Abstract
Background
The potential role of respiratory viruses in the natural history of community-acquired pneumonia (CAP) in adults has not been well described since the advent of nucleic amplification tests (NATs).
Methods
From 2004 to 2006, adults with CAP who were admitted to five hospitals were prospectively enrolled in the study, and clinical data, cultures, serology, and nasopharyngeal swabs were obtained. NATs from swabs were tested for influenza, human metapneumovirus (hMPV), respiratory syncytial virus (RSV), rhinovirus, parainfluenza virus 1–4, coronaviruses (OC43, 229E, and NL63), and adenovirus.
Results
A total of 193 patients were included; the median age was 71 years, 51% of patients were male, and 47% of patients had severe CAP. Overall, 75 patients (39%) had a pathogen identified. Of these pathogens, 29 were viruses (15%), 38 were bacteria (20%), 8 were mixed (4%), and the rest were “unknown.” Influenza (n = 7), hMPV (n = 7), and RSV (n = 5) accounted for most viral infections; other infections included rhinovirus (n = 4), parainfluenza (n = 3), coronavirus (n = 4), and adenovirus (n = 2). Streptococcus pneumoniae was the most common bacterial infection (37%). Compared with bacterial infection, patients with viral infection were older (76 vs 64 years, respectively; p = 0.01), were more likely to have cardiac disease (66% vs 32%, respectively; p = 0.006), and were more frail (eg, 48% with limited ambulation vs 21% of bacterial infections; p = 0.02). There were few clinically meaningful differences in presentation and no differences in outcomes according to the presence or absence of viral infection.
Conclusions
Viral infections are common in adults with pneumonia. Easily transmissible viruses such as influenza, hMPV, and RSV were the most common, raising concerns about infection control. Routine testing for respiratory viruses may be warranted for adults who have been hospitalized with pneumonia.
Key words: community-acquired pneumonia, respiratory viruses
Abbreviations
- CAP
community-acquired pneumonia
- DFA
direct fluorescent antigen test
- hMPV
human metapneumovirus
- IQR
interquartile range
- NAT
nucleic acid amplification test
- NP
nasopharyngeal swab
- PSI
pneumonia severity index
- RSV
respiratory syncytial virus
Community-acquired pneumonia (CAP) is one of the most clinically important diseases in adults, affecting 5 to 20 per 1,000 adults per year.1 Of these, at least 20 to 40% will require hospitalization for the treatment of their pneumonia.2 CAP management guidelines3 have been influenced by older CAP etiology studies,4 which helped to direct empiric therapeutic antimicrobial choices for therapy against bacterial pathogens such as Streptococcus pneumoniae, Haemophilus influenzae, and “atypical” bacteria, including Chlamydophila pneumoniae, Mycoplasma pneumoniae, and Legionella pneumophila. Although CAP guidelines3 acknowledge respiratory viruses as a “cause” of pneumonia, few recommendations are made regarding management, largely due to the paucity of data regarding prevalence, clinical presentation, and outcomes. Furthermore, viral etiology studies in pneumonia are difficult to interpret as noninvasive viral detection methods are often considered to be only markers of infection rather than the cause of pneumonia.5 Clearly, much better knowledge of the potential role of respiratory viruses present in patients with pneumonia is needed.
Most published studies6, 7 of respiratory viruses have relied on tests with relatively poor sensitivity such as serology and direct fluorescent antigen (DFA) tests. Such tests are limited in the sample type to which they can be applied and are not suitable for a broad range of respiratory viruses. More recently, the introduction of highly sensitive nucleic acid amplification tests (NATs) has dramatically improved our ability to detect multiple viral pathogens such as influenza, respiratory syncytial virus (RSV), rhinovirus, parainfluenza, and adenovirus. Such tests can be undertaken using a small single sample of respiratory secretions with results available with rapid turnaround times.7, 8, 9, 10, 11, 12 In addition, these tests have allowed us to detect emerging respiratory viruses such as human metapneumovirus (hMPV) and coronaviruses, viruses that are difficult to grow in cell culture.13, 14, 15 To date, there have been few studies5, 7, 9, 10, 11, 16, 17 reported in patients with pneumonia using NATs to detect viral infection, and these studies have either not included clinical data7, 9, 11 or have not tested for all potentially important respiratory viruses in a comprehensive manner.10, 17
Better knowledge of the role of infection with respiratory viruses in adults with pneumonia may lead to better management. Thus, we performed a prospective study in consecutive adults who had been admitted to the hospital with CAP, and sought to describe their pathogens, clinical presentation, and outcomes.
Materials and Methods
From January 2004 to January 2006, consecutive adults (≥ 18 years of age) who had been admitted to five hospitals in Edmonton, AB, Canada, with CAP were enrolled in a prospective study of pneumonia. Patients were excluded from the study if they had received antibiotics or been hospitalized within the prior 2 weeks, were unable or unwilling to provide informed consent, or had the following conditions: immunocompromised (ie, had received > 10 mg of prednisone per day for > 1 month, other immunosuppressives, had cancer with recent chemotherapy, or had HIV with a CD4 count of < 250 cells/μL); tuberculosis; bronchiectasis; cystic fibrosis; or pregnancy. All patients gave written informed consent, and the Health Research Ethics Board of the University of Alberta approved the study. We did not record data on patients who were unable to provide consent or who did not meet the enrollment criteria.
Data Collection
Pneumonia was defined as an acute lower respiratory tract illness with two or more of the following symptoms or signs: cough; productive cough; fever; chills; dyspnea; pleuritic chest pain; crackles; and bronchial breathing plus an opacity or infiltrate seen on a chest radiograph that was interpreted as pneumonia by the treating physician. To characterize the severity of the pneumonia itself, we calculated the pneumonia severity index (PSI) using the methods of Fine et al.18
Clinical, radiographic, and laboratory data and short-term outcomes were collected by a trained research nurse; the nurse was masked to microbiology results at the time of data collection. Patients were followed up throughout their hospital stay until discharge.
Diagnostic Tests Undertaken
Routine blood culture, sputum specimens, nasopharyngeal (NP) swabs, and serum samples were processed for each patient according to the study protocol. NP swabs that were submitted for the detection of viral pathogens first underwent DFA testing for influenza A and B, RSV, and parainfluenza virus 1–3 (Imagen; Dakocytomation Ltd; Ely, UK). In addition, expanded testing of NP samples was undertaken for a range of respiratory pathogens by NATs using extraction and amplification methods that have been described previously.11 Briefly, NATs were designed to amplify and detect influenza A and B, hMPV, RSV, rhinovirus, parainfluenza 1–4, coronaviruses (OC43, 229E, and NL63), and adenoviruses. All the NATs utilized in this study have been published, and the assay parameters evaluated.11, 12, 19, 20 Laboratory validation of these assays confirmed a limit of detection of ≤ 100 copies (cloned target or synthetic RNA) or one or fewer tissue culture infectious dose of 50% (for culturable viruses). The specificity of all assays was confirmed using samples and spiked materials containing high loads of alternative respiratory pathogens. (Further details on viral NATs are available from J.D.F. on request [also see references 11, 12, 19, 20].)
Bacterial infections were identified using standard laboratory protocols. Acute and convalescent serum samples were collected on the day of hospital admission and were repeated 4 to 6 weeks later. Serum samples were tested for the presence of C pneumoniae and Chlamydia psittaci IgM and IgG by a microimmunofluorescence assay,21 M pneumoniae IgM enzyme immunoassay (Platelia; BioRad; Hercules, CA),22 Coxiella burnetii phase I and phase II titers by indirect immunofluorescence,23 and L pneumophila titers by indirect immunofluorescence.24 M pneumoniae and L pneumophila were also tested using NATs, and were validated as above but with a limit of detection of ≤ 100 copies (cloned target or synthetic RNA) or ≤ 1 cfu.11
Criteria to Establish Presence of Respiratory Pathogens
A diagnosis of respiratory viral infection was made if a virus was detected by NAT or DFA and a coexisting bacterial pathogen was not identified. A diagnosis of bacterial infection was made if a viral pathogen was not detected, and the following criteria were met: (1) isolation of a respiratory pathogen from purulent sputum (defined as an adequate quality sputum sample with > 25 leukocytes and < 10 epithelial cells per × 100 magnification field) or blood culture1, 25; (2) a fourfold rise in IgG titers for C pneumoniae (> 1:32) and C psittaci (> 1:32)1; (3) a single increased IgM titer for M pneumoniae (> 1:64) or C pneumoniae (> 1:16)1; (4) an antibody titer of > 1:1,024 to L pneumophila in a serum specimen obtained during either the acute or convalescent phase1; (5) a fourfold rise in antibody titer to > 1:128 or a fourfold rise in antibodies to C burnetii 1; (6) a single titer of > 1:128 to a phase II C burnetii antigen1; or (7) the detection of M pneumoniae or L pneumophila by NAT.11 A mixed infection was defined as the presence of both respiratory virus and bacteria, as defined above. Last, if no pathogens were detected, based on the tests used in the study protocol, we classified this as “unknown.”
Statistical Analysis
Patient characteristics and outcomes according to pathogen were compared using χ2 test, Fisher exact test, Student t test, or Mann-Whitney U test, as appropriate. Although we present data for all pathogen categories, our primary analyses compare viral infection to bacterial infection. The few viral cases (n = 29) in our sample precluded attempts at multivariable analyses. All data were analyzed using a statistical software package (SPSS, version 15.0; SPSS Inc; Chicago, IL).
Results
Patient Characteristics
Three hundred patients were enrolled into the study, and 193 patients (64%) had evaluable NP swabs. The reasons for nonevaluable NP specimens included insufficient sample (n = 68) and missed collection (n = 39). Because the primary purpose of the study was to evaluate for the presence of viral pathogens, we excluded those patients without NP swabs. There were essentially no differences between those with evaluable NP swabs and those without for either clinical characteristics or outcomes, with the following exceptions: impaired functional status (35% vs 21%, respectively; p = 0.02); lobar pneumonia seen on a chest radiograph (72% vs 57%, respectively; p = 0.009); and median length of stay (7 vs 6 days, respectively; p = 0.02).
We considered the 193 patients with evaluable NP swabs to be our final study sample. Sputum and blood cultures were requested for all patients, but these were not performed in some patients due to their inability to produce a sputum specimen (n = 106), their refusal of a blood draw (n = 61), or death (n = 5). Convalescent serum samples were not obtained in 153 patients because they did not return for follow-up blood work (n = 146) or died (n = 7). Overall, the median age of patients was 71 years (interquartile range [IQR], 58 to 80 years), 51% were male, and 47% had severe CAP (PSI class 4 or 5).
Respiratory Pathogens
In total, 75 patients (39%) had a respiratory pathogen identified, whereas pathogens were classified as “unknown” in 118 patients (61%). Of those patients with a pathogen identified, 29 (39%) had a viral infection, 38 (51%) had a bacterial infection, and 8 (11%) had a mixed viral and bacterial infection.
Of the 29 patients with a viral infection, the most common organisms were influenza A (n = 3), influenza B (n = 4), hMPV (n = 7), and RSV (n = 5). Other organisms included coronavirus (n = 4), rhinovirus (n = 4), parainfluenza (n = 3), and adenovirus (n = 2). Three patients had two viruses detected. Of 29 patients with viral infections, 18 (62%) had influenza, hMPV, or RSV as a cause (Table 1 ). Of the 38 patients with bacterial infection, 14 infections (37%) were caused by S pneumoniae and 9 infections (24%) were caused by common atypical pathogens (Table 1).
Table 1.
Distribution of Viral and Bacterial Respiratory Pathogens
| Pathogens | No. |
|---|---|
| Viral pathogens* (n = 29) | |
| Influenza A | 3 |
| Influenza B | 4 |
| hMPV | 7 |
| RSV | 5 |
| Parainfluenza 1–4 | 3 |
| Rhinovirus | 4 |
| Coronavirus OC43 | 4 |
| Coronavirus 229E | 0 |
| Coronavirus NL63 | 0 |
| Adenovirus | 2 |
| Bacterial pathogens† (n = 38) | |
| Typical pathogens | |
| S pneumoniae | 14 |
| Aerobic Gram-negative bacilli | 6 |
| Moraxella spp | 3 |
| Streptococcal spp | 2 |
| Staphylococcus aureus | 2 |
| Haemophilus spp | 2 |
| Anaerobes | 2 |
| Atypical pathogens | |
| M pneumoniae | 4 |
| C pneumoniae | 4 |
| L pneumophila | 1 |
| C psittaci | 0 |
| C burnetii | 0 |
Three patients had two viruses isolated: parainfluenza plus hMPV; influenza plus RSV; and influenza plus adenovirus.
Two patients had two different bacteria isolated: M pneumoniae and C pneumoniae; and Moraxella spp and aerobic Gram-negative bacilli.
Clinical Presentation of Pneumonia in Patients With Viral Infection
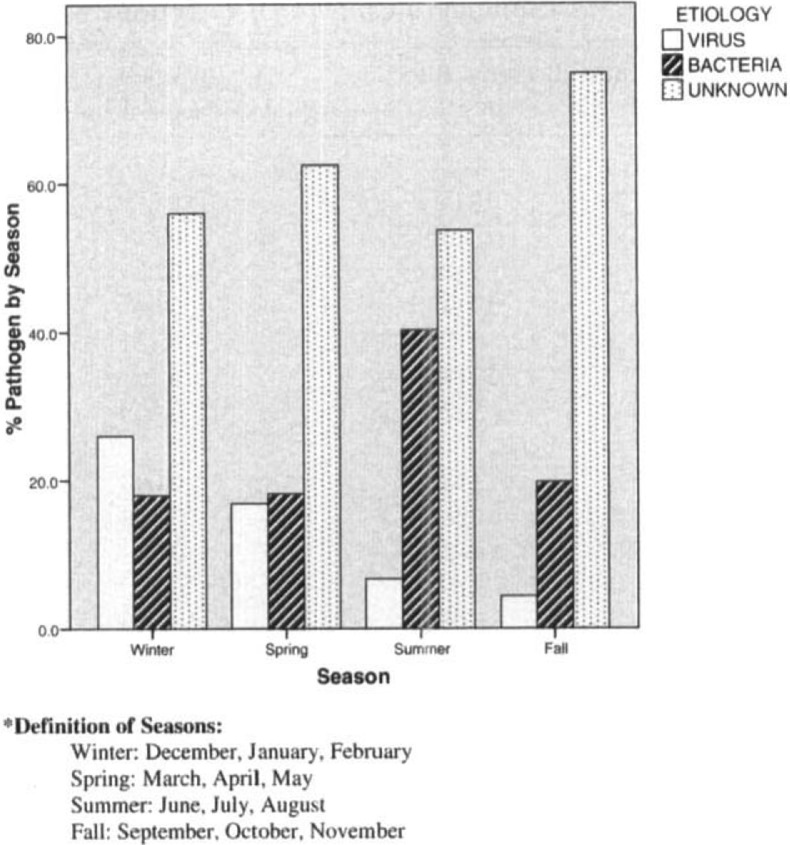
Patients with viral infections were older than those without viral infections (median age, 76 vs 64 years, respectively; p = 0.01), were more likely to have underlying cardiac disease (66% vs 32%, respectively; p = 0.006), and tended to be more frail (eg, 48% had severely limited ambulation vs 21% of those with bacterial pneumonia; p = 0.02). Other differences included the presence of chest pain, which was far less common in those patients with a viral infection than in those with a bacterial infection (7% vs 37%, respectively; p = 0.004) [Table 2 ]. In terms of laboratory findings, those with viral infections were far more likely to have a normal leukocyte count than those without viral infection (74% vs 14% leukocytes, respectively; p < 0.001). All cases of viral infection occurred between the months of October and May, with one exception (one episode of rhinovirus infection occurred in July), whereas bacterial infections occurred year round (Fig 1 ).
Table 2.
Characteristics of Patients With Viral, Bacterial, Mixed, and Unknown Respiratory Infections*
| Characteristics | Unknown (n = 118) | Mixed Infection† (n = 8) | Viral Infection (n = 29) | Bacterial Infection (n = 38) | p Value (Viral vs Bacterial Infection) |
|---|---|---|---|---|---|
| Sociodemographics | |||||
| Age ≥ 65 yr | 79 (67) | 4 (50) | 19 (65) | 19 (50) | 0.20 |
| Male sex | 57 (48) | 3 (38) | 18 (62) | 19 (50) | 0.32 |
| Nursing home | 1 (1) | 0 (0) | 0 (0) | 1 (3) | 0.38 |
| Current smoker | 27 (23) | 5 (63) | 6 (21) | 17 (45) | 0.04 |
| Alcohol use | 4 (3) | 0 (0) | 1 (3) | 2 (5) | 0.72 |
| Influenza vaccination‡ | 67 (57) | 4 (50) | 18 (62) | 17 (45) | 0.16 |
| Pneumococcal vaccination§ | 43 (36) | 2 (25) | 15 (52) | 8 (21) | 0.009 |
| Comorbidities | |||||
| Cardiovascular disease | 45 (38) | 2 (25) | 19 (66) | 12 (32) | 0.006 |
| Respiratory disease | 46 (39) | 4 (50) | 16 (55) | 12 (32) | 0.052 |
| Neurologic disease | 28 (24) | 1 (13) | 6 (21) | 10 (26) | 0.59 |
| Diabetes requiring insulin | 11 (9) | 1 (13) | 1 (3) | 2 (5) | 0.72 |
| Impaired functional status‖ | 43 (36) | 1 (13) | 14 (48) | 8 (21) | 0.019 |
| Symptoms and signs | |||||
| Altered mental status | 8 (7) | 1 (13) | 0 (0) | 3 (8) | 0.12 |
| Cough | 100 (85) | 7 (88) | 26 (90) | 32 (84) | 0.52 |
| Sputum | 75 (64) | 7 (88) | 20 (69) | 22 (58) | 0.35 |
| Dyspnea | 99 (84) | 5 (63) | 28 (97) | 35 (92) | 0.45 |
| Chest pain | 26 (22) | 1 (13) | 2 (7) | 14 (37) | 0.004 |
| Respiratory rate ≥ 30 breaths/min | 20 (17) | 2 (25) | 5 (17) | 9 (24) | 0.48 |
| Abnormal temperature (>38.5 or < 35|SDC) | 8 (7) | 2 (25) | 5 (17) | 10 (26) | 0.38 |
| Hypoxia (oxygen saturation < 92%) | 93 (79) | 6 (75) | 27 (93) | 31 (82) | 0.17 |
| Investigations | |||||
| Abnormal WBC (>12 or < 4 cells/mL) | 69 (59) | 4 (50) | 4 (14) | 28 (74) | <0.001 |
| Lobar infiltrate on chest radiograph | 87 (74) | 3 (38) | 18 (62) | 32 (84) | 0.04 |
| Pleural effusion on chest radiograph | 28 (24) | 0 (0) | 5 (17) | 10 (26) | 0.38 |
| Pneumonia severity | |||||
| PSI class IV or V | 64 (54) | 4 (50) | 16 (55) | 18 (47) | 0.53 |
Values are given as No. (%), unless otherwise indicated.
Mixed cases included the following pathogens:C pneumoniae plus hMPV (two cases);C pneumoniae plus RSV; H influenza plus influenza; L pneumophila plus H influenza plus influenza; S pneumoniae plus RSV; aerobic Gram-negative bacilli plus S pneumoniae plus rhinovirus; and S aureus plus coronavirus.
Influenza vaccination within the current influenza season.
Pneumococcal polysaccharide vaccination within the previous 5 years.
Requiring walking aid or wheelchair, or bed bound.
Figure 1.
Seasonal distribution of pneumonia, arranged by pathogen.
Outcomes According to Pathogen
There were no significant differences in outcomes according to the pathogens identified. Specifically, there were no differences in median length of hospital stay (patients with viral infection, 7 days [IQR, 6 to 10 days]; patients with bacterial infection, 8 days [IQR, 6 to 18 days]; p = 0.37), ICU admission (no patients in either group went to the ICU), or mortality rate (patients with viral infection, 3%; patients with bacterial infection, 3%; p = 0.85).
Discussion
Although the importance of S pneumoniae and atypical bacterial pathogens is well understood in patients with CAP, in this prospective cohort study we have now demonstrated the significant potential contribution of respiratory viruses in patients presenting with pneumonia. Indeed, fully one sixth of all cases (15%) in this cohort of adults who were hospitalized with pneumonia had a respiratory virus identified; alternatively, more than one third of those patients (39%) with a pathogen identified had a respiratory viral infection. Influenza, hMPV, and RSV comprised almost two thirds of all cases of viral infection and, in our study, were acquired during the influenza season. There were some differences in presentation between those patients with a viral infection and those without, including the following: older age; presence of cardiac disease (but in the near absence of chest pain on presentation); and greater frailty. Of note, patients with a viral infection were far more likely than patients with bacterial pneumonia to have a normal leukocyte count. Despite these apparent differences, given that the majority of our cohort never had a respiratory pathogen identified, it is obviously very difficult to distinguish the presence or absence of viral infection in patients with pneumonia. This is further borne out by the fact that outcomes were virtually identical irrespective of the pathogens involved.
Adult CAP etiology studies26 conducted prior to the use of NATs estimated viral involvement in 0.3 to 30% of all CAP cases. Testing was generally based on serologic conversion or positive DFA test results for influenza A or B; RSV; parainfluenza virus 1, 2, and 3; or adenovirus. In our cohort, viral infection without evidence of bacterial coinfection was detected 15% of the time, which falls within the commonly reported range.26 A study by Marcos et al,10 which used NATs, reported a similar prevalence of viral infection in Spain. However, the study by Marcos et al10 differed from ours in several noteworthy ways, as follows: they did not test for hMPV; and they included immunocompromised patients in their study. Our results also differ from those of Jennings et al,5 as follows: they documented a viral infection 29% of the time in their cohort of adults with CAP, but, surprisingly, more than a third of infections were attributed to rhinovirus, and almost one fifth were mixed infections. The impact of rhinoviruses in our study may be underestimated as data27 have indicated that the picornavirus family of viruses is much more variable than originally thought. It is extremely difficult to design and validate assays to pick up all divergent rhinoviruses, and the original assay design that we utilized in this study would not identify all those that have been reported.27 This is an inherent limitation in the type of study undertaken; as we identify more novel respiratory pathogens and variants, it is inevitable that some will have been missed.
Most older etiology studies28 have reported influenza infection in patients with pneumonia 4 to 19% of the time, followed by RSV. Influenza was the most common virus identified in our study, affecting 4% of patients; however, we found hMPV to occur as commonly as influenza, and more frequently than RSV. This important finding has not been widely documented as most respiratory virus studies7, 10, 29, 30 have not included testing for hMPV, largely due to the difficulty in its identification in the past. To our knowledge, only two previous etiology studies5, 17 used NATs for detecting hMPV. One study,17 which was restricted to COPD patients with pneumonia, found hMPV as a pathogen in 4.1% of cases; another study5 from New Zealand found no cases of hMPV.
Strengths and Limitations
The strengths of this study include its prospective nature and the thorough collection of data from a cohort of consecutive patients who had been admitted to the hospital with CAP. There are also several limitations to the study. First and foremost, despite our best efforts and a detailed study protocol, a number of bacterial investigations (ie, blood culture, sputum culture, and convalescent serum specimens) were missed, thereby potentially underestimating the number of cases of bacterial pneumonia and (potentially) underestimating the number of mixed infections. The number of missed bacterial investigations may be the reason for our 61% rate of unknown infections, although our rate of recovery is similar to other studies22, 31, 32, 33, 34 that have reported 47 to 60% unknown infections. Second, we excluded patients without evaluable NP swabs from our analyses. Not obtaining specimens for conducting a NAT was a study protocol violation in 13% of patients (39 of 300 patients). We speculate that either NP swabs were not collected when patients transitioned from the emergency department to the wards, or that there was a miscommunication with the reference laboratory regarding when or where to send the study-related swabs. That said, there were few important clinical differences between patients with and without evaluable NP swabs, with two exceptions. Those patients without evaluable NP swabs were more likely to have lobar pneumonia, which, according to our data, would bias the results toward bacterial infection. Those patients without evaluable NP swabs were also more likely to be functionally impaired, which would bias the results toward viral infection. Third, there is potential for both false-positive and false-negative NP results, although testing with NATs has been reported to have excellent sensitivity and specificity.12 As noted above, sequence divergence for the rhinoviruses (and, potentially for the other virus groups) may also have led to some underestimation of the number of viral infections. Fourth, we detected viruses in the upper respiratory tract using NP specimens, which does not necessarily equate with the causation of pneumonia. However, the purpose of this study was to describe the potential role of respiratory viral infection in those patients with pneumonia; a study describing “confirmed” viral pneumonia would require lung tissue samples from all enrolled patients. Last, our overall sample size might be considered small by some, and our cohort was drawn from only one health region in Canada, which might limit the generalizability of the results to some degree.
Clinical Implications
In our study, patients with pneumonia and respiratory viral infection were older and more frail than those without evidence of viral infection. Differentiating between patients with viral infection and those without based on clinical findings and routine laboratory test results remains a challenge. Indeed, although we were unable to perform a multivariable logistic regression analysis due to the small sample size, it seems unlikely that any constellation of symptoms, signs, and routine laboratory findings will ever reliably differentiate between the presence or absence of a virus.3, 10, 29, 30 Current guidelines3 recommend empiric antibiotic therapy targeted against common bacterial pathogens for patients who are admitted to the hospital with pneumonia. How to manage patients with pneumonia and a respiratory viral infection, without a documented coexisting bacterial pathogen, is far less clear. Future research, similar to that found in the pediatrics literature, is needed to help answer whether empiric therapy with antibiotics can be discontinued in this clinical scenario.35
Perhaps most importantly, the inability to identify patients with a respiratory virus without comprehensive respiratory viral testing is a concern from the perspective of infection control. The presence of a respiratory viral infection can result in nosocomial outbreaks. For instance, outbreaks due to influenza have been well documented,28 and cases of RSV and hMPV nosocomial transmission are increasingly recognized.28, 36, 37 The nosocomial spread of respiratory viruses among adults poses the biggest threat to immunocompromised patients, including frail elderly patients.3, 38, 39, 40 The current infection control guidelines recommend placing patients with a suspected respiratory viral infection in private rooms or cohorting them with patients with the same viral infection as a way to prevent transmission.41 Given the 15% prevalence of viral infection in adults in our study, and the indistinguishable presentation from typical bacterial pneumonia, our results suggest routine isolation (with droplet and contact precautions) of all adults with pneumonia, from the time of hospital admission until respiratory viral infection is ruled out, should be considered to help prevent the nosocomial transmission of respiratory viruses. This suggested approach should become logistically feasible when the turnaround time for NAT results is < 24 h and as the price of testing with NATs decreases over time. This will be facilitated by emerging commercial viral identification assays that are both accurate and relatively inexpensive.42
Conclusion
Infections with respiratory viruses are common in patients who are hospitalized with pneumonia, comprising 39% of all identified pathogens and 15% of all patients in our study. Influenza, hMPV, and RSV were the most common respiratory viruses identified. In patients presenting with pneumonia, it remains difficult to differentiate patients with viral infection from those without viral infection. Our findings suggest that routine testing for common respiratory viruses may be warranted for all adults hospitalized with pneumonia.
Acknowledgment
Thanks to Carol Mangan, our research nurse, for her invaluable contribution, and to the Provincial Laboratory for their comprehensive laboratory help. Thanks also go to Sipi Garg and Meagan Rosenthal for help with preparing the database for analyses.
Footnotes
Dr. Majumdar was supported by the Alberta Heritage Foundation for Medical Research (Health Scholar) and the Canadian Institutes of Health Research (New Investigator). This project was funded in part by an establishment grant (to Dr. Marrie) from the Alberta Heritage Foundation for Medical Research.
Funding sources had no role in study design, data collection, data analysis or interpretation, or writing of the report. All authors participated in the study conception, design, analysis, interpretation of results, and revision of the manuscript, and approved the final version of the manuscript. Dr. Johnstone drafted the initial manuscript. Dr. Marrie acquired the data, obtained funding for the study, and will act as guarantor.
The authors have reported to the ACCP that no significant conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians (www.chestjournal.org/misc/reprints.shtml).
References
- 1.Marrie TJ. Community-acquired pneumonia: clinical features and outcomes. In: Marrie TJ, editor. Community acquired pneumonia. 1st ed. Kluwer Academic/Plenum Publishers; New York, NY: 2001. pp. 29–34. [Google Scholar]
- 2.Macfarlane J, Boswell T, Douglas G. BTS guidelines for the management of community acquired pneumonia in adults. Thorax. 2001;56(suppl):iv1–iv64. doi: 10.1136/thorax.56.suppl_4.iv1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandell LA, Wunderink RG, Anzueto A. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44:S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howard LS, Sillis M, Pasteur MC. Microbiological profile of community-acquired pneumonia in adults over the last 20 years. J Infect. 2005;50:107–113. doi: 10.1016/j.jinf.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Jennings LC, Anderson TP, Beynon KA. Incidence and characteristics of viral community acquired pneumonia in adults. Thorax. 2008;63:42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 6.Casiano-Colon AE, Hulbert BB, Mayer TK. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J Clin Virol. 2003;28:169–174. doi: 10.1016/s1386-6532(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 7.Templeton KE, Scheltinga SA, van den Eeden WC. Improved diagnosis of the etiology of community acquired pneumonia with real-time polymerase chain reaction. Clin Infect Dis. 2005;41:345–351. doi: 10.1086/431588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Elden LJ, van Kraaij MG, Nijhuis M. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis. 2002;34:177–183. doi: 10.1086/338238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Templeton KE, Scheltinga SA, Beersma MF. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and B viruses, respiratory syncytial virus and parainfluenza viruses 1, 2, 3 and 4. J Clin Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcos AM, Camps M, Pumarola T. The role of viruses in the aetiology of community acquired pneumonia in adults. Antivir Ther. 2006;11:351–359. [PubMed] [Google Scholar]
- 11.Lee BE, Robinson JL, Khurana V. Enhanced identification of viral and atypical bacterial pathogens in lower respiratory tract samples with nucleic acid amplification tests. J Med Virol. 2006;78:702–710. doi: 10.1002/jmv.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox JD. Nucleic acid amplification tests for detection of respiratory viruses. J Clin Virol. 2007;40(suppl):S15–S23. doi: 10.1016/S1386-6532(07)70005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahn JS. Newly identified respiratory viruses. Pediatr Infect Dis J. 2007;26:745–746. doi: 10.1097/INF.0b013e3181376428. [DOI] [PubMed] [Google Scholar]
- 14.Van den Hoogen BG, de Jong JC, Groen J. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouchier RA, Hartwig NG, Bestebroer TM. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liolios L, Jenney A, Spelman D. Comparison of multiplex reverse transcription PCR enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J Clin Microbiol. 2001;39:2779–2783. doi: 10.1128/JCM.39.8.2779-2783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamelin ME, Cote S, Laforge J. Human metapneumovirus infection in adults with community acquired pneumonia and exacerbation of chronic obstructive pulmonary disease. Clin Infect Dis. 2005;41:498–502. doi: 10.1086/431981. [DOI] [PubMed] [Google Scholar]
- 18.Fine MJ, Auble TE, Yealy DM. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med. 1997;336:243–250. doi: 10.1056/NEJM199701233360402. [DOI] [PubMed] [Google Scholar]
- 19.Pabbaraju K, Wong S, McMillan T. Diagnosis and epidemiological studies of human metapneumovirus using real-time PCR. J Clin Virol. 2007;40:186–192. doi: 10.1016/j.jcv.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong S, Pabbaraju K, Pang XL. Detection of a broad range of human adenoviruses in respiratory tract samples with a sensitive multiplex real-time PCR assay. J Med Virol. 2008;80:856–865. doi: 10.1002/jmv.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S. The microimmunofluorescence test for Chlamydia pneumoniae infection: technique and interpretation. J Infect Dis. 2000;181(suppl):S421–S425. doi: 10.1086/315622. [DOI] [PubMed] [Google Scholar]
- 22.Petitjean J, Vabret A, Gouarin S. Evaluation of four commercial immunoglobulin G and immunoglobulin M specific enzyme immunoassays for diagnosis of Mycoplasma pneumoniae infections. J Clin Microbiol. 2002;40:165–171. doi: 10.1128/JCM.40.1.165-171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Field PR, Hunt JG, Murphy AM. Detection and persistence of specific IgM antibody to Coxiella burnetii by enzyme-linked immunosorbent assay: a comparison with immuno-fluorescence and complement fixation tests. J Infect Dis. 1983;148:477–487. doi: 10.1093/infdis/148.3.477. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson HW, Reingold AL, Brake BJ. Reactivity of serum from patients with suspected legionellosis against 29 antigens of Legionellaceae and Legionella-like organisms by indirect immunofluorescence assay. J Infect Dis. 1983;147:23–31. doi: 10.1093/infdis/147.1.23. [DOI] [PubMed] [Google Scholar]
- 25.Basi SK, Marrie TJ, Huang JQ. Patients admitted to hospital with suspected pneumonia and normal chest radiographs: epidemiology, microbiology, and outcomes. Am J Med. 2004;117:305–311. doi: 10.1016/j.amjmed.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 26.Falsey A. Community acquired viral pneumonia. Clin Geriatr Med. 2007;23:535–552. doi: 10.1016/j.cger.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamson N, Renwick N, Kapoor V. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falsey AR, Walsh EE. Viral pneumonia in older adults. Clin Infect Dis. 2006;42:518–524. doi: 10.1086/499955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz A, Barria P, Niederman M. Etiology of community-acquired pneumonia in hospitalized patients in Chile. Chest. 2007;131:779–787. doi: 10.1378/chest.06-1800. [DOI] [PubMed] [Google Scholar]
- 30.De Roux A, Marcos MA, Garcia E. Viral community-acquired pneumonia in nonimmunocompromised adults. Chest. 2004;125:1343–1351. doi: 10.1378/chest.125.4.1343. [DOI] [PubMed] [Google Scholar]
- 31.Flamaing J, Engelmann I, Joosten E. Viral lower respiratory tract infection in the elderly: a prospective in-hospital study. Eur J Clin Microbiol Infect Dis. 2003;22:720–725. doi: 10.1007/s10096-003-1042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luna CM, Famiglietti A, Absi R. Community-acquired pneumonia: etiology, epidemiology, and outcomes at a teaching hospital in Argentina. Chest. 2000;118:1344–1354. doi: 10.1378/chest.118.5.1344. [DOI] [PubMed] [Google Scholar]
- 33.El-solh AA, Sikka P, Ramadan F. Etiology of severe pneumonia in the very elderly. Am J Respir Crit Care Med. 2001;163:645–651. doi: 10.1164/ajrccm.163.3.2005075. [DOI] [PubMed] [Google Scholar]
- 34.Bates JH, Campbell D, Barron AL. Microbial etiology of acute pneumonia in hospitalized patients. Chest. 1992;101:1005–1012. doi: 10.1378/chest.101.4.1005. [DOI] [PubMed] [Google Scholar]
- 35.Bonner AB, Monroe KW, Talley LI. Impact of the rapid diagnosis of influenza on physician decision-making and patient management in the pediatric emergency department: results of a randomized, prospective, controlled trial. Pediatrics. 2003;112:363–367. doi: 10.1542/peds.112.2.363. [DOI] [PubMed] [Google Scholar]
- 36.Boivin G, De Serres G, Hamelin M. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long-term care facility. Clin Infect Dis. 2007;44:1152–1158. doi: 10.1086/513204. [DOI] [PubMed] [Google Scholar]
- 37.Louie JK, Schnurr DP, Pan CY. A summer outbreak of human metapneumovirus infection in a long-term care facility. J Infect Dis. 2007;196:705–708. doi: 10.1086/519846. [DOI] [PubMed] [Google Scholar]
- 38.Ison MG, Hayden FG. Viral infections in immunocompromised patients: what's new with respiratory viruses? Curr Opin Infect Dis. 2002;15:355–367. doi: 10.1097/00001432-200208000-00002. [DOI] [PubMed] [Google Scholar]
- 39.Williams JV, Martino R, Rabella N. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. J Infect Dis. 2005;192:1061–1065. doi: 10.1086/432732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Englund JA, Boeckh M, Kuypers J. Brief communication: fatal human metapneumovirus infection in stem-cell transplant recipients. Ann Intern Med. 2006;144:344–349. doi: 10.7326/0003-4819-144-5-200603070-00010. [DOI] [PubMed] [Google Scholar]
- 41.Centers for Disease Control and Prevention Guidelines for preventing health-care–associated pneumonia, 2003. MMWR Morb Mortal Wkly Rep. 2003;53:1–36. [Google Scholar]
- 42.Fox JD. Nucleic acid amplification tests for detection and analysis of respiratory viruses: the future for diagnostics? Future Microbiol. 2007;2:199–211. doi: 10.2217/17460913.2.2.199. [DOI] [PubMed] [Google Scholar]