Abstract
SARS 8b is one of the putative accessory proteins of the severe acute respiratory syndrome‐associated coronavirus (SARS‐CoV) with unknown functions. In this study, the cellular localization and activity of this estimated 9.6 kDa protein were examined. Confocal microscopy results indicated that SARS 8b is localized in both nucleus and cytoplasm of mammalian cells. Functional study revealed that overexpression of SARS 8b induced DNA synthesis. Coexpression of SARS 8b and SARS 6, a previously characterized SARS‐CoV accessory protein, did not elicit synergistic effects on DNA synthesis.
Keywords: Severe acute respiratory syndrome, Coronavirus, SARS ORF 8, SARS 8b, X5
1. Introduction
Severe acute respiratory syndrome (SARS) is a newly discovered infectious disease that has caused more than 7900 cases and 800 deaths worldwide in 2003 [1]. The SARS‐CoV, which is a novel coronavirus, was identified as the causative agent of SARS. It is an enveloped, positive‐stranded RNA virus with a genome of approximately 30 kb, encompassing five major open reading frames (ORFs) which encode the replicase polyproteins, the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. Besides these major proteins, SARS‐CoV genome also encodes five to eight putative accessory proteins that share little homology with any known proteins of other coronaviruses [2, 3].
Although accessory proteins from other coronaviruses are often dispensable for viral replication, they may play vital roles in virulence and pathogenesis by affecting host innate immune responses, encoding pro‐ or anti‐apoptotic activities, or impacting other signaling pathways that might influence disease outcomes [4]. For instance, group‐specific ORFs of murine coronavirus were found to be non‐essential for viral replication, yet their deletions were attenuating in natural hosts [5]. Recent research on the SARS‐CoV have also indicated that accessory proteins SARS 3a, 3b and 7a could induce apoptosis [6, 7, 8]. On the other hand, coronavirus infection in the early stages had been reported to stimulate epithelial cells, causing cellular proliferation and squamous metaplasia in the lungs [9]. Interestingly, SARS 6 has been shown to increase DNA synthesis in mammalian cells of epithelium origins [10].
SARS 8b, also known as X5, is predicted to be a soluble protein with 84 amino acids and an estimated size of 9.6 kDa. It shows minor homology to the human coronavirus E2 glycoprotein precursor [2]. Several studies had suggested that human SARS‐CoV evolved from SARS‐CoV‐like virus harbored in exotic animals such as palm civet cats [11]. Evolutionary studies of zoonotic and human SARS‐CoV have indicated that in the early phase of the SARS epidemic, ORF8 was encoded as a single ORF while isolates of human SARS‐CoV at a later stage of the epidemic contained a 29‐nucleotide deletion which resulted in two ORFs designated as ORF8a and ORF8b [11]. This deletion might be associated with transmission of the virus during the epidemic [12]. In this study, the cellular localization and functional roles of SARS 8b were examined.
2. Materials and methods
2.1. Cell culture
Vero E6 (African green monkey kidney fibroblasts) and CHO (Chinese hamster ovary) cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured at 37 °C in 5% CO2 in Dulbecco modified eagle medium (Gibco BRL Life Technologies, Inc., Carlsbad, CA, USA) containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin.
2.2. Plasmid construction
The SARS 8b cDNA (CUHK‐Su10, GenBank Accession No. http://AY282752) [13] was subcloned into pEGFP‐N1 (BD Bioscience, Clontech, Palo Alto, CA, USA) and pcDNA3.1 (Invitrogen, Carlsbad, CA, USA) vectors, respectively. Primers SARS 8b‐GFP‐F (5′ GGC GCC GAG CTC ATG TGC TTG AAG ATC CTT 3′) and SARS 8b‐GFP‐R (5′ CCG GGG TAC CTC ATT TGT TCG TTT ATT TAA 3′) containing SacI and KpnI restriction cut sites were used for polymerase chain reaction (PCR) of the SARS 8b cDNA. Amplicons that had been cut with SacI and partially digested with KpnI were subcloned into pEGFP‐N1 vectors, yielding the SARS 8b‐pEGFP vector that encodes the SARS 8b‐EGFP (enhanced green fluorescent protein). For constructing the untagged plasmid of SARS 8b, primers SARS 8b‐F (5′ GGC GCC GGT ACC ATG TGC TTG AAG ATC CTT 3′) and SARS 8b‐R (5′ GGC GCC TCT AGA TTA ATT TGT TCG TTT ATT 3′) which contain KpnI and XbaI cut sites were used for PCR amplification of the SARS 8b cDNA. Amplicons that had been cut with XbaI and paritally digested with KpnI were cloned into pcDNA3.1 at the appropriate cut sites, generating pcDNA3.1‐SARS 8b construct which encodes the SARS 8b proteins.
The SARS 6 cDNA were subcloned into pDsRed‐N1 vectors (BD Biosciences, Clontech). Primers SARS 6‐RFP‐F (5′ GGC GCC GAG CTC ATG TTT CAT CTT GTT GAC 3′) and SARS 6‐RFP‐R (5′CCG GGG TAC CTC TGG ATA ATC TAA CTC CAT 3′) containing SacI and KpnI restriction cut sites were used for PCR of the SARS 6 cDNA and amplicons cut with the appropriate enzymes were subcloned into pDsRed‐N1 vectors, yielding the SARS 6‐pDsRed vector that encodes the SARS 6‐RFP (red fluorescent protein). The SARS 6‐pcDNA3.1 vectors were constructed as described elsewhere [10].
The sequences of all constructs in the present study were confirmed by DNA sequencing (Macrogen Inc., Seoul, Korea).
2.3. Western blot analysis
Western blotting was performed as described elsewhere [10].
2.4. Cellular localization of SARS 8b and SARS 6 proteins
Cells plated on sterile coverslips in 12‐well plates were allowed to attach overnight followed by transient transfection with SARS 8b‐EGFP fusion constructs and/or SARS 6‐pDsRed vectors by using Lipofectamine™ 2000 reagent (Invitrogen). Prior to transfection, the cells were washed with PBS and 500 μl of medium with no serum. 1.6 μg of the appropriate vector was diluted in 100 μl of medium with no serum and incubated for 5 min before mixing with 4 μl of Lipofectamine™ 2000. The complex was then added onto the cells after incubation at room temperature for 20 min. Maintenance medium was replaced 5.5 h after transfection.
Cells were washed with phosphate‐buffered saline (PBS) 30 h posttransfection. Coverslips were mounted and fluorescent images were captured at 2000× magnification with a laser scanning confocal microscope (Leica Model TCS‐NT, Leica Microsystems, Heidelberg, Germany). Green fluorescence (excitation wavelength: 476–488 nm; emission wavelength: 525/50 nm) and red fluorescence (excitation wavelength: 568 nm; emission wavelength: 600/30 nm) were detected through FITC and rhodamine filters, respectively.
2.5. RT‐PCR
Cells were transiently transfected with appropriate vectors and total RNA was extracted using an RNA extraction kit (Molecular Research Center, Inc., Cincinnati, OH, USA). cDNAs were synthesized with 8.5 μl of reaction mixture containing 5 μg of RNA, 0.5 μl of 10 mM dNTP, 4 μl of 5× reaction buffer, 2 μl of 100 μM oligo dT, 0.5 μl of RNAse inhibitor and 0.5 μl of M‐MuLV reverse transcriptase (Amersham Biosciences, Buckinghamshire, UK) at 37 °C for 1 h followed by 70 °C for 10 min. PCR amplification was performed with primer sets β‐actin‐F (5′ AGC GGG AAA TCG TGC GTG AC 3′) and β‐actin‐R (5′ GAC TCG TCA TAC TCC TGC TTG 3′), SARS 6‐F and SARS 6‐R [10] and SARS 8b‐F and SARS 8b‐R on the resulting cDNAs with an initial denaturation of 5 min at 95 °C followed by 30 cycles of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 90 s.
2.6. DNA synthesis analysis
Cells were plated in 24‐well plates (9 × 104 cells/well) and allowed to attach overnight. 0.8 μg of the appropriate vectors were transiently transfected into the cells using the Lipofectamine™ 2000 reagent (Invitrogen). Thirty‐six hours posttransfection, the cells were synchronized for 40 h in DMEM containing 0.1% FBS. Cells were then incubated in media containing 10% FBS and 1 μCi/ml [3H]‐thymidine (Amersham Biosciences) for 24 h at 37 °C. [3H]‐thymidine incorporation assay was performed as described elsewhere [10].
2.7. Statistical analysis
Data were expressed as means ± SD. Mean values were compared by Student's t‐tests or Kruskal–Wallis one‐way analysis of variance (ANOVA) on Ranks. All the data were analyzed with the SigmaStat 2.03 software (SPSS Inc., Chicago, IL, USA). A statistical significant difference was defined as p < 0.05.
3. Results and discussion
3.1. Expression of SARS 8b in Vero E6 and CHO cells
To express the SARS 8b‐EGFP fusion proteins, Vero E6 and CHO cells were transfected with SARS 8b‐pEGFP. The expression of the EGFP and SARS 8b‐EGFP proteins in both cell types were detected by Western blotting using anti‐GFP primary antibodies (Fig. 1 ). The expected bands of EGFP (∼27 kDa) and SARS8b‐EGFP (∼36.6 kDa) were observed, indicating the proteins were expressed in the cells. Interestingly, a smaller band with size of about 30 kDa was observed in both transfected Vero E6 and CHO cells (Fig. 1, lanes 2 and 5). This band was speculated to be a truncated form of the SARS 8b‐EGFP protein due to proteolytic cleavages, which are common processing steps of viral proteins. For instance, Ito et al. reported that the 3a protein of SARS‐CoV might undergo specific proteolytic processing [14]. Studies have also shown that the multiple cleavages of the precore protein of the hepatitis B virus resulted in different sizes of smaller proteins which could be translocated from the cytosol to other compartments of the cell [15]. SARS 8b may possibly employ similar mechanism as the 3a or the precore protein and the smaller band might be a processed form of the protein. However, further investigation is required to verify this hypothesis.
Figure 1.
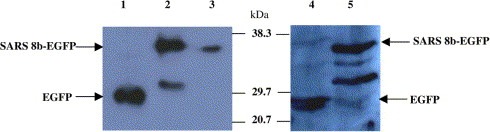
Expression of recombinant SARS 8b‐EGFP protein in Vero E6 and CHO cells. Total cell proteins were extracted and 50–150 μg of protein were resolved by 12% SDS–PAGE. Western blots of these proteins were probed with anti‐GFP antibodies. Lane 1: 50 μg of proteins from Vero E6 cells transfected with pEGFP vectors. Lane 2: 150 μg of proteins from Vero E6 cells transfected with SARS 8b‐EGFP vectors. Lane 3: 50 μg of proteins from Vero E6 cells transfected with SARS 8b‐EGFP vectors. Lane 4: 30 μg of proteins from CHO cells transfected with pEGFP vectors. Lane 5: 30 μg of proteins from CHO cells transfected with SARS 8b‐EGFP vectors.
3.2. Cellular localization of SARS 8b protein
EGFP fusion proteins have been widely used for studying localization and monitoring the expression of proteins [16, 17]. Hence, the SARS 8b‐EGFP fusion protein was used in this study for the cellular localization of SARS 8b analysis to circumvent the problem of the lack of SARS 8b antibodies. Results of confocal microscopy showed that the fusion SARS 8b‐EGFP possessed a similar fluorescent pattern as the EGFP control because signals for both proteins were observed in the cytosol and nuclei in Vero E6 and CHO cells (Fig. 2 A, B, D and E).
Figure 2.
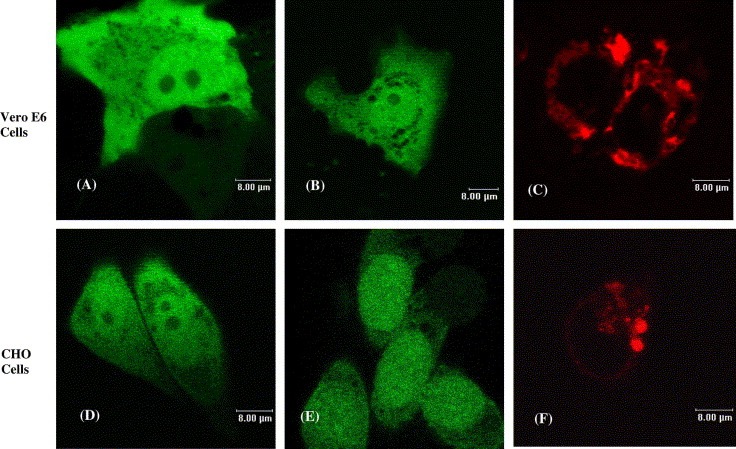
Subcellular localization of individually expressed recombinant SARS 8b‐EGFP and SARS 6‐RFP proteins. Images shown are Vero E6 cells transfected with vectors encoding (A) EGFP, (B) SARS 8b‐EGFP, and (C) SARS 6‐RFP; and CHO cells transfected with vectors encoding (D) EGFP, (E) SARS 8b‐EGFP, and (F) SARS 6‐RFP.
Recently, our group has reported that SARS 6 is localized at the endoplasmic reticulum (ER) [10]. Fig. 2C and F reconfirm such results with SARS 6‐RFP fusion protein expression. As SARS 8b was shown to be distributed throughout the cell, the possibility that the two viral accessory proteins interact and redistribute in the cell was investigated. Confocal microscopy results showed that the coexpression of these two proteins do not incur observable changes of localization relative to individually expressed SARS 6 or SARS 8b (Fig. 3 ). Such observation, however, does not rule out the possible interaction of the two proteins.
Figure 3.
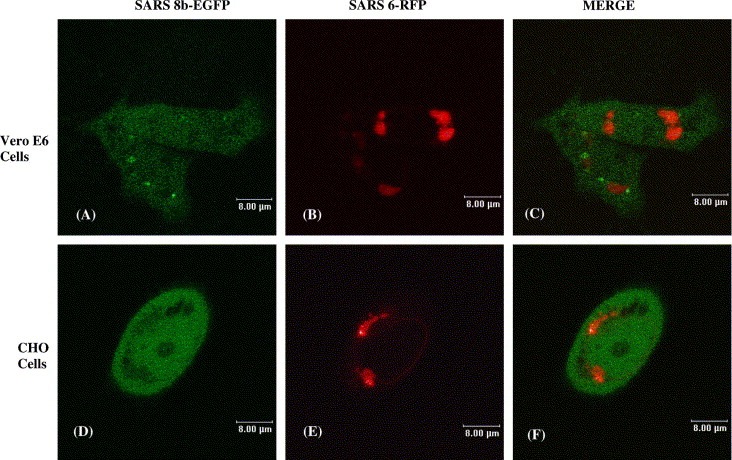
Subcellular localization of co‐expressed SARS 8b‐EGFP and SARS 6‐RFP proteins. Localization of cotransfected SARS 8b‐EGFP and SARS 6‐RFP in Vero E6 cells: (A) SARS 8b‐EGFP, (B) SARS 6‐RFP, (C) overlay image of (A) and (B). Localization of cotransfected SARS 8b‐EGFP and SARS 6‐RFP in CHO cells: (D) SARS 8b‐EGFP, (E) SARS 6‐RFP, (F) overlay image of (D) and (E).
3.3. Thymidine incorporation studies
In the present study, we examined the effect of SARS 6 and SARS 8b on DNA synthesis following transient transfection of SARS 6‐ and SARS 8b‐encoding vectors. Studies have shown that some viruses such as the murine leukemia virus induce host cell proliferation for productive infection [18]. Coronavirus infection in the early stages had been reported to stimulate epithelial cells, causing cellular proliferation and squamous metaplasia in the lungs [9]. SARS pathology of the lung has also been associated with diffuse alveolar damage in addition to epithelial cell proliferation [9, 19]. We have previously reported that SARS 6 affects cell‐proliferative function by inducing DNA synthesis in vitro [10]. We questioned if SARS 8b has a similar effect. RT‐PCR was used to confirm the expression of the mRNA of these untagged SARS‐CoV genes. Fig. 4 showed that mRNAs of untagged SARS 8b and SARS 6 proteins were expressed when transfected alone or together into the Vero E6 cells. Similar results were obtained in CHO cells (data not shown).
Figure 4.
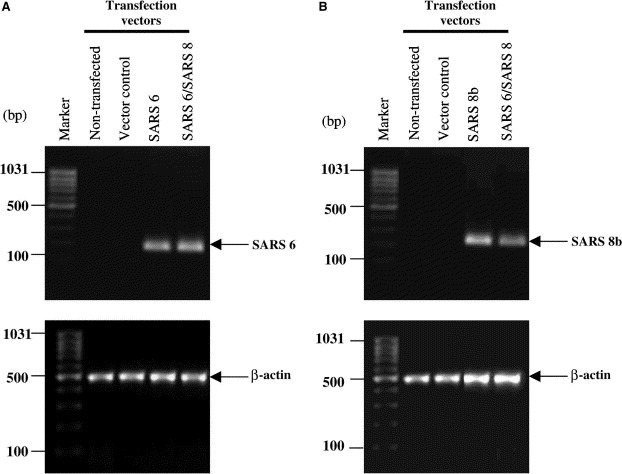
Expression of untagged SARS 6 and SARS 8b mRNAs in Vero E6 cells. RT‐PCR was performed with β‐actin, SARS 6, and/ or SARS 8b primers on mRNAs extracted from control cells and cells transiently transfected with SARS 6 and/or SARS 8b vectors. PCR products for SARS 6, SARS 8b, β‐actin were 192, 255, and 481 bp, respectively.
[3H]‐thymidine incorporation assays were performed to measure DNA synthesis in cells expressing the untagged SARS 8b. Results indicated that SARS 8b expression induced DNA synthesis in both cell lines, with an approximate of 18% and 36% increase of DNA synthesis in Vero E6 and CHO cells, respectively (Fig. 5 ). [3H]‐thymidine incorporation assays done on CHO cells expressing SARS 8b‐EGFP fusion protein also induced DNA synthesis (data not shown). These results further support the active expression of SARS 8b in the cells.
Figure 5.
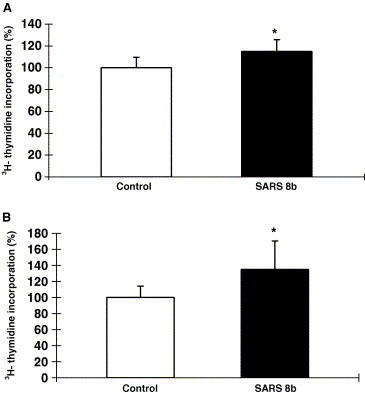
Stimulation of DNA synthesis by untagged SARS 8b expression. [3H]‐thymidine incorporation assays were performed on (A) Vero E6 cells and (B) CHO cells transiently transfected with vector control and SARS 8b vectors. Each bar represents the means ± S.D. of three experiments in four‐ to six‐replicate setup. ∗Significant difference between the control and SARS 8b‐expressing cells detected by Student's t test (p < 0.05).
Thymidine incorporation assays on cotransfected cells indicated that the co‐existence of untagged SARS 6 and SARS 8b proteins do not elicit additional or synergistic effects on DNA synthesis (Fig. 6 ). These results raise the possibility that SARS 6 and SARS 8b induce DNA synthesis via shared pathways. Although the ability of SARS 8b in cell proliferation may seem counterintuitive to the apoptotic characteristics of SARS 3a, 3b and 7a [6, 7, 8], past examples have shown that accessory proteins produced by a virus can be multifunctional and sometimes elicit counteracting effects. For instance, p12I, one of the accessory proteins of the human T‐cell lymphotropic virus type 1 (HTLV‐1) which localizes at the ER and cis‐Golgi compartments, is able to promote T‐lymphocytes proliferation and enhances the effectiveness of viral transmission during the early stage of infection [20, 21, 22]. Interestingly, HTLV‐1 p13II, which is another accessory protein of HTLV‐1 that localizes in the inner membranes of mitochondria, displays apoptotic effect on both animal model and human Jurkat T‐cells [23, 24]. Therefore, it is possible that SARS 8b work in a similar fashion as the HTLV‐1 p12I, causing cell proliferation that helps in viral transmission in the early stage of the infection. In fact, the cell proliferative effect of SARS 8b in epithelial cell lines used in this study is consistent to the pathology of epithelial proliferation observed in SARS patients [9].
Figure 6.
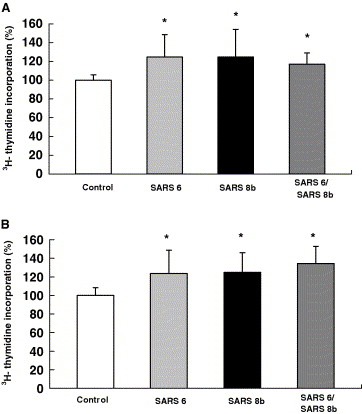
Stimulation of DNA synthesis by untagged SARS 6 and SARS 8b expression. [3H]‐thymidine incorporation assays were performed on (A) Vero E6 cells and (B) CHO cells transiently transfected with vector controls, SARS 6 and/or SARS 8b, respectively. Each bar represents the means ± S.D. of three to five experiments in four‐ to six‐replicate setup. ∗Significant differences among the controls and SARS proteins‐expressing cells detected by Kruskal–Wallis ANOVA on Ranks (p < 0.05).
Although viral accessory proteins are often not involved in viral replication, their roles in viral pathogenesis should not be overlooked. For example, the HIV accessory protein Vpr is essential for inducing cell cycle arrest and apoptosis [25]. Studies have also shown that SARS 3a, 3b and 7a could cause apoptosis in mammalian cells [6, 7, 8]. In this study, we examined the effects of SARS 8b on DNA synthesis in the context of mammalian cell lines as a prelude. The possibility that in the context of the complete SARS‐CoV genome, the function of the SARS 8b may not be related to an increase in DNA synthesis cannot not be excluded. For instance, a combination of SARS‐CoV 8b and some other accessory proteins may function to shut off host macromolecular synthesis. Further analyses will be required to verify the significance of the cell proliferative function of SARS 8b in the context of SARS‐CoV infection and the whole viral genome. Nevertheless, consistent with the previous report of the presence of SARS 6 and SARS 8b mRNA associated with SARS‐CoV infection [26], the results in this study further substantiate the compatible co‐expression of the two viral proteins in the cells. The distribution of SARS 8b throughout the cell suggests its potential to interact with multiple cellular partners as well as the products of the SARS‐CoV genome located both in the cytoplasm and nucleus which may in turn modify cellular function, i.e. DNA synthesis.
Acknowledgements
This research was supported in part by the Research Fund for the Control of Infectious Disease, Health, Welfare and Food Bureau, Hong Kong (Ref. No. 01030592); the RGC Special SARS Grant (Ref. No. CUHK 4536/03M); Direct Grant, Faculty of Medicine, The Chinese University of Hong Kong, and the Croucher Foundation.
Law Pui Ying Peggy,Liu Yuet-Man,Geng Hua,Kwan Ka Ho,Waye Mary Miu-Yee and Ho Yuan-Yuan(2006), Expression and functional characterization of the putative protein 8b of the severe acute respiratory syndrome-associated coronavirus, FEBS Letters, 580, doi: 10.1016/j.febslet.2006.05.051
References
- 1. World Health Organization (WHO). http://www.who.int/csr/sars/en/.
- 2. Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science, 300, (2003), 1394– 1399. [DOI] [PubMed] [Google Scholar]
- 3. Marra M.A., Jones S.J.M., Astell C.R., The genome sequence of the SARS-associated coronavirus. Science, 300, (2003), 1399– 1404. [DOI] [PubMed] [Google Scholar]
- 4. Alcami A., Koszinowski J.H., Viral mechanisms of immune evasion. Immunol. Today, 21, (2000), 445– 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Haan C.A.M., Masters P.S., Shen X., Weiss S., Rottier P.J.M., The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology, 296, (2002), 177– 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K., To K.F., Lo A.W.I., Chan J.Y.W., Suen Y.K., Chan H.Y., Fung K.P., Waye M.M.Y., Sung J.J.Y., Lo Y.M., Tsui S.K.W., The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol., 86, (2005), 1921– 1930. [DOI] [PubMed] [Google Scholar]
- 7. Yuan X., Shan Y., Zhao Z., Chen J., Cong Y., G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J., 2, (2005), 66– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan Y.J., Burtram C.F., Goh P.Y., Shen S., Tan T.H.P., Lim S.G., Hong W., Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol., 78, (2005), 14043– 14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nicholls J.M., Poon L.L.M., Lee K.C., Lung pathology of fatal severe acute respiratory syndrome. Lancet, 361, (2003), 1773– 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geng H., Liu Y.M., Chan W.S., Lo A.W.I., Au D.M.Y., Waye M.M.Y., Ho Y.Y., The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett., 579, (2005), 6763– 6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guan Y., Zheng B.J., He Y.Q., Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science, 302, (2003), 276– 278. [DOI] [PubMed] [Google Scholar]
- 12. Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol., 79, (2005), 11892– 11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsui S.K.W., Chim S.S.C., Lo Y.M.D., Coronavirus genomic-sequence variations and the epidemiology of the severe acute respiratory syndrome. N. Engl. J. Med., 349, (2003), 187– 188. [DOI] [PubMed] [Google Scholar]
- 14. Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang Ch., Inoue T., Peters C.J., Makino S., Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol., 79, (2005), 3182– 3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang S.Q., Walter M., Standring D.N., Hepatitis B virus p25 precore protein accumulates in Xenopus oocytes as an untranslocated phosphoprotein with an uncleaved signal peptide. J. Virol., 66, (1992), 37– 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gubin A.N., Reddy B., Njoroge J.M., Miller J.L., Long-term, stable expression of green fluorescent protein in mammalian cells. Biochem. Biophys. Res. Commun., 236, (1997), 347– 350. [DOI] [PubMed] [Google Scholar]
- 17. Degreve B., Johansson M., De Clercq E., Karlsson A., Balzarini J., Differential intracellular compartmentalization of herpetic thymidine kinaeses (TKs) in TK gene-transfected tumor cells: molecular characterization of the nuclear localization signal of herpes simplex virus type 1 TK. J. Virol., 72, (1998), 9535– 9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamashita M., Emerman M., Retroviral infection of non-dividing cells: old and new perspectives. Virology, 344, (2006), 88– 93. [DOI] [PubMed] [Google Scholar]
- 19. Weiss S.R., Navas-Martin S., Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev., 69, (2005), 635– 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding W., Albrecht B., Kelley R.E., Muthusamy N., Kim S.J., Altschuld R.A., Laimore M., Human T-Cell lymphotropic virus type 1 p12I expression increases cytoplasmic calcium to enhance the activation of nuclear factor of activated T cells. J. Virol., 76, (2002), 10374– 10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ding W., Kim S.J., Nair A.M., Michael B., Boris-Lawrie K., Tripp A., Feuer G., Lairmore M.D., Human T-cell lymphotropic virus type 1 p12I enhances interleukin-2 production during T-cell activation. J. Virol., 20, (2003), 11027– 11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Albrecht B., Lairmore M.D., Critical role of human T-lymphotrophic virus type 1 accessory proteins in viral replication and pathogenesis. MMBR, 66, (2002), 396– 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hiraragi H., Michael B., Nair A., Silic-Benussi M., Ciminale V., Lairmore M.D., Human T-cell lymphotropic virus type 1 mitochondrion-localizing protein p13II sensitizes Jurkat T cells to Ras-mediated apoptosis. J. Virol., 79, (2005), 9449– 9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silic-Benussi M., Cavallari I., Zorzan T., Ciminale V., Suppression of tumor growth and cell proliferation by p13II, a mitochondrial protein of human T cell leukemia virus type 1. Proc. Natl. Acad. Sci. USA, 101, (2004), 6629– 6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mahalingam S., Ayyavoo V., Patel M., Kieber-emmons T., Weiner D.B., Nuclear import, virion incorporation, and cell cycle arrest/ differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J. Virol., 71, (1997), 6339– 6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziehuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E., Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol., 331, (2003), 991– 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]


