due to HAdV-55 were about 10 years older (P = .027) and had higher PSI scores (P = .030)
Our study has several limitations. First, our case series mainly represents the relatively mild and moderate end of the disease. We believe a wider surveillance study is needed to evaluate the spectrum of the disease caused by this emerging pathogen in an affected area in China. In addition, virus isolates were typed only by amplification and sequencing of the hexon gene, and no seroneutralization was performed. More laboratory tests should be carried out to understand the genomics and pathogenic characteristics.
Conclusions
In conclusion, our data provide new insight into the epidemiology of HAdV-55 infection in China. Patients with HAdV-55 infection were about 10 years older and had higher PSI scores than did patients infected by other types (HAdV-3, HAdV-7, HAdV-14). Furthermore, because it is difficult to discern HAdV pneumonia from clinical symptoms and signs, viral cause determination and a good surveillance system are important.
Acknowledgments
Author contributions: Drs Cao and Wang had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Dr Cao: contributed to the design of the study, care of the adenovirus pneumonia cases, data gathering, analysis of clinical data, writing of the manuscript, and the decision to publish.
Dr Huang: contributed to the PCR analysis and genotyping and writing of the manuscript.
Dr Pu: contributed to data gathering and manuscript revision.
Dr Qu: contributed to the care of the adenovirus pneumonia cases, data gathering, clinical specimen collection, PCR analysis, and manuscript revision.
Dr Yu: contributed to the care of the adenovirus pneumonia cases, data gathering, analysis of clinical data, and manuscript revision.
Dr Zhu: contributed to the PCR analysis and genotyping and manuscript revision.
Dr Dong: contributed to the care of the adenovirus pneumonia cases, data gathering, and manuscript revision.
Dr Gao: contributed to the care of the adenovirus pneumonia cases, data gathering, and manuscript revision.
Dr Zhang: contributed to the care of the adenovirus pneumonia cases, data gathering, and manuscript revision.
Dr X-H. Li: contributed to the care of the adenovirus pneumonia cases, data gathering, analysis of clinical data, and manuscript revision.
Dr Liu: contributed to the care of the adenovirus pneumonia cases, data gathering, and manuscript revision.
Dr H. Wang: contributed to the care of the adenovirus pneumonia cases, data gathering, and manuscript revision.
Dr Q. Xu: contributed to the care of the adenovirus pneumonia cases, data gathering, and manuscript revision.
Dr H. Li: contributed to the care of the adenovirus pneumonia cases, data gathering, and manuscript revision.
Dr W. Xu: contributed to the design of the study and the decision to publish and manuscript revision.
Dr C. Wang: contributed to the design of the study and the decision to publish and manuscript revision.
Financial/nonfinancial disclosures: The authors have reported to CHEST that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Role of sponsors: The sponsors had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Other contributions: We thank all the doctors who have joined the BNACAP network but are not listed in the authorship. We also thank Yingmei Liu, MD, and Zhenjia Liu, MD, for their contribution to the specimen collection and PCR analysis.
Additional information: The e-Figure and e-Table can be found in the “Supplemental Materials” area of the online article.
Footnotes
Drs. Cao, Huang, and Pu contributed equally to this article.
Funding/Support: This work was supported by the Beijing Science and Technology Project [Grant D101100049810002], the National Natural Science Foundation of China [Grants 81070005/H0104, 81030032/H19, and 81271840], China’s Key Technologies Research and Development Program of the National Ministry of Science [Grants 2013ZX10004-202 and 2012ZX10004201-003], and the Program for New Century Excellent Talents in University [Grant NCET-09-0006].
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians. See online for more details.
Supplementary Material
References
- 1.Woodhead M. Community-acquired pneumonia in Europe: causative pathogens and resistance patterns. Eur Respir J Suppl. 2002;36:20s–27s. doi: 10.1183/09031936.02.00702002. [DOI] [PubMed] [Google Scholar]
- 2.Cao B, Ren LL, Zhao F. Viral and Mycoplasma pneumoniae community-acquired pneumonia and novel clinical outcome evaluation in ambulatory adult patients in China. Eur J Clin Microbiol Infect Dis. 2010;29(11):1443–1448. doi: 10.1007/s10096-010-1003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandell LA, Wunderink RG, Anzueto A, Infectious Diseases Society of America, American Thoracic Society Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(suppl 2):S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jennings LC, Anderson TP, Beynon KA. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax. 2008;63(1):42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 5.Johansson N, Kalin M, Tiveljung-Lindell A, Giske CG, Hedlund J. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50(2):202–209. doi: 10.1086/648678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Templeton KE, Scheltinga SA, van den Eeden WCJFM, Graffelman AW, van den Broek PJ, Claas EC. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis. 2005;41(3):345–351. doi: 10.1086/431588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angeles Marcos M, Camps M, Pumarola T. The role of viruses in the aetiology of community-acquired pneumonia in adults. Antivir Ther. 2006;11(3):351–359. [PubMed] [Google Scholar]
- 8.Hohenthal U, Vainionpää R, Nikoskelainen J, Kotilainen P. The role of rhinoviruses and enteroviruses in community acquired pneumonia in adults. Thorax. 2008;63(7):658–659. [PubMed] [Google Scholar]
- 9.Johnstone J, Majumdar SR, Fox JD, Marrie TJ. Viral infection in adults hospitalized with community-acquired pneumonia: prevalence, pathogens, and presentation. Chest. 2008;134(6):1141–1148. doi: 10.1378/chest.08-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman D, Shimoni A, Shemer-Avni Y, Keren-Naos A, Shtainberg R, Lieberman D. Respiratory viruses in adults with community-acquired pneumonia. Chest. 2010;138(4):811–816. doi: 10.1378/chest.09-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu L, Liu Z, Li X. Severe community-acquired pneumonia caused by adenovirus type 11 in immunocompetent adults in Beijing. J Clin Virol. 2012;54(4):295–301. doi: 10.1016/j.jcv.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, Zhang Y, Xu S. Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J Clin Microbiol. 2009;47(3):697–703. doi: 10.1128/JCM.01769-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J Clin Microbiol. 2010;48(3):991–993. doi: 10.1128/JCM.01694-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Seto D, Cao B, Zhao S, Wan C. Genome sequence of human adenovirus type 55, a re-emergent acute respiratory disease pathogen in China. J Virol. 2012;86(22):12441–12442. doi: 10.1128/JVI.02225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chmielewicz B, Benzler J, Pauli G, Krause G, Bergmann F, Schweiger B. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J Med Virol. 2005;77(2):232–237. doi: 10.1002/jmv.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Zhu Z, Tang L. Genomic analyses of recombinant adenovirus type 11a in China. J Clin Microbiol. 2009;47(10):3082–3090. doi: 10.1128/JCM.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fine MJ, Auble TE, Yealy DM. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med. 1997;336(4):243–250. doi: 10.1056/NEJM199701233360402. [DOI] [PubMed] [Google Scholar]
- 18.Binn LN, Sanchez JL, Gaydos JC. Emergence of adenovirus type 14 in US military recruits–a new challenge. J Infect Dis. 2007;196(10):1436–1437. doi: 10.1086/522969. [DOI] [PubMed] [Google Scholar]
- 19.Kajon AE, Dickson LM, Metzgar D, Houng HS, Lee V, Tan BH. Outbreak of febrile respiratory illness associated with adenovirus 11a infection in a Singapore military training cAMP. J Clin Microbiol. 2010;48(4):1438–1441. doi: 10.1128/JCM.01928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu-ying T, Xiu-yun L, Qi L. Genotype characterization of adenoviruses that caused children pneumonia in Beijing [in Chinese] Chin J Virol. 2006;22(4):286–291. [Google Scholar]
- 21.Deng J, Qian Y, Zhao LQ. Identification and typing of adenoviruses from pediatric patients with acute respiratory infections in Beijing from 2003 to 2008 [in Chinese] Zhonghua Er Ke Za Zhi. 2010;48(10):739–743. [PubMed] [Google Scholar]
- 22.Guo L, Gonzalez R, Zhou H. Detection of three human adenovirus species in adults with acute respiratory infection in China. Eur J Clin Microbiol Infect Dis. 2012;31(6):1051–1058. doi: 10.1007/s10096-011-1406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li QG, Hambraeus J, Wadell G. Genetic relationship between thirteen genome types of adenovirus 11, 34, and 35 with different tropisms. Intervirology. 1991;32(6):338–350. doi: 10.1159/000150218. [DOI] [PubMed] [Google Scholar]
- 24.Metzgar D, Osuna M, Kajon AE, Hawksworth AW, Irvine M, Russell KL. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J Infect Dis. 2007;196(10):1465–1473. doi: 10.1086/522970. [DOI] [PubMed] [Google Scholar]
- 25.Gray GC, Chorazy ML. Human adenovirus 14a: a new epidemic threat. J Infect Dis. 2009;199(10):1413–1415. doi: 10.1086/598522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tate JE, Bunning ML, Lott L. Outbreak of severe disease associated with emergent human adenovirus serotypes 14 at a US air force training facility in 2007. J Infect Dis. 2009;199(10):1419–1426. doi: 10.1086/598520. [DOI] [PubMed] [Google Scholar]
- 27.Lewis PF, Schmidt MA, Lu X. A community-based outbreak of severe respiratory illness caused by human adenovirus serotype 14. J Infect Dis. 2009;199(10):1427–1434. doi: 10.1086/598521. [DOI] [PubMed] [Google Scholar]
- 28.Esposito DH, Gardner TJ, Schneider E. Outbreak of pneumonia associated with emergent human adenovirus serotype 14—Southeast Alaska, 2008. J Infect Dis. 2010;202(2):214–222. doi: 10.1086/653498. [DOI] [PubMed] [Google Scholar]
- 29.Vento TJ, Prakash V, Murray CK. Pneumonia in military trainees: a comparison study based on adenovirus serotype 14 infection. J Infect Dis. 2011;203(10):1388–1395. doi: 10.1093/infdis/jir040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.